Abstract
Background
Elevated numbers of regulatory T cells (Tregs) have been implicated in certain cancers. Depletion of Tregs has been shown to increase anti-tumor immunity. Tregs also play a critical role in the suppression of autoimmune responses. The study of Tregs has been hampered by a lack of adequate surface markers. Leucine Rich Repeat Containing 32 (LRRC32), also known as Glycoprotein A Repetitions Predominant (GARP), has been postulated as a novel surface marker of activated Tregs. However, there is limited information regarding the processing of LRRC32 or the regulatory phenotype and functional activity of Tregs expressing LRRC32.
Results
Using naturally-occurring freshly isolated Tregs, we demonstrate that low levels of LRRC32 are present intracellularly prior to activation and that freshly isolated LRRC32+ Tregs are distinct from LRRC32- Tregs with respect to the expression of surface CD62L. Using LRRC32 transfectants of HEK cells, we demonstrate that the N-terminus of LRRC32 is cleaved prior to expression of the protein at the cell surface. Furthermore, we demonstrate using a construct containing a deleted putative signal peptide region that the presence of a signal peptide region is critical to cell surface expression of LRRC32. Finally, mixed lymphocyte assays demonstrate that LRRC32+ Tregs are more potent suppressors than LRRC32- Tregs.
Conclusions
A cleaved signal peptide site in LRRC32 is necessary for surface localization of native LRRC32 following activation of naturally-occurring freshly-isolated regulatory T cells. LRRC32 expression appears to alter the surface expression of activation markers of T cells such as CD62L. LRRC32 surface expression may be useful as a marker that selects for more potent Treg populations. In summary, understanding the processing and expression of LRRC32 may provide insight into the mechanism of action of Tregs and the refinement of immunotherapeutic strategies aimed at targeting these cells.
Background
Leucine Rich Repeat Containing 32 (LRRC32), also known as Glycoprotein A Repetitions Predominant (GARP), is a member of the leucine rich repeat family that exhibits evolutionary similarity to Toll-like receptors [1]. It was initially localized to chromosome 11q13-14 but has since been further defined and mapped to the 11q13.5-14 region [2-5]. The Lrrc32 gene consists of two coding exons and is expressed as two different transcripts, 4.4 and 2.8 kb in length [3]. The homologous mouse gene has been found in the 7F chromosomal region and shares high sequence homology to human LRRC32 [2,6]. The homologous gene in grass carp (Ctenopharyn-godonidellus) shares 37% homology with human Lrrc32, and it has been found to contain transcription factor binding sites for AP1, IRF-1, IRF4, IRF-7, and NFAT, which are critical for the expression of many cytokines, suggesting a role for LRRC32 in the immune system [7].
Lrrc32 has been shown, via Northern blot, to be expressed in placenta, lung, kidney, heart, liver, skeletal muscle, and pancreas but not brain [3]. Furthermore, Lrrc32 mRNA is also highly expressed in activated Tregs and appears to mediate FoxP3 expression, enabling Tregs to suppress effector cell activation [8-10]. With respect to the structural aspects of LRRC32, a sequence analysis of the human 662 amino acid protein product initially suggested that it was almost entirely extracellular, with 20 leucine rich repeats in the extracellular portion of the protein followed by a hydrophobic stretch of proteins thought to be a transmembrane domain, followed by a short cytoplasmic domain consisting of 15 amino acids [3]. Sequence analysis by SignalP 3.0 revealed a putative N terminal signal peptide with a likely cleavage site after residue 17 of the pro-peptide. Surface localization of this protein in transfected cells has been confirmed [8,9,11]. N-linked glycosylation has also been reported to play a role in post-translational processing of this protein [3].
LRRC32 is expressed on the surface of expanded Tregs, and IL-2-expanded LRRC32-positive CD25hi cells are more suppressive than their IL-2-expanded LRRC32-negative counterparts [10]. Inhibition of LRRC32 expression using lentiviral or siRNA strategies in expanded Treg populations results in decreased suppressive capacity of the targeted cells [10]. The studies summarized above used previously-expanded Tregs. Therefore, they do not address the suppressive capacity of freshly isolated naturally-occurring LRRC32+ and LRRC32- subpopulations of CD25hi regulatory cells. Although addition of TGF-β to LRRC32-CD25- cells induced FoxP3 expression, LRRC32 was not upregulated, and cells treated in this manner were unable to suppress the proliferation of T effectors, suggesting that the upregulated expression of FoxP3 was not sufficient to confer suppressive capacity on effector cells [10]. Furthermore, over-expression of FoxP3 on activated CD4+ T cells did not induce expression of LRRC32 on the cell surface [10]. Finally, it has also recently been reported that LRRC32 binds latency-associated protein (LAP) and that surface expression of LAP, in turn, is upregulated on activated Tregs in conjunction with LRRC32 upregulation [11-13]. As previously reported, Tregs may also use cell-surface bound transforming growth factor beta (TGF-β) to directly inhibit Teff growth in a cell to cell contact dependent manner [14].
Naturally-occurring Tregs are currently defined by the phenotypic expression of numerous surface markers including CD4, CD25, CD127, CD49, GITR, CTLA4, and the intracellular transcription factor FoxP3 [15-22]. Since no single marker identifies the Treg subset, the potential use of LRRC32 as an additional surface marker for potent Tregs is of interest. We surmised that LRRC32 surface expression on Tregs might have utility for the selection of Tregs for functional studies as well as differentiation and activation studies. Although previous studies have looked at the functional suppressive capacity of Tregs that were expanded with cytokines such as IL-2, we chose to study the suppressive capacity of naturally-occurring freshly isolated activated Tregs in the absence of long-term culture or repeated rounds of stimulation.
A previous study utilized a signal peptide deletion construct to show that naive T cells transfected with an LRRC32 signal peptide deletion construct lacked protein upregulation of CD25, CD62L, and FoxP3 compared to transfection with wildtype LRRC32 [8]. This study utilized GFP-tagged signal peptide deletion constructs that were transfected into HEK293 cells to study the contribution of signal peptide to surface expression of LRRC32 [8]. Surface expression was only evaluated by phase contrast and DAPI confocal microscopy of single cells. These images suggested that some of the LRRC32 signal traffic to the cell surface, in contrast with our prediction. Because experiments by Wang et al. were unclear and did not conclusively show that deletion of the signal peptide region affects cell surface expression of conformationally intact native LRRC32 using antibodies capable of recognizing the extracellular domain of LRRC32, we decided to characterize the functional and phenotypic properties of Tregs expressing LRRC32 by immunuohistochemistry.
We show that LRRC32 is a marker for a more potent subset of freshly isolated activated Tregs. We further characterize Treg subsets with respect to the expression of other Treg markers in the context of LRRC32 expression. We examine the intracellular processing of LRRC32 and conclusively demonstrate in multiple cells that the N-terminal portion of LRRC32 is cleaved prior to expression of the protein on the cell surface and that cleavage of this signal peptide is necessary for translocation of the mature protein to the cell surface. Furthermore, we directly confirm, using antibodies specific for native LRRC32, that the signal peptide region of LRRC32 is critical for its surface expression. We also demonstrate low levels of intracellular LRRC32 prior to activation via the T cell receptor (TCR) and CD28, suggesting that low levels of LRRC32 are sequestered intracellularly and that T cell activation is necessary for the synthesis and surface expression of LRRC32. Expression of LRRC32 may enhance Treg function. Therefore, refinement of immunotherapeutic strategies aimed at targeting LRRC32 may improve strategies for Treg isolation and yield more potent Tregs.
Methods
Isolation of CD4+ cells
Peripheral blood was donated by healthy human volunteers coordinated by the Skin Diseases Research Center at University Hospitals Case Medical Center. Signed informed consents were obtained from volunteers prior to their participation in the study. The study protocol was approved by the Institutional Review Board of University Hospitals Case Medical Center. Peripheral blood mononuclear cells (PBMCs) were prepared by Histopaque-1077 (Sigma-Aldrich) density gradient separation in accordance with manufacturer's protocols. Negatively selected CD4+ T cells were purified using magnetic bead technology, per manufacturer's instructions (Miltenyi Biotec).
Generation of Constructs
Plasmid DNA encoding the cDNA of full length human LRRC32 protein (TrueClone pCMV6-XL6 Human Full-Length cDNA Clones, OriGene) was used to transform competent One Shot Top 10 E. coli (Invitrogen), according to the manufacturer's instructions. Selection of positive clones was performed on kanamycin (Invitrogen) LB agar plates. Plasmids were recovered and purified using a Qiaquick MaxiPrep purification kit (Qiagen). The following primers encoding regions flanking the entire Lrrc32 sequence were used to amplify the Lrrc32 cDNA sequence via PCR and insert a TOPO cloning site for pENTR/D-TOPO (Invitrogen): pCMV6-LRRC32 reverse (TTAG GCTTTATACTGTTGGTTAAACTT), pCMV6-LRRC32 reverse readthrough (GGCTTTATA CTGTTGGTTAAACTTCTG), and pCMV6-LRRC32 forward (CACCATGAGACCCCAGA TCCTGCT).
The following 1x PCR buffer conditions were used: AccuPrime Pfx 1x reaction buffer (Invitrogen), 10 mM of forward primer, 10 mM of reverse or reverse readthrough primer, 50 ng of template DNA, and 1 unit of AccuPrime Pfx DNA (Invitrogen). PCR conditions were as follow: initial denaturation at 95 C for 2 minutes followed by 36 cyles of denaturation, annealing, and extension at 95 C, 55 C, and 68 C for 15 s, 30, and 2 minutes, respectively, followed by final extension at 68 C for 30 minutes.
PCR products containing the Lrrc32 sequence derived from reactions utilizing either the forward and reverse primers or forward and reverse readthrough primers were then inserted into pENTR/D-TOPO, per the manufacturer's instructions to generate two different respective constructs: pENTR/D-TOPO/C-terminus LRRC32 (containing a stop codon at the end of the LRR32 sequence) and pENTR/D-TOPO/N-terminus LRRC32 (lacking a stop codon at the end of the LRR32 sequence). One Shot Top 10 E. coli were transformed as described above, and positive clones were selected using LB Agar plus kanamycin plates. Plasmid DNA was isolated using a Qiaquick MiniPrep (Qiagen) kit, and M13 forward and reverse primers were subsequently used to facilitate sequencing of the inserted PCR products within each of the screened and purified plasmids via the Sequencing Core at Case Western Reserve University. The reported sequence was aligned with the reported cDNA sequence of LRRC32 obtained from OriGene using the Vector NTI (Invitrogen) software system, and sequence alignments were then confirmed prior to further utilization of the LRRC32 sequence-verified plasmids.
LRRC32 sequence-verified pENTR/D-TOPO/N-terminus LRRC32 or pENTR D-TOPO/C-terminus LRRC32 plasmids were then used in a Gateway (Invitrogen) cloning strategy utilizing either the Vivid Colors pcDNA6.2/N-EmGFP or pcDNA6.2/C-EmGFP (Invitrogen) as destination vectors, per manufacturer's instructions to generate two products, pcDNA6.2 N-terminus LRRC32/C-EmGFP and pcDNA6.2 C-terminus LRRC32/N-EmGFP, encoding "C-GFP/LRRC32" and "N-GFP/LRRC32" respect-ively. Products were used to transform One Shot Top 10 E. coli., and selection of positive clones was performed on LB Agar plus ampicillan plates. Purified plasmids derived from expanded clones were screened again using restriction enzyme analysis to confirm expected restriction sites in the sequence.
To generate an Lrrc32 signal peptide deletion construct, we utilized the newly created pcDNA6.2 C-terminus LRRC32/N-EmGFP vector as a template for PCR amplification. The pCMV6-LRRC32 reverse readthrough primer, described above, as well as a newly created pCMV6-LRRC32ΔSP primer (CACCATGGCACAACA CCAAGACAAAGT), designed to anneal optimally to bases just distal to the presumed signal peptide cleavage site of LRRC32, were utilized to generate a PCR product containing the Lrrc32 sequence without a putative signal peptide sequence. The PCR product was then inserted into pENTR/D-TOPO, and an identical strategy as that described above was utilized to create a pcDNA6.2 N-terminus LRRC32ΔSP/C-EmGFP product coding for a LRRC32 signal peptide deletion construct ("C-GFP/LRRC32ΔSP") tagged at the C-terminus end with GFP. The primer "C-EmGFP TOPO Internal Rev" (TGAACTTCAGGGTCAGCTTGCCGTA) was utilized to confirm that the GFP and LRRC32 (minus the signal peptide) coding regions were in frame.
Realtime PCR Analysis
RNA was extracted using the RNeasy kit (Qiagen). 100 ng of total RNA was then processed using the SuperScript III First-Strand Synthesis System (Invitrogen) and random hexamers as primers and to generate cDNA, per manufacturer's protocol. The following gene expression assays were subsequently used for RT-PCR analysis: FoxP3 (HS00203958_ml, Applied Biosystems), LRRC32 (HS00194136_ml, Applied Biosystems), 18S (HS99999901_sl, Applied Biosystems), and GFP (Applied Biosystems). RT-PCR analysis was performed in accordance with the manufacturer's suggested protocol (Applied Biosystems) with an Applied Biosystems 7500 Realtime PCR system, and StepOne 2.0 analysis software was used to analyze the data.
Transfection of and Establishment of Stable Cell Lines
HEK293 cells (ATCC, Manassas, VA) were transfected with plasmid containing either pENTR D-TOPO/C-terminus LRRC32 ("C-GFP/LRRC32") or N-terminus LRRC32 ("N-GFP/LRRC32"), utilizing a 3:1 ratio of FuGene 6 (microliters, Roche) to plasmid (micrograms) according to the manufacturer's protocol. Stable transfectants were selected using blastocidin (Invitrogen) according to the manufacturer's protocol, and individual cells were sorted based upon their GFP expression using a FACS Aria cell sorting system (Becton Dickinson) into 96 well plates. Stable clones expressing N-GFP/LRRC32, C-GFP/LRRC32, or C-GFP/LRRC32ΔSP were thus derived from sorted single cells with the highest GFP expression.
Biotinylation of cell surface proteins, Immunoprecipitation and Western Blotting
Sulfo-NHS-LC-Biotin (#21327, Pierce) was utilized to label cell surface proteins, in accordance with the manufacturer's protocol. Biotinylated cells were lysed in Glo Lysis Buffer (#E266A, Promega) per manufacturer protocols. GFP-tagged proteins derived from the cell lysates were then immunoprecipitated with mouse anti-GFP (#A11120, Invitrogen) using an ExactaCruz E system, per manufacturer's instructions (#sc-45042, Santa Cruz Biotechnology). Immunoprecipitated proteins were electrophoresed on precast NuPAGE gels (Invitrogen) with their corresponding premade MOPS SDS buffers (Invitrogen). Proteins were transferred to polyvinylidene difluoride (PVDF) membranes (#LC2002, Invitrogen), per manufacturer's protocol, for subsequent probing using anti-GFP antibody (#A11121, Invitrogen; #33-2600, Zymed) or isotype control antibody (Invitrogen). A SuperSignal PICO (Pierce) kit containing HRP-coupled rabbit anti-mouse antibody was used for detection of GFP-tagged proteins. HRP-coupled goat polyclonal to mouse IgG (#ab6789, Abcam) was also used to recognize bound murine antibody. Rabbit polyclonal antibodies to Foxp3 (#ab10563, Abcam) were used to probe for human FoxP3 on membranes, and HRP-coupled goat polyclonal antibodies to rabbit IgG were used to recognize bound rabbit antibodies. For detection of biotinylated proteins, streptavidin-HRP (#21126, Pierce) was utilized.
For the assessment of cellular LRRC32 expression in the context of the signal peptide deletion constructs, an identical protocol as above was used except that cells were lysed using the M-PER Mammalian Protein Extraction Reagent (#78503, Pierce). Plato-1(ALX-804-867-C100, Axxora) was used to detect LRRC32, and the Pierce Fast Western Blot Kit (#35050, Pierce) was used to visualize immunoblots.
Flow Cytometry Analysis of CD4+ Cells
CD4+ cells were placed in culture media (RPMI, 10% FBS, penicillin, streptomycin, L-glutamine, and β-mercaptoethanol) and rested overnight in 96-well plates or stimulated in anti-CD3-coated plates supplemented with soluble murine anti-human CD28 (1 microgram/ml, Becton Dickenson) [23]. Cells were subsequently stained using antibodies to CD25 (clone 2A3, Becton Dickenson), LRRC32 (Axxora), FoxP3 (eBioscience), and a panel of antibodies specific for CD69 (Becton Dickinson), CD62L (Becton Dickinson), GITR (R & D Biosystems), CTLA4 (Becton Dickinson), HLA-DR (Becton Dickinson), and CD45RO (Becton Dickinson), per manufacturer's protocol for staining cells for flow cytometry (eBioscience). Cells were fixed and then permeabilized using a fixation permeabilization kit (eBioscience) after staining surface antigens in order to study the intracellular expression of certain proteins (LRRC32, FoxP3). For assessment of intracellular expression of LRRC32, permeabilized cells were incubated with 2.5 micrograms/ml of IgG2b control isotype antibody (Invitrogen) for 30 minutes prior to incubation with labeled anti-LRRC32 antibody to decrease non-specific binding. Isotype controls were performed for compensation. Cells were analyzed on a Becton Dickenson LSR II flow cytometer. In all instances, with the exception of the mRNA studies, CD25hi cells represented the top 5% of CD25+ cells. In the case of the mRNA studies in which CD25hi and mid populations were identified, CD25hi++ cells represented the top 1.5% of CD25+ cells, and CD25hi+ cells represented the next highest 2-3% of CD25+ cells as depicted in Figure 1a.
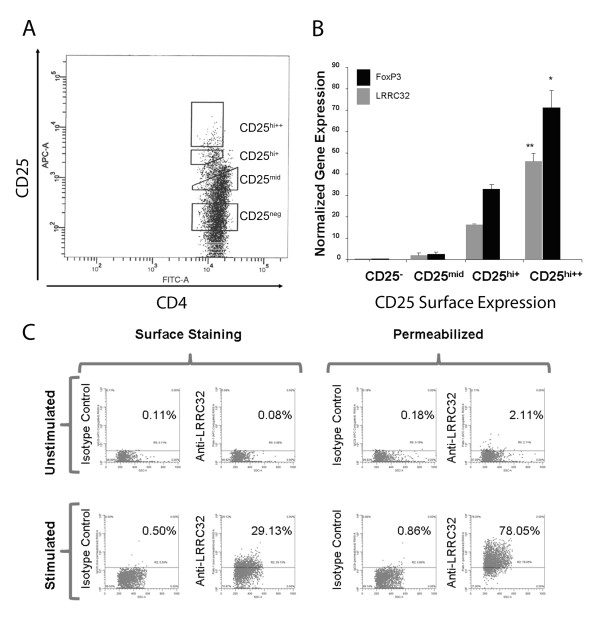
Figure 1.
Lrrc32 mRNA is preferentially expressed in naturally-occurring freshly-isolated non-expanded human Tregs but is not observed on the surface of these cells. a) T cell sorting gates based upon CD25 surface expression. b) Lrrc32 mRNA expression comports with Foxp3 mRNA expression and CD25 surface expression, and Lrrc32 mRNA is preferentially expressed in Tregs, compared to Teffs. Relative expressions of 18SrRNA-normalized Foxp3 and Lrrc32 genes were determined using real-time PCR. Data summarize four independent experiments. Results are expressed as the mean ± SEM. *p = 0.01 compared to CD25mid cells, **p = 0.005 compared to CD25mid cells. c) Flow cytometric analysis of freshly-isolated activated CD4+ human T cells shows that the CD25hi subgroup (composed of the CD25hi+ and CD25hi++ subgroups denoted in figure 1b) demarcating Tregs expresses LRRC32 on the cell surface (top right panel) but that the CD25- subgroup demarcating Teffs expresses negligible amounts of LRRC32 on the cell surface (bottom right panel). Analysis strategy is indicated via the arrows. d) Flow cytometric analysis of sorted resting CD4+ human Tregs shows that they lack surface expression of LRRC32 protein (top left panel). However, after permeabilization of the cell membrane, a low expression of LRRC32 protein can be observed intracellularly (top right panel). After activation of the sorted Tregs with a Treg expansion kit, LRRC32 can be seen on the surface of these activated Tregs (bottom left panel), and evidence of intracellular expression of LRRC32 can also be observed after permeabilization of the cell membrane (bottom right panel).
Expansion and stimulation of Tregs
CD4+ cells were isolated as described above. Subsequently, the cells were stained with anti-CD25 (Becton Dickenson). The top 5% of CD25hi cells were sorted, and cells were expanded and stimulated for 2 weeks using beads coated with anti-CD3 and anti-CD28 according to manufacturer directions (Dynabeads Human Treg Expander, Catalog #111.29, Invitrogen) prior to characterization. Unstimulated sorted CD25hi cells derived from isolated CD4+ cells from the same patient were used as a control for unsimulated Tregs.
Proliferation Assay
Negatively selected CD4+ cells were stained with anti-CD25, and the top 5% of CD25 cells (CD25hi) were sorted using a Becton Dickenson FACS Aria cell sorter. Isolated cells were stimulated overnight on anti-CD3-coated plates (Becton Dickinson) supplemented with soluble murine anti-human CD28 (Pharmingen) as described above. CD25- effector T cells were maintained in culture media. Stimulated CD25hi cells were subsequently stained with antibody specific for LRRC32 (Axxora), and LRRC32+ and LRRC32- subpopulations were further sorted. LRRC32+ positive and LRRC32- Treg subpopulations were then co-cultured with CD25- effector T cells plus irradiated allogeneic antigen presenting cells (APCs) at various Treg:Teff ratios ranging from 1:1 to 1:16 in a 96 well round bottomed tissue culture plate (Costar) as previously described [23]. Each well contained 20,000 Teffs and 50,000 APCs which had been previously irradiated at 3,000 Rad. Mixed lymphocytes were cultured for 6 days, and cells were pulsed with 1 μCi/well [3H]thymidine (NET-027A, Perkin-Elmer) for the last 16 h. Proliferation was measured using a TopCount scintillation counter (Perkin-Elmer). Maximum proliferation was calculated by measuring the proliferation of cells in wells lacking Tregs and containing only Teffs and APCs. Background proliferation was ascertained via measurement of proliferation of Teffs only in the absence of APCs.
Statistical Analysis
Statistical analysis was performed using Student's t test (surface phenotype analysis) or a three way ANOVA (mixed lymphocyte response assays) as indicated in the figure legends. A value of p = 0.05 was considered significant, unless otherwise indicated.
For the ANOVA, the overall p value reported is reflective of the difference between the average proliferation of Tregs using LRRC32 as an independent variable while controlling for the individual assays as well as the titration. The R2 value represents the amount of variability in the ANOVA analysis that is accounted for by the presence or absence of LRRC32 on the surface of a Treg, the individual assays, as well as the titration.
Results
Lrrc32 Is Preferentially Expressed in Tregs
A preliminary microarray analysis demonstrated upregulated Lrrc32 gene expression in Tregs [9]. To confirm these findings, we performed RT PCR analysis examining LRRC32 mRNA expression in sorted subpopulations of CD25-expressing cells as depicted (Figure 1a) representing 1.5% of the CD4+ cell population with the highest expression of CD25 (CD25hi++), 2-3% of the CD4+ cell population with high expression of CD25 (CD25hi+), 17-20% of the CD4+ cell population with reduced expression of CD25 (CD25mid), or no expression of CD25 (CD25-). These analyses revealed that Lrrc32 expression increased with surface CD25 expression in a manner similar to Foxp3 expression (Figure 1b), confirming that Lrrc32 is preferentially expressed in Tregs. To confirm that LRRC32 is specifically upregulated in activated Tregs, we compared the surface expression of LRRC32 in the CD25hi population and the CD25- Teff population of CD4+ cells stimulated overnight with plate bound anti-CD3 and soluble anti-CD28 and found that LRRC32 is preferentially expressed on the surface of activated Tregs but not activated Teffs (Figure 1c), in accordance with a previous study [10]. Furthermore, in accordance with this previous study, we also confirmed that surface LRRC32 is present on sorted CD4+CD25hi Tregs representing 5% of the CD4+ cell population with the highest expression of CD25 (i.e. including both the CD25hi++ and CD25hi+ subgroups described above, Figure 1d, surface expression, stimulated, bottom left panel set) that had been activated for 2 weeks with beads coated with anti-CD28 and anti-CD3 but not on unstimulated sorted CD4+CD25hi Tregs (Figure 1d, surface expression, unstimulated, top left panel set) [10]. A low intracellular expression of LRRC32 in resting sorted CD4+CD25hi Tregs was noted (Figure 1d, permeabilized expression, unstimulated, top right panel set). This suggests that low levels of LRRC32 may be sequestered in Tregs or might require additional processing prior to surface expression. Increased expression of LRRC32 could be seen in activated CD4+CD25hi sorted Tregs that had been permeabilized (Figure 1d, permeabilized expression, stimulated, bottom right panel set) and was higher than that seen on the surface only (Figure 1d, surface expression, stimulated, bottom left panel set), as would be expected for a surface protein that is initially produced intracellularly upon cell activation before trafficking to the cell surface.
Lrrc32 Is Cleaved After Processing
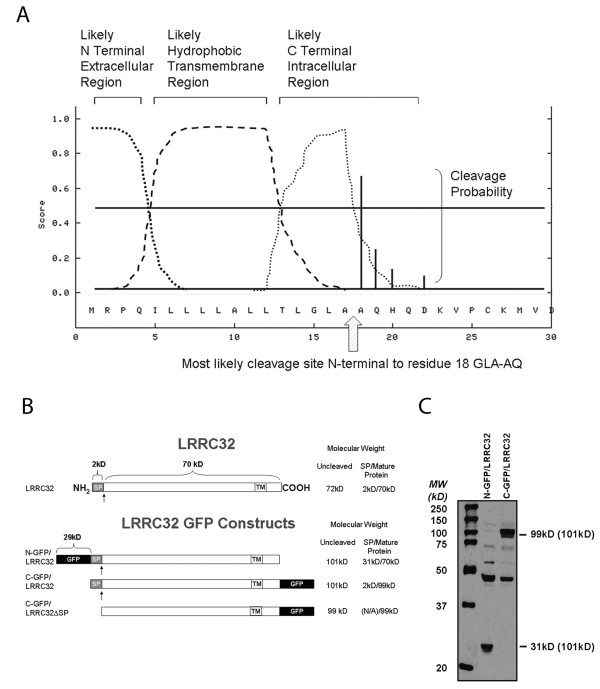
Sequence analysis of murine LRRC32 predicted that LRRC32 would be expressed as a transmembrane protein [6]. We analyzed the sequence of human LRRC32 using the transmembrane domain prediction server at Stockholm Bioinformatics Center with a dense alignment method and confirmed the existence of a putative 19 amino acid transmembrane domain incorporating residues 628 through 646 (data not shown), in agreement with prior reports that this protein was expressed at the cell surface [10,11]. Using the SignalP 3.0 server, we also identified a putative 17 amino acid signal peptide sequence and cleavage site between residues 17 and 18 (Figure 2a), consistent with the requirement for a signal peptide to be present in a protein destined for expression at the cell surface [24]. To further understand protein processing and surface expression of human LRRC32, we generated LRRC32 constructs incorporating green fluorescent protein (GFP) at either the N- or C- terminus, thereby allowing us to examine the tagged signal peptide or the mature protein, respectively (Figure 2b). Using anti-GFP antibodies, we examined total lysates from HEK-transfected C- and N- terminus tagged LRRC32-expressing clones (Figure 2c). Here, we show that 99kD (29 kD GFP+70 kD LRRC32) and 31 kD (29 kD GFP+2 kD LRRC32) fusion proteins, respectively, are generated in HEK transfectants overexpressing LRRC32, consistent with a cleaved N-terminus signal peptide generated prior to surface expression of LRRC32.
Figure 2.
LRRC32 contains an N-terminal signal peptide and a transmembrane region. a) Analysis of the amino acid sequence of LRRC32 by SignalP 3.0 software predicted a putative N-terminal cleavage site between alanines 17 and 18. Alternative potential cleavage sites are predicted and indicated as vertical solid lines on amino acids 19, 20, and 22. b) To address the actual cleavage site, N-terminal and C-terminal GFP-tagged constructs were designed as depicted and generated to facilitate further analysis of surface expression and cleavage. SP = signal peptide. TM = transmembrane region. GFP = green fluorescent protein. Arrow = putative cleavage site. c) Anti-GFP immunoblot analysis of total lysates from C-and N- terminal expressing clones revealed 99 kD (29 kD GFP + 70 kD LRRC32) and 31 kD (29 kD GFP + 2 kD LRRC32 signal peptide) fusion proteins respectively. Expected sizes of uncleaved fusion proteins, based upon predicted protein sequences, are shown in parentheses. The difference in protein size between the C- and N-terminal fusion proteins confirms a cleavage site in the N-terminus of the protein between alanines 17 and 18.
Lrrc32 on Transfected HEK293 Cells Is Expressed at the Cell Surface Regardless of Stimulation
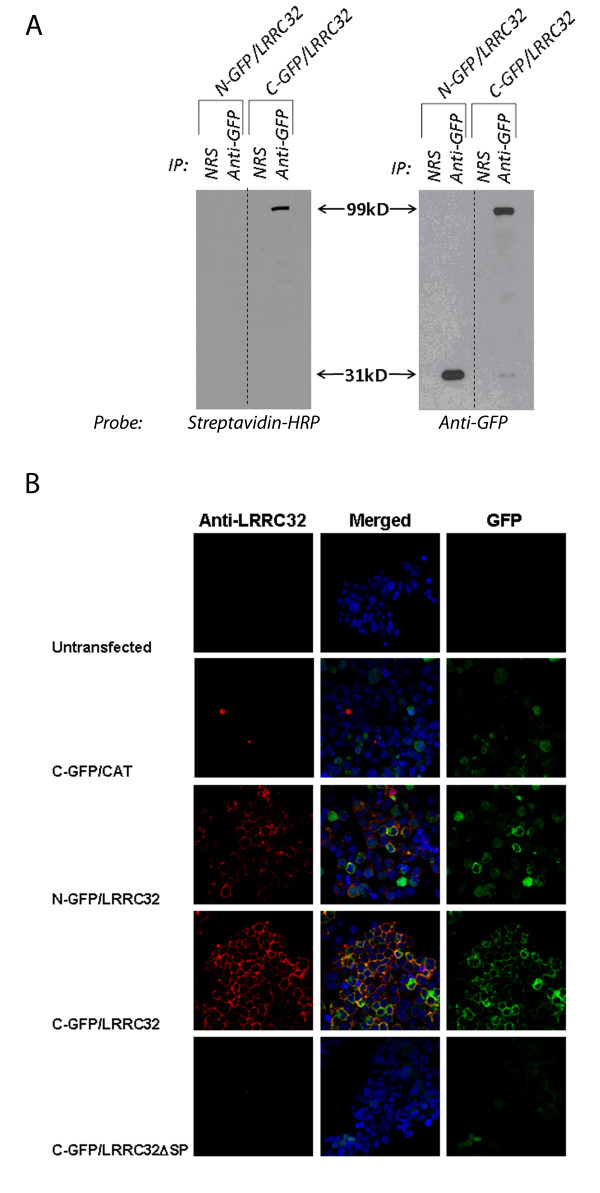
Although Tregs appear to require activation prior to expressing LRRC32 on their surface, the same is not true for transfected cell lines such as HEK293, and the use of the HEK293 cell line allowed us to easily study factors affecting the surface expression of LRRC32 in a system that would not require constant T cell isolation and activation [10,11,25]. We used C- and N-terminal GFP-tagged LRRC32 (C-GFP/LRRC32 and N-GFP/LRRC32, respectively) expressing HEK293 cell clones for surface biotinylation and analysis. Following surface biotinylation, cell lysates were prepared and anti-GFP antibodies were used to immunoprecipitate the fusion proteins. Precipitated protein was transferred to a PVDF membrane. The membrane was then probed for biotin using streptavidin-HRP (Figure 3a, left panel) or for GFP using an anti-GFP antibody (Figure 3a, right panel). Our results demonstrate biotinylated surface protein only in C-GFP/LRRC32-expressing clones (Figure 3a, left panel, rightmost lane), consistent with both surface membrane expression of LRRC32 and intracellular N-terminal processing (removal of the GFP-N-terminal signal peptide) prior to membrane localization. Probing the membrane with anti-GFP (Figure 3a, right panel) demonstrates the surface and intracellular portions of LRRC32 after cleavage.
Figure 3.
LRRC32 is a cell surface protein. a) C-and N- terminus GFP-tagged LRRC32 expressing HEK293 cell clones were surface biotinylated, and cell lysates were immunoprecipitated using antibody specific for GFP or using normal rabbit serum (NRS) as a control (left and right panels). Protein lysates were then electrophoresed, transferred to membrane PDVF, and probed for the presence of biotinylation using streptavidin-HRP (left panel only). Blots were also probed with anti-GFP (right panel only). b) Confocal analysis of untransfected (top row) HEK293 cells, C-GFP/CAT-transfected HEK293 cells (second row), N-GFP/LRRC32-transfected HEK293 cells (third row), C-GFP/LRRC32-transfected HEK293 cells (fourth row), and C-GFP/LRRC32ΔSP-transfected HEK293 cells (last row); Green = GFP, Red = anti-LRRC32 antibody, Blue = nuclear counterstain. The left column shows anti-LRRC32 only. The right column shows GFP only. The middle column shows the merged composite confocal picture (anti-LRRC32 + GFP) with the nuclear counterstain.
Confocal studies with C-terminal GFP-tagged LRRC32 (C-GFP/LRRC32)-transfected HEK293 cells confirm that GFP expression is found on the cell surface without stimulation, in accordance with predictions that LRRC32 would be found on the cell surface (Figure 3b, fourth row, right column). In contrast, N-terminal GFP-tagged LRRC32 (N-GFP/LRRC32)-transfected HEK293 cells exhibit GFP expression diffusely within cells, and GFP expression is not concentrated at cell surfaces, presumably since the GFP-tagged cleaved signal peptide of N-GFP/LRRC32 remains intracellularly after cleavage, prior to the translocation of the mature LRRC32 protein to the cell surface (Figure 3b, third row, right column). Antibody staining of surface LRRC32 confirmed that mature C-GFP/LRRC32 and N-GFP/LRRC32 traffic to the cell surface (Figure 3b, third and fourth rows, left column). Our untransfected control cell line did not express GFP or LRRC32 staining, as expected (Figure 3b, top row). Furthermore, our C-terminal GFP-tagged chloramphenicol acetyltransferase (C-GFP/CAT) control cell line did not express LRRC32 staining, as expected (Figure 3b, second row).
A putative signal peptide region corresponding to the first 17 amino acids of Lrrc32 is required for surface protein expression of Lrrc32
Since signal peptides are generally necessary to direct surface expression of proteins, we next decided to see whether deletion of the putative signal peptide region would inhibit surface expression of LRRC32 [26-28]. HEK293 cells transfected with a C-terminus GFP-tagged LRRC32 construct lacking the signal peptide (C-GFP/LRRC32ΔSP) did not express surface LRRC32 by confocal microscopy (Figure 3b, bottom row). Furthermore, they did not express surface LRRC32 by flow cytometry but were GFP positive, compared to HEK293 cells transfected with the full length C-terminus GFP-tagged LRRC32 construct, expressing both GFP and surface LRRC32 (Figure 4a). Control untransfected cells did not express GFP or surface LRRC32, as expected (Figure 4a, leftmost panel).
Figure 4.
A 17 AA signal peptide is required for the cell surface expression of LRRC32. a) Untransfected HEK293 cells or cells transfected with either C-terminus GFP-tagged LRRC32 or C-terminus GFP-tagged LRRC32 with a deleted signal peptide region were analyzed by flow cytometry for surface expression of LRRC32 or GFP expression. b) Anti-LRRC32 immunoblot analysis of total lysates from C-and N- terminus GFP-tagged LRRC32 expressing clones revealed intact LRRC32 expression at 99 kD (fusion protein: 29 kD GFP + 70 kD LRRC32) and 70 kD, respectively. However, immunoblot analysis of total lysates from C- terminus GFP-tagged LRRC32 expressing clones lacking an intact signal peptide did not detect the presence of LRRC32 (rightmost lane). c) RT-PCR analysis of HEK293 cell lysates utilizing untransfected, C-CAT, C-terminus GFP-tagged LRRC32, or C-terminus GFP-tagged LRRC32 lacking an intact signal peptide was performed using primers for Lrrc32 (left panel) or GFP (right panel).
Given the absence of surface expression of LRRC32 in cells transfected with C-GFP/LRRC32ΔSP, we concluded that the signal peptide portion of LRRC32 was critical for surface expression of LRRC32. It was unclear, however, whether the absence of surface expression of LRRC32 was due to intracellular sequestration of LRRC32. We therefore conducted a western blot analysis of lysates from HEK293 cells transfected with our various constructs using an anti-LRRC32 antibody. Our results showed that while LRRC32 can be detected in cell lysates from cells transfected with full-length LRRC32 constructs (C-GFP/LRRC32 and N-GFP/LRRC32 at 99 and 70 kD, respectively), LRRC32 could not be detected in cell lysates of HEK293 cells transfected with the C-GFP/LRRC32ΔSP, suggesting that LRRC32 is either produced at levels that were undetectable or is rapidly broken down following translation (Figure 4b).
To confirm that Lrrc32 was being transcribed, we utilized RT-PCR to analyze mRNA from lysates of HEK293 cells transfected with our various constructs. Our analysis showed that Lrrc32 mRNA was detected in the C-GFP/LRRC32 as well as the C-GFP/LRRC32ΔSP-transfected HEK293 cell lysates (Figure 4c, left panel). In contrast, untransfected HEK293 cells as well as the control C-terminus tagged chloramphenicol acetyltransferase (C-CAT)-transfected HEK293 cell lysates had undetectable levels of Lrrc32 mRNA. Furthermore, as expected, GFP expression was observed in all of the HEK293 transfected cells examined (Figure 4c, right panel). As we utilized stable clones derived from sorted single cells with the highest GFP expression, as described above, the different efficiencies of C-GFP/LRRC32 vs. C-GFP/LRRC32ΔSP may be due to differential stability/integration of the plasmids in the selected clones. Furthermore, it is possible that the signal peptide mutant may be less stable, and as a result, cells transfected with GFP/LRRC32ΔSP may compensate by producing more mRNA to produce more protein.
Characterization of CD62L expression and functional status of Lrrc32+ and Lrrc32- naturally-occurring freshly isolated human Tregs
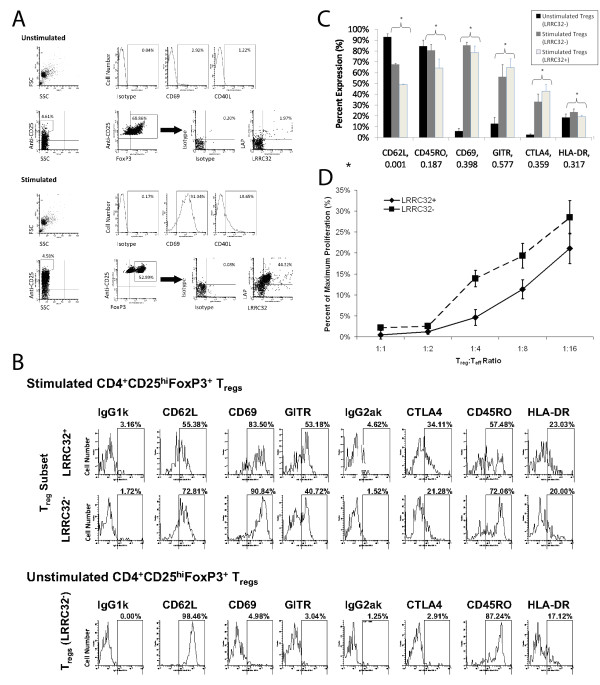
Using polyclonal activation via the TCR in combination with anti-CD28 co-stimulation, we confirmed that LRRC32 is expressed on the surface of naturally-occurring freshly-isolated activated Tregs compared to unstimulated Tregs (24.02% ± 1.73% (n = 6) vs. 2.30% ± 1.05% (n = 6), data not shown), respectively. We subsequently confirmed that surface LAP expression is also observed in this cell population following activation with plate-bound anti-CD3 and soluble anti-CD28, in agreement with a recently published report (Figure 5a) [11].
Figure 5.
LRRC32+ CD4+CD25hiFoxP3 Tregs appear to be more potent suppressors than LRRC32- CD4+CD25hiFoxP3 and exhibit decreased CD62L upon activation. a) Expression of LRRC32 and LAP in CD4+ T cells rested overnight (top panel) or stimulated with plate bound anti-CD3 and soluble anti-CD28 (bottom panel). Tregs were selected from the top 5% CD25-expressing and FoxP3+ populations, as previously described. Confirmation of activation by expression of the surface markers CD40L and CD69 are also shown (top of each panel). b) The expression patterns of various Treg and activation surface markers (CD62L, CD69, GITR, CTLA4, CD45RO, and HLA-DR) in FoxP3+ and LRRC32+-gated populations of CD25hi cells were studied using flow cytometry. Stimulated CD4+FoxP3+CD25hi Tregs (top panel) & unstimulated CD4+FoxP3+CD25hiTregs (bottom panel). c) Composite summary of phenotypic analysis of unstimulated LRRC32-CD4+CD25hiFoxP3+ Tregs and stimulated LRRC32+ and LRRC32- CD4+CD25hiFoxP3+ Tregs. Black bars = unstimulated LRRC32- Tregs. Dark grey bars = stimulated LRRC32- Tregs. Light grey bars = stimulated LRRC32+ Tregs. Data are expressed as the mean ± SEM from 3 individuals. Heteroscedastic variances and an independent t-test comparing stimulated LRRC32+ and LRRC32- subsets were used for calculations of the p values which are reported along the x-axis, below each surface marker (*). d) CD25hi cells were sorted and activated overnight using anti-CD3-coated plates and soluble anti-CD28 (1 microgram/ml). Cells were then resorted based upon LRRC32 expression. The suppressive capacities of these LRRC32+ and LRRC32- Tregs were subsequently tested in a mixed lymphocyte reaction utilizing syngeneic effectors (Teff, 20,000/well) and allogenic antigen presenting cells (50,000/well). Treg:Teff ratios are depicted above. Data summarize 3 independent experiments. Results are expressed as the mean ± SEM. p = 0.0001 and R2 = 0.7244. Absolute proliferation values for the 3 experiments were as follow: Teffs alone: average of 31094 cpm to average of 47483 cpm (at least 6 replicates per assay), background: average of 24 cpm to 35 cpm (at least 6 replicates per assay); Treg:Teff ratio of 1:1: 89 cpm to 346 cpm. When titrating Tregs vs. Teffs, 3 replicates were performed at each titration for the LRRC32+ and LRRC32- Tregs except for in one assay set in which there was limited number of LRRC32+ Tregs. In this case, only one replicate was performed at the 1:1 and 1:2 titrations, and two replicates were performed for the other titrations (0:1, 1:4, 1:8, and 1:16). We performed 3 replicates for each titration utilizing the LRRC32- Tregs.
To date, no single surface marker is sufficient for identifying naturally-occurring Tregs. In order to address the expression of LRRC32 in the context of previously described surface markers, we analyzed LRRC32+ and LRRC32- subsets of CD4+CD25hiFoxP3+LRRC32+ Tregs, as well as unactivated Tregs, with respect to the surface co-expression of CD62L, CD45RO, CD69, GITR, CTLA4, and HLA-DR (Figures 5b and 5c). Stimulated Tregs demonstrated expected increases in the surface expression of GITR, CD69, and CTLA4. Of interest, however, stimulated LRRC32+ Tregs exhibited less CD62L and CD45RO than resting and stimulated LRRC32- Tregs, suggesting that LRRC32+ Tregs may represent a subset of activated or differentiated Tregs.
To address whether or not functional differences in LRRC32+ and LRRC32- subsets of naturally-occurring freshly isolated Tregs exist, we examined the suppressive capacity of LRRC32+ and LRRC32- Tregs. Given previous reports that transfection of Lrrc32-bearing constructs into "pre-regulatory T cells" could induce them to upregulate FoxP3 expression, we hypothesized that the LRRC32+ subset of naturally-occurring freshly-isolated CD4+CD25hi Tregs would be more suppressive than the LRRC32- subset of CD4+CD25hi Tregs [8]. As described above, our data demonstrates that LRRC32 expression comports with FoxP3+ expression. Therefore, we first sorted CD25hi CD4+-purified cells and then activated these and sorted again for Tregs expressing surface LRRC32. Isolated LRRC32+ or LRRC32- Tregs were then used in mixed lymphocyte response assays to assess the relative suppressive capabilities of each isolated population of Tregs. Our results confirmed that naturally-occurring freshly-isolated LRRC32+ Tregs are more suppressive than LRRC32- Tregs (Figure 5d), exhibiting significant increases in suppression at Treg:Teff ratios of 1:4, 1:8, and 1:16 (p = 0.0324, 0.0142, and 0.0430, respectively).
Discussion
The isolation of naturally-occurring functional Tregs will be a prerequisite for successful adoptive immunotherapy techniques in humans. Murine adoptive immunotherapy models have demonstrated proof of concept that isolated Tregs can be used as therapy for several autoimmune disorders [29-31]. The isolation of functional Tregs in humans, however, is problematic, in part due to a paucity of specific surface markers. In this report, we demonstrate direct evidence of the surface expression of LRRC32 on freshly-isolated, naturally-occurring, and non-expanded CD4+ CD25hi human Tregs following TCR activation. We furthermore have characterized LRRC32 processing and demonstrate that sorted subsets of freshly isolated Tregs bearing this marker appear more suppressive than subsets lacking this marker.
Previous studies have shown that constitutive over-expression of Lrrc32 in CD25- Teffs can lead to Foxp3 upregulation and that these cells subsequently acquire a suppressive phenotype [8,25]. Similarly, overexpression of Foxp3 has been shown to result in increased mRNA levels of Lrrc32, suggesting positive feedback between FoxP3 and LRRC32 [8,25]. Although other groups have shown that surface LRRC32 is highly elevated in expanded activated Tregs compared to CD25- Teffs, we demonstrate here that low levels of intracellular LRRC32 are detectable in naturally-occurring freshly-isolated unstimulated Tregs (Figure 1d) [8,25].
Previous studies utilizing an antibody generated against amino acids 296-308 of LRRC32 failed to detect LRRC32 on transfected Jurkat cells or on native CD4+CD25hiTregs [25]. However, as shown here, a commercially available anti-LRRC32 monoclonal antibody does recognize surface LRRC32 on transfected HEKs and naturally-occuring freshly-derived Tregs that have undergone stimulation. Furthermore, it detects low intracellular expression of LRRC32 in naturally-occurring freshly-derived Tregs. Failure to detect surface LRRC32 by the antibody raised against peptide 296-308 may be due to competitive occupation by a ligand as this region of LRRC32 corresponds to a loop and has been hypothesized to correspond to a ligand binding site [25]. One proposed ligand that could occupy this site may be LAP, as recently published work has demonstrated an interaction between LAP and LRRC32 [11,13,32]. If residues 296-308 of LRRC32 act as a binding site for LAP, occupation of this site may account for the failure of previous LRRC32-specific antibodies to recognize surface LRRC32 expression.
To determine if LRRC32 is sequestered in Tregs, we examined the intracellular expression of LRRC32 in naturally-occurring freshly-isolated unstimulated Tregs by flow cytometry. We show that unstimulated Tregs contain low levels of intracellular LRRC32 protein. However, coupled with our RT-PCR studies showing high levels of Lrrc32 mRNA in unstimulated Tregs relative to Teffs, these data suggest that post-transcriptional mechanisms may play a role in controlling LRRC32 production and expression in non-activated Tregs. Such post-transcriptional controls may be diminished upon stimulation via TCR/CD28 signaling, as indeed, upon stimulation, evidence of increased intracellular LRRC32 protein was evident in Tregs as assessed by flow cytometry (Figure 1d).
In addition, our signal peptide deletion construct studies reveal that the putative signal peptide in LRRC32 is critical for the cell surface expression of LRRC32, consistent with other reports that signal peptides are necessary for surface protein expression [33]. Our data showing that LRRC32ΔSP is transcribed (Figure 4c) but not detected intracellularly (Figure 4b) suggest that LRRC32ΔSP is rapidly broken down in the cytosol or is not translated at detectable levels following transcription. However, mechanisms for the rapid degradation of misfolded proteins exist to maintain cell viability, and as such, cytosolic LRRC32ΔSP, unable to enter the endoplasmic reticulum owing to the lack of a signal peptide, may be translated but rapidly degraded afterwards by processes such as ubiquitination [34].
Finally, using freshly isolated, non-expanded CD4+CD25hiLRRC32+ Tregs, we show that such cells expressing surface LRRC32 appear to be functionally more suppressive than CD4+CD25hiLRRC32- Tregs. Previous reports have shown that upon activation of Teffs, surface CD62L is usually decreased [35-37]. However, Tregs normally maintain CD62L expression and functional phenotype [38]. Furthermore, previous reports have shown that CD62L+CD4+CD25hi Tregs are more suppressive than their CD62L- counterparts [39,40].We show here that expression of surface CD62L appears to decrease significantly on LRRC32+ Tregs compared to LRRC32- Treg populations.
Differences in CD62L processing may be responsible for the observed difference in CD62L expression between LRRC32+ and LRRC32- activated Tregs. It has been shown that 90% of CD62L is rapidly cleaved from the surface within 4 hours of T cell activation prior to increasing over the next 48 hours, due to enhanced message stability, before ultimately decreasing due to downregulation of gene transcription [41]. Furthermore, it has been reported that CD62L is rapidly shed in T cells, including Tregs, after activation [42,43]. In accordance, our data show that unstimulated LRRC32- CD4+CD25hiFoxP3+ Tregs expressed more surface CD62L than stimulated LRRC32+ or LRRC32- CD4+CD25hiFoxP3+ Tregs. However, upon activation, decreases in surface CD62L expression of LRRC32+ versus LRRC32- cells were noted, suggesting that LRRC32+ cells are more activated compared to LRRC32- Tregs. Given that an overnight stimulation is sufficient to induce LRRC32 expression on the cell surface of Tregs, we chose this as our timepoint for phenotypic analysis. However, altering the time course of stimulation may also alter surface Treg marker expression.
Activated Tregs that express LRRC32 may also represent a distinct population of more highly activatable Tregs compared to LRRC32- Tregs [41]. Indeed, our phenotypic studies using LRRC32+ and LRRC32- subsets of Tregs in the context of CD62L expression would appear to support the interpretation that LRRC32+ Tregs, relative to LRRC32- Tregs, are more prone to activation, as shown by increased cleavage of surface CD62L, and that this more highly activated state may translate into increased suppressive activity. Notably, although only a fraction of Tregs expressed LRRC32 upon activation overnight, these cells appeared to be more functionally suppressive than their LRRC32- counterparts.
Most natural FoxP3+ adult Tregs are CD45RO+, and the expression of CD45RO is typically a marker of T cell activation [44,45]. As LRRC32+ Tregs appear to be more suppressive than LRRC32- Tregs, it is possible that lower expression of CD45RO on LRRC32+ Tregs relative to LRRC32- Tregs may be due to increased auto-suppressive activity by LRRC32+ Tregs compared to LRRC32- Tregs. As noted above, stimulated Tregs demonstrated expected increases in the surface expression of GITR, CD69, and CTLA4. GITR, or glucocorticoid-induced tumor necrosis factor receptor, was originally shown to be highly expressed on unactivated Tregs but relative to Teffs, and its expression was increased upon cell activation [46-48]. It appears that GITR is a co-stimulatory molecule, and although it is preferentially expressed on CD25hi cells, it is also expressed at lower levels on Teffs, and upon activation, Teffs can also upregulate GITR [48,49]. Hence, the use of GITR as a specific marker for Tregs appears to be limited. CD69 has been described in the context of a CD69+CD4+CD25- Treg subset that does not express Foxp3 but does express surface-bound TGF-β1 in an ERK-dependent manner [50]. Normally, CD69 is upregulated upon T cell activation, and thus expected on our Tregs [50-52]. Since LRRC32 also binds LAP, thereby helping to concentrate TGF-β1 at the cell surface, it is interesting to speculate whether CD69 in Tregs may play a role in helping to upregulate TGF-β1 surface expression in the context of LRRC32 when the latter is available. Although our data did not find any significant differences in the CD69 expression in stimulated LRRC32+ and LRRC32- Treg subsets (Figure 5c), it is possible that part of the reason for the observed difference in suppressive activity between the LRRC32+ and LRRC- Treg subsets may be in part due to synergy between CD69 and LRRC32 via increased surface expression of TGF-β1. CTLA4, or cytotoxic T lymphocyte antigen-4, can inhibit Teff activation via 1) binding B7.1 and B7.2, thereby depriving CD28 on Teffs of the ability to bind these co-stimulatory ligands, 2) inhibiting IL-2 transcription and progression of cells through the cell cycle via inhibition of cyclin D3, cdk4, and cdk6 production, and 3) decreasing the amount of time the TCR is engaged [53-57]. As we did not see any significant differences in CTLA4 expression in the LRRC32+ and LRRC32- Treg subsets, we do not have data to suggest that differences in CTLA4 expression might have contributed to the observed differences in suppressive function in the LRRC32+ and LRRC32- Treg subsets. Clearly, the results in this set of experiments raise many more interesting questions and suggest that the role of LRRC32 in the context of these other cell activation markers is complex.
Previous studies examining LRRC32 and T cell regulation have utilized Teffs transfected with constructs containing either wildtype Lrrc32 or Lrrc32 lacking leucine rich repeat regions, the signal peptide, the cytoplasmic domain, or Lrrc32 with a mutated cytoplasmic residue postulated to be part of a PDZ domain and thus thought to bind an intracellular protein [8,25]. These experiments were performed to address how LRRC32 may be processed and ultimately function in Treg cells. PDZ domain mutation studies have suggested that the intracellular portion of LRRC32 is critical for surface expression, and other studies examined the LRRC32 deletion mutants in the context of downstream effector molecules such as FoxP3 [8,25]. These studies concluded that because FoxP3 expression was markedly decreased upon deletion of the leucine rich regions or the signal peptide, these regions were critical for LRRC32 function [8,25]. However, these studies never demonstrated actual cleavage of the putative signal peptide [8]. Here, we demonstrate via immunoprecipitation and confocal studies that LRRC32 encodes a signal peptide that is cleaved, and upon cleavage, allows mature LRRC32 to reach the cell surface.
It is likely that the LRRC32 signal peptide is cleaved from the newly translocated preprotein by type I eukaryotic endoplasmic reticulum signal peptidase, based upon the amino acid sequence of LRRC32 [58]. The initial amino acid sequence of LRRC32 incorporating a charged N-terminal domain followed by a hydrophobic domain is consistent with published reports of the consensus motif for eukaryotic type I endoplasmic reticulum signal peptidase [58,59]. Furthermore, the sequence G-L-A at positions 15 though 17 of the preprotein is consistent with the -3, -1 rule, stating that residues at the -3 and -1 positions, relative to the cleavage site, must be neutral and have small side chains [58].
Conclusions
In summary, we have demonstrated a cleaved signal peptide site in LRRC32 is necessary for surface localization of native LRRC32 following activation of naturally-occurring freshly-isolated regulatory T cells. We show that LRRC32+ CD4+CD25hiFoxP3+ Tregs express lower levels of surface CD62L compared to LRRC32- CD4+CD25hiFoxP3+ Tregs, suggesting that LRRC32 expression may alter surface expression of other activation markers of T cells such as CD62L. Finally, functional data demonstrate that LRRC32+ Tregs appear more suppressive compared to LRRC32- Tregs, suggesting that LRRC32 surface expression may be useful as a marker that selects for more potent Treg populations, although our data suggest that the functional difference in suppression between these two populations is not markedly robust. Hence, LRRC32 selection may be most useful when used in combination with other Treg markers [60].
Authors' contributions
DVC, AKS, ABY, JVM, JO, HS, and EG performed experiments represented in this manuscript. DB provided statistical support. KDC, HS, and TSM provided valuable designed the study. DVC, TSM, and KDC drafted the manuscript. All authors read and approved the final manuscript.
Contributor Information
Derek V Chan, Email: derek.chan@uhhospitals.org.
Ally-Khan Somani, Email: somania@iupui.edu.
Andrew B Young, Email: aby3@case.edu.
Jessica V Massari, Email: jessmassari@yahoo.com.
Jennifer Ohtola, Email: jao7@case.edu.
Hideaki Sugiyama, Email: sugiyamahm@gmail.com.
Edina Garaczi, Email: egaraczi@yahoo.com.
Denise Babineau, Email: denise.babineau@case.edu.
Kevin D Cooper, Email: kevin.cooper@uhhospitals.org.
Thomas S McCormick, Email: tsm4@case.edu.
Acknowledgements
DVC was supported by NIH 2T32AR007569-16A (Program Director: KDC) and NIH 3P50AR055508-03S309 (DVC, TSM, and KDC). AKS was supported in part by the Dermatology Foundation. This research was also supported by NIH P30AR039750 (KDC). Flow cytometry was also supported by NIH P30CA43703 awarded to the Case Comprehensive Cancer Center. The authors wish to thank Christianne Sykes and Christy Malbasa, M.D. for their efforts in helping to procure patient samples, David Soler, Ph.D. for help in preparing cells, and Wendy Goodman, Ph.D. for critical review of the manuscript.
References
- Bottcher RT, Pollet N, Delius H, Niehrs C. The transmembrane protein XFLRT3 forms a complex with FGF receptors and promotes FGF signalling. Nat Cell Biol. 2004;6(1):38–44. doi: 10.1038/ncb1082. [DOI] [PubMed] [Google Scholar]
- Ollendorff V, Szepetowski P, Mattei MG, Gaudray P, Birnbaum D. New gene in the homologous human 11q13-q14 and mouse 7F chromosomal regions. Mamm Genome. 1992;2(3):195–200. doi: 10.1007/BF00302877. [DOI] [PubMed] [Google Scholar]
- Ollendorff V, Noguchi T, deLapeyriere O, Birnbaum D. The GARP gene encodes a new member of the family of leucine-rich repeat-containing proteins. Cell Growth Differ. 1994;5(2):213–219. [PubMed] [Google Scholar]
- Bekri S, Adelaide J, Merscher S, Grosgeorge J, Caroli-Bosc F, Perucca-Lostanlen D, Kelley PM, Pebusque MJ, Theillet C, Birnbaum D. et al. Detailed map of a region commonly amplified at 11q13-->q14 in human breast carcinoma. Cytogenet Cell Genet. 1997;79(1-2):125–131. doi: 10.1159/000134699. [DOI] [PubMed] [Google Scholar]
- Merscher S, Bekri S, de Leeuw B, Pedeutour F, Grosgeorge J, Shows TB, Mullenbach R, Le Paslier D, Nowak NJ, Gaudray P. et al. A 5.5-Mb high-resolution integrated map of distal 11q13. Genomics. 1997;39(3):340–347. doi: 10.1006/geno.1996.4460. [DOI] [PubMed] [Google Scholar]
- Roubin R, Pizette S, Ollendorff V, Planche J, Birnbaum D, Delapeyriere O. Structure and developmental expression of mouse Garp, a gene encoding a new leucine-rich repeat-containing protein. Int J Dev Biol. 1996;40(3):545–555. [PubMed] [Google Scholar]
- Chang MX, Nie P, Xie HX, Sun BJ, Gao Q. Characterization of two genes encoding leucine-rich repeat-containing proteins in grass carp Ctenopharyngodon idellus. Immunogenetics. 2005;56(10):710–721. doi: 10.1007/s00251-004-0737-3. [DOI] [PubMed] [Google Scholar]
- Wang R, Wan Q, Kozhaya L, Fujii H, Unutmaz D. Identification of a regulatory T cell specific cell surface molecule that mediates suppressive signals and induces Foxp3 expression. PLoS ONE. 2008;3(7):e2705. doi: 10.1371/journal.pone.0002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somani A, Young A, Sugiyama H, Bookout A, Lam M, Cooper K, McCormick T. Characterization of glycoprotein A repetition predominant protein (GARP) identified as a novel gene marker of human CD4+CD25high regulatory T cells (Treg) 2008 International Investigative Dermatology Meeting. 2008.
- Wang R, Kozhaya L, Mercer F, Khaitan A, Fujii H, Unutmaz D. Expression of GARP selectively identifies activated human FOXP3+ regulatory T cells. Proc Natl Acad Sci USA. 2009;106(32):13439–13444. doi: 10.1073/pnas.0901965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran DQ, Andersson J, Wang R, Ramsey H, Unutmaz D, Shevach EM. GARP (LRRC32) is essential for the surface expression of latent TGF-beta on platelets and activated FOXP3+ regulatory T cells. Proc Natl Acad Sci USA. 2009;106(32):13445–13450. doi: 10.1073/pnas.0901944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran DQ, Andersson J, Hardwick D, Bebris L, Illei GG, Shevach EM. Selective expression of latency-associated peptide (LAP) and IL-1 receptor type I/II (CD121a/CD121b) on activated human FOXP3+ regulatory T cells allows for their purification from expansion cultures. Blood. 2009;113(21):5125–5133. doi: 10.1182/blood-2009-01-199950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockis J, Colau D, Coulie PG, Lucas S. Membrane protein GARP is a receptor for latent TGF-beta on the surface of activated human Treg. Eur J Immunol. 2009;39(12):3315–3322. doi: 10.1002/eji.200939684. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Kitani A, Strober W. Cell contact-dependent immunosuppression by CD4(+)CD25(+) regulatory T cells is mediated by cell surface-bound transforming growth factor beta. J Exp Med. 2001;194(5):629–644. doi: 10.1084/jem.194.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T. Immunologic self-tolerance maintained by CD25+CD4+ regulatory T cells constitutively expressing cytotoxic T lymphocyte-associated antigen 4. J Exp Med. 2000;192:303–310. doi: 10.1084/jem.192.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka Y, Sakakura T. Thymus and reproduction: sex-linked dysgenesia of the gonad after neonatal thymectomy in mice. Science. 1969;166:753–755. doi: 10.1126/science.166.3906.753. [DOI] [PubMed] [Google Scholar]
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor [alpha]-chains (CD25) J Immunol. 1995;155:1151–1164. [PubMed] [Google Scholar]
- Schwartz RH. Natural regulatory T cells and self-tolerance. Nat Immunol. 2005;6(4):327–330. doi: 10.1038/ni1184. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol. 2003;4:337–342. doi: 10.1038/ni909. [DOI] [PubMed] [Google Scholar]
- Bachmann MF. Cutting edge: lymphoproliferative disease in the absence of CTLA-4 is not T cell autonomous. J Immunol. 1999;163:1128–1131. [PubMed] [Google Scholar]
- Sugiyama H, Gyulai R, Toichi E, Garaczi E, Shimada S, Stevens SR, McCormick TS, Cooper KD. Dysfunctional Blood and Target Tissue CD4+CD25high Regulatory T Cells in Psoriasis: Mechanism Underlying Unrestrained Pathogenic Effector T Cell Proliferation. J Immunol. 2005;174(1):164–173. doi: 10.4049/jimmunol.174.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyrløv Bendtsen J, Nielsen H, von Heijne G, Brunak S. Improved Prediction of Signal Peptides: SignalP 3.0. Journal of Molecular Biology. 2004;340(4):783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Probst-Kepper M, Geffers R, Kroger A, Viegas N, Erck C, Hecht HJ, Lunsdorf H, Roubin R, Moharregh-Khiabani D, Wagner K. et al. GARP: a key receptor controlling FOXP3 in human regulatory T cells. J Cell Mol Med. 2009;13:13. doi: 10.1111/j.1582-4934.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2(4):953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Gilmore R, Blobel G, Walter P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J Cell Biol. 1982;95(2 Pt 1):463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore R, Walter P, Blobel G. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J Cell Biol. 1982;95(2 Pt 1):470–477. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Cava A. T-regulatory cells in systemic lupus erythematosus. Lupus. 2008;17(5):421–425. doi: 10.1177/0961203308090028. [DOI] [PubMed] [Google Scholar]
- Salomon B, Lenschow DJ, Rhee L, Ashourian N, Singh B, Sharpe A, Bluestone JA. B7/CD28 costimulation is essential for the homeostasis of the CD4+CD25+ immunoregulatory T cells that control autoimmune diabetes. Immunity. 2000;12(4):431–440. doi: 10.1016/S1074-7613(00)80195-8. [DOI] [PubMed] [Google Scholar]
- Janine LC, Nicholas JR, Kevin JM, Holm HU, Fiona P. Regulatory T cells and intestinal homeostasis. Immunological Reviews. 2005;204(1):184–194. doi: 10.1111/j.0105-2896.2005.00250.x. [DOI] [PubMed] [Google Scholar]
- Tran DQ, Shevach EM. Therapeutic potential of FOXP3+ regulatory T cells and their interactions with dendritic cells. Human Immunology. 2009;70(5):294–299. doi: 10.1016/j.humimm.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvineau A, Rouyer-Fessard C, Laburthe M. Presence of a N-terminal signal peptide in class II G protein-coupled receptors: crucial role for expression of the human VPAC1 receptor. Regulatory Peptides. 2004;123(1-3):181–185. doi: 10.1016/j.regpep.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426(6968):895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Mascarell L, Truffa-Bachi P. T lymphocyte activation initiates the degradation of the CD62L encoding mRNA and increases the transcription of the corresponding gene. Immunol Lett. 2004;94(1-2):115–122. doi: 10.1016/j.imlet.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Gerberick GF, Cruse LW, Miller CM, Sikorski EE, Ridder GM. Selective Modulation of T Cell Memory Markers CD62L and CD44 on Murine Draining Lymph Node Cells Following Allergen and Irritant Treatment. Toxicology and Applied Pharmacology. 1997;146(1):1–10. doi: 10.1006/taap.1997.8218. [DOI] [PubMed] [Google Scholar]
- Gomes-Pereira S, Rodrigues OR, Santos-Gomes GM. Dynamics of CD62L/CD45RB CD4+ and CD8+ lymphocyte subsets in hepatic and splenic tissues during murine visceral leishmaniasis. Immunology Letters. 2004;95(1):63–70. doi: 10.1016/j.imlet.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Strauss L, Bergmann C, Whiteside TL. Functional and phenotypic characteristics of CD4+CD25highFoxp3+ Treg clones obtained from peripheral blood of patients with cancer. Int J Cancer. 2007;121(11):2473–2483. doi: 10.1002/ijc.23001. [DOI] [PubMed] [Google Scholar]
- Lepault F, Gagnerault MC. Characterization of Peripheral Regulatory CD4+ T Cells That Prevent Diabetes Onset in Nonobese Diabetic Mice. J Immunol. 2000;164(1):240–247. doi: 10.4049/jimmunol.164.1.240. [DOI] [PubMed] [Google Scholar]
- Fu S, Yopp AC, Mao X, Chen D, Zhang N, Mao M, Ding Y, Bromberg JS. CD4+ CD25+ CD62+ T-regulatory cell subset has optimal suppressive and proliferative potential. Am J Transplant. 2004;4(1):65–78. doi: 10.1046/j.1600-6143.2003.00293.x. [DOI] [PubMed] [Google Scholar]
- Chao CC, Jensen R, Dailey MO. Mechanisms of L-selectin regulation by activated T cells. J Immunol. 1997;159(4):1686–1694. [PubMed] [Google Scholar]
- Guo Z, Jang MH, Otani K, Bai Z, Umemoto E, Matsumoto M, Nishiyama M, Yamasaki M, Ueha S, Matsushima K. et al. CD4+CD25+ regulatory T cells in the small intestinal lamina propria show an effector/memory phenotype. Int Immunol. 2008;20(3):307–315. doi: 10.1093/intimm/dxm143. [DOI] [PubMed] [Google Scholar]
- Tang Q, Bluestone JA. The Foxp3+ regulatory T cell: a jack of all trades, master of regulation. Nat Immunol. 2008;9(3):239–244. doi: 10.1038/ni1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbar AN, Terry L, Timms A, Beverley PC, Janossy G. Loss of CD45R and gain of UCHL1 reactivity is a feature of primed T cells. J Immunol. 1988;140(7):2171–2178. [PubMed] [Google Scholar]
- Booth NJ, McQuaid AJ, Sobande T, Kissane S, Agius E, Jackson SE, Salmon M, Falciani F, Yong K, Rustin MH. et al. Different proliferative potential and migratory characteristics of human CD4+ regulatory T cells that express either CD45RA or CD45RO. J Immunol. 2010;184(8):4317–4326. doi: 10.4049/jimmunol.0903781. [DOI] [PubMed] [Google Scholar]
- McHugh RS, Whitters MJ, Piccirillo CA, Young DA, Shevach EM, Collins M, Byrne MC. CD4(+)CD25(+) immunoregulatory T cells: gene expression analysis reveals a functional role for the glucocorticoid-induced TNF receptor. Immunity. 2002;16(2):311–323. doi: 10.1016/S1074-7613(02)00280-7. [DOI] [PubMed] [Google Scholar]
- Shimizu J, Yamazaki S, Takahashi T, Ishida Y, Sakaguchi S. Stimulation of CD25(+)CD4(+) regulatory T cells through GITR breaks immunological self-tolerance. Nat Immunol. 2002;3(2):135–142. doi: 10.1038/ni759. [DOI] [PubMed] [Google Scholar]
- Ermann J, Fathman CG. Costimulatory signals controlling regulatory T cells. Proc Natl Acad Sci USA. 2003;100(26):15292–15293. doi: 10.1073/pnas.0307001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tone M, Tone Y, Adams E, Yates SF, Frewin MR, Cobbold SP, Waldmann H. Mouse glucocorticoid-induced tumor necrosis factor receptor ligand is costimulatory for T cells. Proc Natl Acad Sci USA. 2003;100(25):15059–15064. doi: 10.1073/pnas.2334901100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Guo Q, Zhang M, Chen Z, Cao X. CD69+ CD4+ CD25- T cells, a new subset of regulatory T cells, suppress T cell proliferation through membrane-bound TGF-beta 1. J Immunol. 2009;182(1):111–120. doi: 10.4049/jimmunol.182.1.111. [DOI] [PubMed] [Google Scholar]
- De Maria R, Cifone MG, Trotta R, Rippo MR, Festuccia C, Santoni A, Testi R. Triggering of human monocyte activation through CD69, a member of the natural killer cell gene complex family of signal transducing receptors. J Exp Med. 1994;180(5):1999–2004. doi: 10.1084/jem.180.5.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho D, Gomez M, Sanchez-Madrid F. CD69 is an immunoregulatory molecule induced following activation. Trends Immunol. 2005;26(3):136–140. doi: 10.1016/j.it.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Brunner MC, Chambers CA, Chan FK, Hanke J, Winoto A, Allison JP. CTLA-4-Mediated inhibition of early events of T cell proliferation. J Immunol. 1999;162(10):5813–5820. [PubMed] [Google Scholar]
- Friedline RH, Brown DS, Nguyen H, Kornfeld H, Lee J, Zhang Y, Appleby M, Der SD, Kang J, Chambers CA. CD4+ regulatory T cells require CTLA-4 for the maintenance of systemic tolerance. J Exp Med. 2009;206(2):421–434. doi: 10.1084/jem.20081811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrov DA, Shi W, Schwartz JC, Almo SC, Nathenson SG. Structure of murine CTLA-4 and its role in modulating T cell responsiveness. Science. 2000;290(5492):816–819. doi: 10.1126/science.290.5492.816. [DOI] [PubMed] [Google Scholar]
- van der Merwe PA, Bodian DL, Daenke S, Linsley P, Davis SJ. CD80 (B7-1) binds both CD28 and CTLA-4 with a low affinity and very fast kinetics. J Exp Med. 1997;185(3):393–403. doi: 10.1084/jem.185.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider H, Downey J, Smith A, Zinselmeyer BH, Rush C, Brewer JM, Wei B, Hogg N, Garside P, Rudd CE. Reversal of the TCR stop signal by CTLA-4. Science. 2006;313(5795):1972–1975. doi: 10.1126/science.1131078. [DOI] [PubMed] [Google Scholar]
- Tuteja R. Type I signal peptidase: An overview. Archives of Biochemistry and Biophysics. 2005;441(2):107–111. doi: 10.1016/j.abb.2005.07.013. [DOI] [PubMed] [Google Scholar]
- von Heijne G. The signal peptide. J Membr Biol. 1990;115(3):195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- Liu W, Putnam AL, Xu-Yu Z, Szot GL, Lee MR, Zhu S, Gottlieb PA, Kapranov P, Gingeras TR, Fazekas de St Groth B. et al. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med. 2006;203(7):1701–1711. doi: 10.1084/jem.20060772. [DOI] [PMC free article] [PubMed] [Google Scholar]