Abstract
The roles of DNA and Mcm1p interactions in determining the overlapping and distinct functions of the yeast cell cycle regulatory transcription factors Fkh1p and Fkh2p were examined. Full-length recombinant Fkh1p and Fkh2p were purified and their binding to bona fide promoters examined in vitro. Each protein bound a variety of target promoters with similar specificity in vitro, consistent with the observation that these proteins bind common promoters in vivo. However, in vivo, the Fkh1p and Fkh2p occupied different target promoters to different extents, suggesting that each was primarily responsible for controlling a different set of genes. Additional in vitro studies provided a mechanistic explanation for this differential promoter-occupancy. Specifically, the Fkh2p, but not the Fkh1p, was capable of binding cooperatively with Mcm1p. The Mcm1p–Fkh2p cooperative binding was enhanced by, but did not require, the presence of a Mcm1p-binding site within a target promoter. Consistent with these data, Mcm1p was present at Fkh-controlled promoters in vivo regardless of whether they contained Mcm1p-binding sites, suggesting a role for Mcm1p at promoters not thought previously to be under Mcm1p control. Analysis of Fkh1p and Fkh2p binding to promoter targets in vivo by use of mutant strains indicated that the two proteins compete for promoter-occupancy at a number of target promoters. We postulate that Fkh1p and a stable Fkh2p/Mcm1p complex compete for binding to target promoters and that the levels and/or binding activity of Fkh1p, but not Fkh2p, are most limiting for promoter-occupancy in vivo. Interestingly, the in vitro DNA-binding assays, using a variety of promoter targets, revealed that bona fide Fkh target promoters contained two or more Fkh-binding sites that allowed the Fkh1p and Fkh2p proteins to form multiple protein–DNA complexes in vitro. Multiple Fkh-binding sites may be a distinguishing feature of bona fide Fkh promoters in yeast and other organisms.
Keywords: Forkhead, Mcm1p, cell cycle, SFF, transcription, yeast
Elucidating the rules regulating promoter-occupancy by sequence-specific DNA-binding proteins is critical to understanding transcription control in vivo. These rules can be relatively simple if a cell expresses only one version of a transcription factor with specificity for a particular target site. However, some cells, particularly mammalian cells, simultaneously express several different members of a transcription-factor family, each of which is capable of binding to the same target site (Amati and Land 1994; Levrero et al. 2000). In such examples, the rules regulating promoter-occupancy in vivo are more complex but critically important, as each member of a transcription factor family can have a different impact on transcription by a given promoter (Amati and Land 1994; Levrero et al. 2000).
Recently, two forkhead proteins, Fkh1p and Fkh2p (forkhead homolog), have been shown to share promoter targets and overlapping biological roles in cell cycle progression and differentiation in the single-celled yeast Saccharomyces cerevisiae (for review, see Breeden 2000). FKH1 and FKH2 are named for their homology to an evolutionarily conserved family of transcription factors classified on the basis of their forkhead (winged-helix) DNA-binding domains (Clark et al. 1993; Lai et al. 1993; Kaufmann and Knochel 1996; Kaestner et al. 2000). Transcription factors in this family have roles in the cell cycle and differentiation in a wide variety of eukaryotes including humans (Medema et al. 2000; Nakamura et al. 2000; Tanaka et al. 2001), and there are many cell types in which multiple forkhead family members with similar DNA-binding domains are expressed simultaneously (Lai et al. 1993; Hromas and Costa 1995; Kaufmann and Knochel 1996). Significantly, Fkh1p and Fkh2p are exceptional examples of yeast transcription factors that have clear tissue-specific homologs in mammals (Kaufmann and Knochel 1996; Yang et al. 1997), suggesting that their biological roles and the mechanisms regulating their functions are conserved. The identification of a large number of yeast Fkh-controlled genes by DNA microarray analysis (Zhu et al. 2000) and the availability of all yeast gene regulatory sequences will allow the mechanisms regulating promoter-occupancy by Fkh1p and Fkh2p to be addressed rigorously.
Yeast Fkh1p and Fkh2p share 47% identity and 82% similarity across the length of Fkh1p. Of particular relevance to this study, Fkh1p and Fkh2p contain DNA-binding domains that are 72% identical and, for the phenotypes tested, interchangeable (Hollenhorst et al. 2000), suggesting that the two proteins share the same binding-site specificity in vivo. Consistent with these overlapping structural features, several recent studies indicate that the Fkh1p and Fkh2p have overlapping functional roles (for review, see Breeden 2000). Specifically, deletion of both FKH1 and FKH2, but not either gene alone, leads to pseudohyphal differentiation and significant reductions in transcription of CLB2 and similarly regulated genes collectively referred to as the CLB2-cluster. CLB2-cluster genes are transcribed in late S and G2/M phases of the cell cycle (Spellman et al. 1998) and encode many proteins, including the G2/M phase cyclin Clb2p, that are necessary for normal cell cycle progression in yeast (for review, see Futcher 1996). Slowing progression through the G2/M phase of the cell cycle is required for the pseudohyphal morphology (Lew and Reed 1993; Kron et al. 1994). Therefore, loss of both FKH1 and FKH2 is required for the pseudohyphal phenotype, at least in part, because loss of both genes is required to dramatically reduce cell cycle-regulated transcription of CLB2-cluster genes. Because deletion of both FKH1 and FKH2 is required for these dramatic phenotypic and transcriptional changes, a simple view is that these proteins have redundant roles and in the absence of one, the other simply provides the same function by regulating the same genes.
However, despite this apparently simple picture, other data indicate that Fkh1p and Fkh2p actually have distinct roles in regulating the cell cycle and transcription in yeast. For example, transcription of CLB2 is affected differently by loss of either Fkh1p or Fkh2p alone (Hollenhorst et al. 2000). Specifically, deletion of FKH1 enhances CLB2 transcription during most stages of the cell cycle, whereas high-copy expression of FKH1 or deletion of FKH2 reduces it. Because Fkh1p and Fkh2p can have measurably different effects on the transcription of CLB2 in vivo, and because both proteins can bind this promoter in vivo (Koranda et al. 2000; Kumar et al. 2000), the relative occupancy of the CLB2 promoter by Fkh1p and Fkh2p is crucial for proper CLB2 expression. In addition, Fkh2p, but not Fkh1p, is a component of SFF (Swi five factor) (Koranda et al. 2000; Kumar et al. 2000; Pic et al. 2000; Zhu et al. 2000), a partially characterized protein complex that binds to promoters of genes in the CLB2 cluster at sites required for their cell cycle-regulated transcription (Lydall et al. 1991; Althoefer et al. 1995; Maher et al. 1995). Consistent with this observation, Fkh2p occupies CLB2-cluster promoters in vivo more efficiently than Fkh1p (Koranda et al. 2000; Kumar et al. 2000). These data indicate that Fkh1p and Fkh2p, despite their similarities, may have different inherent DNA-binding properties and/or interactions with other gene regulatory proteins.
To address the outstanding questions relevant to promoter-occupancy by Fkh1p and Fkh2p, we compared promoter binding by both proteins in vitro and in vivo. Our in vitro analysis revealed two biochemical differences between Fkh1p and Fkh2p. First, purified Fkh2p, but not Fkh1p, could bind cooperatively with itself, and second, Fkh2p, but not Fkh1p, could bind cooperatively with Mcm1p, a second gene regulatory protein required for transcription of genes in the CLB2 cluster (Lydall et al. 1991; Althoefer et al. 1995; Maher et al. 1995). The latter difference provided an explanation for the differences in relative promoter-occupancy by Fkh1p and Fkh2p at a variety of promoters in vivo. Together, the in vitro and in vivo data presented here support a model in which Fkh1p is in competition with a stable Fkh2p/Mcm1p complex for promoter-occupancy at Fkh-controlled promoters, and Fkh1p levels and/or binding activity are limiting. This model supports a simple mechanism for how transcription and cell cycle progression can be attenuated by regulation of Fkh1p and may be applicable to mammalian promoters controlled by forkhead proteins.
Results
Purified Fkh1p and Fkh2p bound similarly to SWI5 promoter DNA in vitro
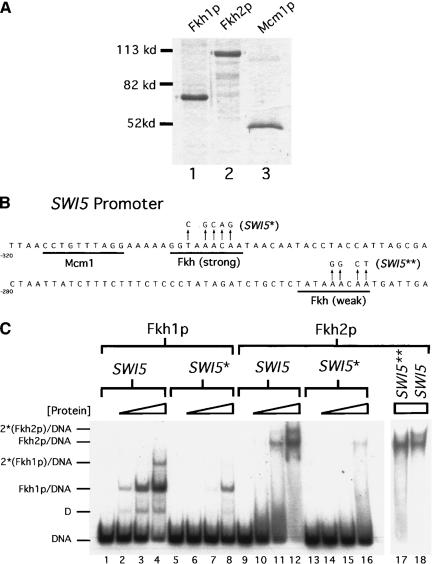
One obvious explanation for the ability of one Fkh protein to compensate for the loss of the other in vivo is that the two proteins have similar DNA-binding properties. However, differences between the amino acid sequence of Fkh1p and Fkh2p, including some notable differences within their conserved forkhead DNA-binding domains, could modulate the DNA-binding properties of these two proteins uniquely (Overdier et al. 1994; Kaufmann et al. 1995). Therefore, full-length recombinant yeast Fkh1p and Fkh2p were each purified substantially from baculovirus-infected insect cells (Fig. 1A, lanes 1 and 2) and used in electrophoretic-mobility shift assays (EMSA) with relevant radiolabeled substrate DNAs. Recombinant Fkh1p and Fkh2p migrated with molecular weights of 67 kD and 106 kD, respectively, close to the molecular weights predicted by their primary sequence.
Figure 1.
Purified Fkh1p and Fkh2p bound similarly to SWI5 promoter DNA in vitro. (A) Recombinant Fkh1p–6xHis (lane 1), Fkh2p–6xHis (lane 2), and Mcm1p–6xHis (lane 3) were expressed in insect Sf9 cells and substantially purified. A total of 1 μg of purified Fkh1p and 0.5 μg of Fkh2p or Mcm1p were analyzed on a 10% SDS–gel stained with Coomassie Brilliant Blue. (B) The relevant region of the 200-bp SWI5 promoter DNA that was analyzed for Fkh1p and Fkh2p binding by an EMSA. The strong and weak Fkh-binding sites and the Mcm1p-binding site in this promoter are underlined. The nucleotide changes in the two mutant SWI5 promoters used are indicated by arrows (SWI5*, strong site mutant; SWI5**, weak site mutant). (C) A radiolabeled 200-bp SWI5 promoter fragment (SWI5; lanes 1–4,9–12,18) or a mutant SWI5 promoter fragment containing changes in either the strong Fkh-binding site (SWI5*; lanes 5–8,13–16) or the weak Fkh-binding site (SWI5**; lane 17) as indicated in B was analyzed for binding to Fkh1p–6xHIS (lanes 1–8) or Fkh2p–6xHIS (lanes 9–18). SWI5 DNA was analyzed for binding to either Fkh1p or Fkh2p at a final concentration of 0 nM (lanes 1,5,9,13), 2.5 nM (lanes 2,6,10,14), 10 nM (lanes 3,7,11,15), and 40 nM (lanes 4,8,12,16,17–18). Free DNA and Fkh/DNA and 2(Fkh)/DNA complexes are indicated for both Fkh1p and Fkh2p (lower mobility complexes are labeled with an asterisk because the actual protein stoichiometry in these complexes has not been determined. Only the simplest possibility has been shown.). The complex labeled D is due to the binding of a minor Fkh1p degradation product to the SWI5 promoter DNA.
To test whether the recombinant Fkh1p and Fkh2p bound a target promoter similarly and with specificity for a Fkh sequence in vitro, a 200-bp region of the SWI5 promoter was used in an EMSA (Fig. 1B). SWI5 is one of ∼30 genes within the CLB2 cluster of genes whose cell cycle-regulated transcription is abolished in strains containing deletions of both FKH1 and FKH2 (Zhu et al. 2000). Significantly, both Fkh1p and Fkh2p bound to the SWI5 promoter DNA with similar apparent affinities as determined by quantitative analysis of an EMSA (Fig. 1C, wild type, lanes 1–4, 9–12); Fkh1p and Fkh2p bound the SWI5 promoter with apparent Kds of ∼12 and ∼5 nM, respectively. The binding of both proteins to a SWI5-promoter fragment containing a mutation of the best match to an Fkh sequence was reduced significantly [Fig. 1B, SWI5* and C, lanes 5–8 (Fkh1p), 13–16 (Fkh2p)]. Residual binding of both proteins to the mutant SWI5 fragment was observed at the highest concentrations of protein, consistent with the presence of a second lower affinity Fkh site in the SWI5 promoter [Fig. 1B and C, lanes 8 (Fkh1p) and 16 (Fkh2p)]. Taken together, these data indicate that recombinant Fkh1p and Fkh2p bind the SWI5 promoter similarly in vitro.
The presence of a second lower affinity Fkh site in the SWI5 fragment could explain the faint low-mobility species observed for the wild-type SWI5 promoter at the highest concentrations of Fkh1p and Fkh2p (Fig. 1C, lanes 4 and 12). To test whether the lower affinity site was responsible for the lower mobility complex observed in lane 12, this same concentration of Fkh2p was used in a second EMSA with a SWI5 probe containing a mutation in the predicted low-affinity Fkh site [Fig. 1C, lanes 17–18; see B for mutant sequence (SWI5**)]. The second lower mobility complex observed with the wild-type SWI5 promoter (Fig. 1C, lane 18) was not observed with a SWI5 promoter containing a mutation in the low-affinity site (Fig. 1C, lane 17). Thus, the SWI5 promoter contained at least two Fkh-binding sites capable of binding Fkh proteins.
Fkh1p and Fkh2p showed similar specificity for a number of different CLB2-cluster gene promoters in vitro
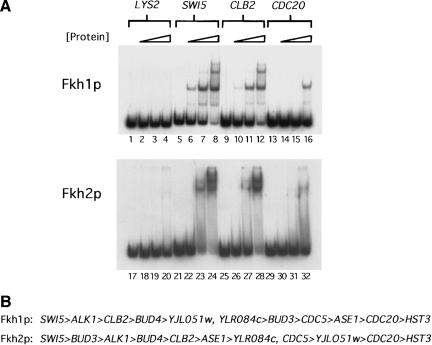
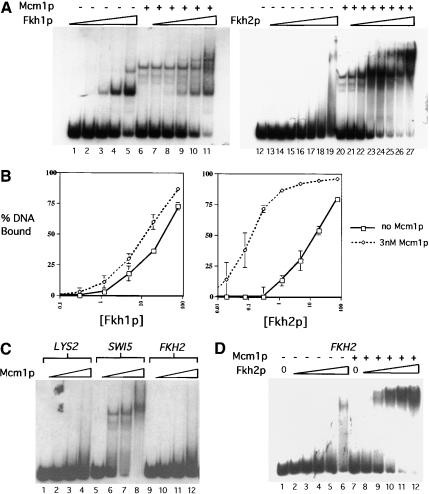
In vivo, Fkh2p has been shown to occupy CLB2-cluster promoters to a greater degree than Fkh1p, as measured by chromatin immunoprecipitation experiments (Koranda et al. 2000; Kumar et al. 2000). Although our in vitro data indicated that the Fkh1p and Fkh2p could bind the SWI5 promoter DNA fragment similarly, it was possible that Fkh2p might bind some CLB2-cluster promoters more efficiently than Fkh1p, thus explaining, at least partially, the in vivo differences observed for the two proteins. Therefore, we measured in vitro DNA binding by Fkh1p and Fkh2p to a variety of CLB2-cluster promoters (Fig. 2A, for summary, see B). Fkh1p and Fkh2p bound to a variety of CLB2-cluster promoters including SWI5, CLB2, and CDC20 with similar apparent affinities (Fig. 2A). In general, the two different proteins exhibited the same relative preferences for a wide variety of promoters within the CLB2 cluster (Fig. 2B). However, two differences between the behavior of the two proteins in an EMSA were observed. First, Fkh1p/DNA complexes migrated as tight bands, whereas Fkh2p/DNA complexes dissociated during electrophoresis and generated a smear of radioactivity in the gel lanes (Fig. 2A, cf. lanes 7 and 8 with lanes 23 and 24, and lane 16 with 32). Second, and perhaps more significantly, Fkh2p but not Fkh1p appeared to bind some CLB2-cluster targets, most notably the CLB2 promoter itself, via a cooperative mechanism (Fig. 2A, lanes 26–28). Note the significantly increased efficiency in binding to the CLB2 promoter for a relatively small increase in the concentration of Fkh2p. This behavior was not observed for Fkh1p (lanes 10–12). Thus, purified Fkh1p and Fkh2p had similar target-site specificity in vitro, but Fkh2p exhibited cooperative binding characteristics that, in principle, could stabilize its interactions with some target promoters.
Figure 2.
Fkh1p and Fkh2p showed a similar range of specificity for CLB2-cluster gene promoters in vitro. (A) Radiolabeled 200-bp fragments from the coding region of LYS2 (lanes 1–4, Fkh1p; lanes 17–20, Fkh2p) and the promoter regions of SWI5 (lanes 5–8, Fkh1p; lanes 21–24, Fkh2p), CLB2 (lanes 9–12, Fkh1p; lanes 25–28, Fkh2p), and CDC20 (lanes 13–16, Fkh1p; lanes 29–32, Fkh2p) were analyzed in an EMSA for binding to Fkh1p or Fkh2p as described in Figure 1. The LYS2-coding region served as a control for nonspecific binding by Fkh1p and Fkh2p. (B) A summary of relative promoter preferences of Fkh1p and Fkh2p as determined by an EMSA for a number of Fkh-controlled promoters. The promoters are arranged, left to right, from those that were bound most efficiently to least efficiently by each Fkh protein.
Interestingly, the CLB2-promoter DNA, as well as a number of other promoter DNA fragments from the CLB2 cluster (for summary, see Fig. 2B), formed a second lower mobility complex in an EMSA with the highest concentrations of either Fkh1 or Fkh2 protein [Fig. 2A, lane 12 (Fkh1p) and lane 28 (Fkh2p)]. Thus, these promoter fragments contained at least one additional lower affinity Fkh site in addition to a strong predicted match to the Fkh consensus, as was observed for the SWI5-promoter DNA (Fig. 1B, lanes 4 and 12, and Fig. 2A, lanes 8 and 16). Thus, multiple Fkh-binding sites may be a distinguishing feature of bona fide Fkh-controlled promoters.
Additional Fkh target sites identified with an extended Fkh-consensus binding site: Fkh1p and Fkh2p bound to the FKH1 and FKH2 promoters in vitro
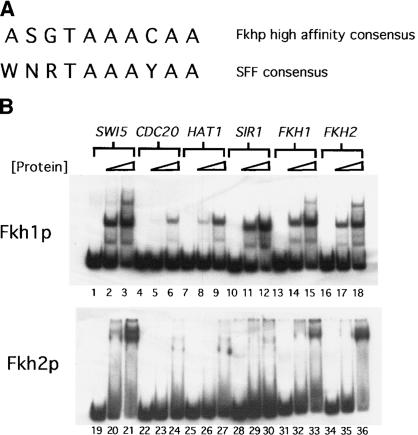
To identify additional Fkh target sites that could provide insights into the mechanisms that govern promoter-occupancy by Fkh1p and Fkh2p, a more restrictive Fkh consensus-binding site was determined by visual examination of the CLB2-cluster promoters that bound Fkh1p and Fkh2p with the highest apparent affinities (Fig. 3A). This sequence was used to search regions upstream of predicted ORFs within the yeast genome. A total of 156 potential Fkh-binding sites with exact matches to this consensus were identified, including one each within the FKH1 and FKH2 promoters. A small subset of 200-bp fragments from this pool of 156 was tested for the ability to bind Fkh1p and Fkh2p in vitro and 4 of those fragments tested are shown (Fig. 3B). Fkh1p bound the fragments upstream of HAT1, SIR1, FKH1, and FKH2 as well or better than it bound the CDC20 promoter (Fig. 3B). Thus, additional potential Fkh targets were identified on the basis of a search of the yeast intergenic regions for high-affinity target sites.
Figure 3.
Additional Fkh target sites identified with an extended Fkh consensus-binding site. (A) A high-affinity Fkh-binding site determined by visual examination of CLB2-cluster promoters bound most efficiently by Fkh1p and Fkh2p in vitro is shown above the SFF consensus sequence reported previously (Spellman et al. 1998). The nucleotide codes are as follows: N, any base; S, G or C; W, A or T; R, A or G; Y, C or T. (B) Radiolabeled 200-bp promoter fragments for SWI5 (lanes 1–3, Fkh1p; lanes 19–21, Fkh2p), CDC20 (lanes 4–6, Fkh1p; lanes 22–24, Fkh2p), HAT1 (lanes 7–9, Fkh1p; lanes 25–27, Fkh2p), SIR1 (lanes 10–12, Fkh1p; lanes 28–30, Fkh2p), FKH1 (lanes 13–15, Fkh1p; lanes 31–33, Fkh2p), and FKH2 (lanes 16–18, Fkh1p; lanes 34–36, fkh2p); were analyzed in an EMSA for binding to Fkh1p or Fkh2p as described in Figure 1. Binding of each promoter by a Fkh protein was examined at three different concentrations of Fkh1p (0, 5, and 20 nM) or Fkh2p (0, 10, and 40 nM).
Interestingly, although Fkh1p and Fkh2p bound most targets similarly in vitro, their binding behaviors with the SIR1 fragment were quite different. Whereas Fkh1p bound the SIR1 fragment efficiently, consistent with the presence of a high-affinity Fkh site, Fkh2p bound this fragment extremely inefficiently [Fig. 3B, cf. lanes 10–12 (Fkh1p) with lanes 28–30 (Fkh2p)]. Examination of the Fkh1p/SIR1 interaction on an EMSA indicated that a lower mobility complex did not form [Fig. 3B, cf. lanes 10–12 (SIR1) with lanes 1–3 (SWI5), lanes 13–15 (FKH1), and lanes 16–18 (FKH2)]. These data suggested that the presence of a second low-affinity binding site might contribute to stable Fkh2p/DNA interactions. However, this cannot be the sole explanation for stable binding by Fkh2p, as Fkh2p bound reasonably well to the SWI5 promoter that contained a mutation in the low-affinity Fkh site (Fig. 1C, lanes 17–18). Regardless, it is noteworthy that both the FKH1 and FKH2-promoter fragments bound the Fkh1p and Fkh2p similarly and both contained a second low-affinity Fkh-binding site (Fig. 3B, lanes 15 and 17).
Differential promoter-occupancy by Fkh1p and Fkh2p in vivo
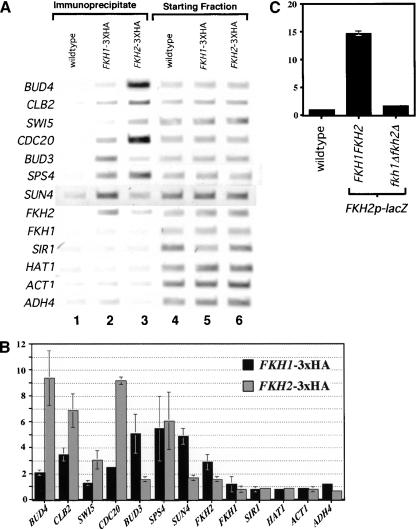
If the promoter fragments that bound Fkh1p and Fkh2p efficiently in vitro represented bona fide Fkh-binding sites in vivo, then these promoter regions might be enriched in a protein–DNA fraction prepared by immunoprecipitation of either Fkh1p–3xHA or Fkh2p–3xHA from a yeast whole-cell extract. Therefore, yeast cells harboring a chromosomal copy of either FKH1–3xHA or FKH2–3xHA and yeast cells containing untagged versions of both genes (wild type) were used in chromatin immunoprecipitation experiments with an antibody to the Hemagglutinin epitope (α-HA). As judged by enrichment of CLB2-cluster promoter DNA fragments in a chromatin immunoprecipitation experiment, both Fkh proteins bound to CLB2-cluster promoters in at least a fraction of the yeast cells in an asynchronously growing population (Koranda et al. 2000; Kumar et al. 2000).
To test whether Fkh1p and/or Fkh2p bound in vivo to the potential Fkh-controlled promoters identified in vitro as described above (Fig. 3B), these promoter targets were analyzed by chromatin immunoprecipitation of Fkh1p–3xHA or Fkh2p–3xHA. Although the HAT1, SIR1, and FKH1 promoter regions contained Fkh consensus sites and bound Fkh proteins in vitro, none of these promoters was occupied measurably by Fkh1p or Fkh2p in vivo (Fig. 4). Thus, a high-affinity Fkh-binding site within a promoter may not be functional for binding by either Fkh1p or Fkh2p in vivo. Significantly, however, both Fkh1p and Fkh2p bound to the FKH2 promoter in at least a fraction of the yeast cells in an asynchronously growing population (Fig. 4). Therefore, in vitro binding analysis revealed at least one new gene, FKH2, whose promoter was occupied by the Fkh1 and Fkh2 proteins.
Figure 4.
Differential promoter-occupancy by Fkh1p and Fkh2p in vivo. (A) Occupancy of a variety of promoters by either Fkh1p or Fkh2p was measured in vivo by performing chromatin immunoprecipitation experiments with an α-HA antibody. Three isogenic strains were analyzed after immunoprecipitation (lanes 1–3) and prior to immunoprecipitation (starting fraction, lanes 4–6) for the presence of promoter fragments listed at left by use of PCR and the appropriate promoter-specific primers (Table 1). The strains analyzed were isogenic except for the presence of a 3xHA-epitope tagged FKH gene [wild type, lanes 1,4 (CFY145, FKH1 FKH2), FKH1–3xHA, lanes 2,5 (CFY482, FKH1–3xHA FKH2), and FKH2–3xHA, lanes 3,6 (CFY854, FKH1 FKH2–3xHA)]. (B) Quantitative analysis of chromatin immunuprecipitation that were performed as in A except that PCR was done in the presence of [α-32P]dCTP and PCR products were analyzed after polyacrylamide gel electrophoresis by quantifying with a PhosphorImager. Data are reported as fold enrichment of promoter fragments in the FKH1–3xHA (CFY482) or FKH2–3xHA (CFY854) strains relative to the untagged wild-type strain (CFY145). Data and error from an average of three separate chromatin immunoprecipitation experiments are reported. (C) Analysis of β-galactosidase expression of a FKH2-promoter/lacZ/FKH2-terminator (FKH2p–lacZ) gene fusion cloned into pRS304 and integrated at the TRP1 locus (TRP1∷FKH2p–lacZ). The wild-type control strain was FKH1 FKH2 and did not contain an integrated copy of FKH2p–lacZ (wild type; CFY145). Two additional isogenic strains were compared that were each TRP1∷FKH2p–lacZ but differed in their FKH genotype. One strain was wild type (FKH1 FKH2, TRP1∷FKH2p–lacZ; CFY1122) and the other contained deletions of both FKH1 and FKH2 (fkh1Δ fkh2Δ, TRP1∷FKH2p–lacZ; CFY1120).β-galactosidase assays were performed on ∼2.5 O.D. of cells as described (Miller 1972). The units reported in C are arbitrary relative to the wild-type control that was given a unit value of 1.0. Each lane represents the average from four independent experiments.
To test whether binding by Fkh1p to the FKH2 promoter reflected a role for the Fkh proteins in controlling transcription of FKH2, a FKH2-promoter–lacZ gene fusion (FKH2p–lacZ) was integrated into a wild-type yeast strain and an isogenic mutant strain containing deletions of both FKH1 and FKH2 (fkh1Δ fkh2Δ). The expression of lacZ was monitored by measuring β-galacotosidase activity (Fig. 4C). Significantly, the level of β-galactosidase produced by the wild-type strain was 10-fold higher than that produced by the fkh1Δ fkh2Δ strain, indicating that the Fkh1p and Fkh2p were required for the function of the FKH2 promoter. Thus, the forkhead proteins regulate the expression of the FKH2 gene in vivo.
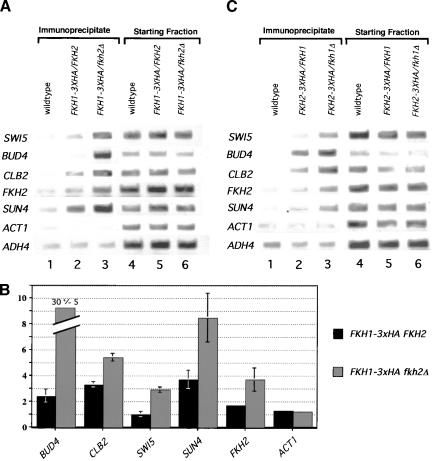
With the exception of BUD3, Fkh2p–3xHA enriched the CLB2-cluster promoter DNAs more effectively than Fkh1p–3xHA, suggesting that Fkh2p bound these targets more efficiently in vivo than Fkh1p (Koranda et al. 2000; Kumar et al. 2000). However, in contrast to these CLB2-cluster promoters that contain predicted strong Mcm1p-binding sites, Fkh1p–3xHA bound the BUD3, FKH2, and SUN4 promoters more efficiently than Fkh2p–3xHA, and it bound the SPS4 gene with a similar efficiency in vivo (Fig. 4). Mcm1p is required for DNA binding by SFF to CLB2-cluster promoters and proper expression of genes within the CLB2 cluster (Lydall et al. 1991; Althoefer et al. 1995; Maher et al. 1995; Kumar et al. 2000). The SUN4 and SPS4 genes were identified as potential non-CLB2-cluster gene targets of Fkh1p/Fkh2p by genome expression analysis of a fkh1Δ fkh2Δ strain (Zhu et al. 2000); neither gene contains a predicted match to a Mcm1p-binding site. Interestingly, the BUD3 promoter, a CLB2-cluster promoter that was occupied more efficiently by Fkh1p–3xHA than Fkh2p–3xHA, has a mutation within its Mcm1p-binding site. The BUD3-promoter fragment failed to bind Mcm1p in vitro (data not shown). Thus, in general, greater occupancy of target promoters by Fkh2p compared with Fkh1p correlated with the presence of a strong Mcm1p-binding site. In contrast, those promoters without Mcm1p-binding sites (FKH2, SUN4, and SPS4), or with a mutant Mcm1p-binding site (BUD3), were occupied equally well or more efficiently by Fkh1p–3xHA. Therefore, the relative amounts of Fkh1p and Fkh2p present at target sites in vivo was modulated significantly by promoter context, and in particular, the presence of a match to a Mcm1p-binding site.
Fkh2p, but not Fkh1p, bound to target promoters cooperatively with Mcm1p in vitro
One simple mechanistic explanation for the differences in Fkh1p and Fkh2p DNA binding in vivo was that each protein was modulated differently by interactions with Mcm1p. To test whether Mcm1p modulated the DNA-binding properties of either Fkh1p and Fkh2p in vitro, full-length recombinant Mcm1p was purified substantially from baculovirus-infected insect cells (Fig. 1A, lane 3) and used in EMSAs with either Fkh1p or Fkh2p (Fig. 5). Fkh1p bound with a similar affinity to the SWI5-promoter DNA fragment in the absence or presence of a constant amount of Mcm1p, (Fig. 5A, lanes 1–11, and B). In striking contrast, Fkh2p bound the SWI5-promoter fragment with a 100-fold higher affinity in the presence of a constant amount of Mcm1p (Fig. 5A, lanes 12–27, and B). These data indicated clearly that Fkh2p, but not Fkh1p, bound cooperatively with Mcm1p to the SWI5 promoter in vitro.
Figure 5.
Fkh2p, but not Fkh1p, bound to Fkh promoters cooperatively with Mcm1p in vitro. (A) A radiolabeled 200-bp SWI5 promoter was analyzed for binding to Fkh1p (lanes 1–11) or Fkh2p (lanes 12–27) as described in Figure 1 in the absence (lanes 1–5,12–19) or presence of a constant final concentration of Mcm1p (3 nM; lanes 6–11,20–27). The final concentrations of Fkh1p or Fkh2p were 0 nM (lanes 6,12,20), 0.019 nM (lanes 13,21), 0.075 nM (lanes 14,22), 0.30 nM (lanes 1,7,15,23), 1.2 nM (lanes 2,8,16,24), 4.8 nM (lanes 3,9,17,25), 19 nM (lanes 4,10,18,26), or 77 nM (lanes 5,11,19,27). (B) The data from three separate experiments, including the experiment shown in A were plotted as percent DNA bound by a Fkh protein (y-axis) vs. concentration of Fkh protein (x-axis). Free DNA was quantified by PhosphorImager and percent DNA bound was determined by subtracting the percent of free DNA from 100% (% bound = 100% − % free). The 100% value was determined as the total radioactivity in each lane. Each data point is the average of three independent experiments except for [Fkh2p] = 0.019 nM and [Fkh2p] = 0.075 nM, which are the average of two data points. (C) Mcm1p binding to a radiolabeled 200-bp region of the LYS2-coding region (lanes 1–4), the SWI5 promoter fragment (lanes 5–8), or the FKH2 promoter fragment (lanes 9–12) was analyzed by an EMSA as described in Figure 1. The final concentrations of Mcm1p used were 0 nM (lanes 1,5,9), 49 nM (lanes 2,6,10), 147 nM (lanes 3,7,11), and 393 nM (lanes 4,8,12). (D) A radiolabeled 200-bp FKH2-promoter fragment was analyzed for binding by Fkh2p in the absence (lanes 1–6) or presence (lanes 7–12) of a high concentration of Mcm1p (196 nM). The final concentrations of Fkh2p were 0 nM (lanes 1,7), 1.2 nM (lanes 2,8), 4.8 nM (lanes 3,9), 19 nM (lanes 4,10), 77 nM (lanes 5,11), and 308 nM (lanes 6,12).
The ability of Mcm1p to enhance the binding of Fkh2p, but not Fkh1p, to the SWI5 promoter in vitro explains why Fkh2p occupies most CLB2-cluster promoters to a greater extent than Fkh1p in vivo. However, Fkh2p does bind promoters in vivo that lack Mcm1p-binding sites, albeit less efficiently. For example, the FKH2 promoter, which lacks a Mcm1p-binding site and failed to bind Mcm1p efficiently in vitro (Fig. 5C), bound Fkh2p to some extent in vivo (Fig. 4). Therefore, to test whether Mcm1p could enhance Fkh2p binding to this promoter, we analyzed binding of Fkh2p to the FKH2 promoter in vitro in the absence and presence of Mcm1p (Fig. 5D). Mcm1p enhanced binding of Fkh2p to the FKH2 promoter significantly (Fig. 5D, lanes 1–10), albeit less effectively than it enhanced binding of Fkh2p to a promoter that contained an Mcm1p-binding site, because it required a higher concentration of Mcm1p. Thus, in vitro Mcm1p enhanced binding by Fkh2p to a promoter that lacked a Mcm1p-binding site, indicating that Mcm1p might play a role in stabilizing Fkh2p/DNA interactions in vivo at promoters that lack Mcm1p-binding sites.
Fkh2p stabilized Mcm1p–DNA interactions at Fkh-controlled promoters in vivo
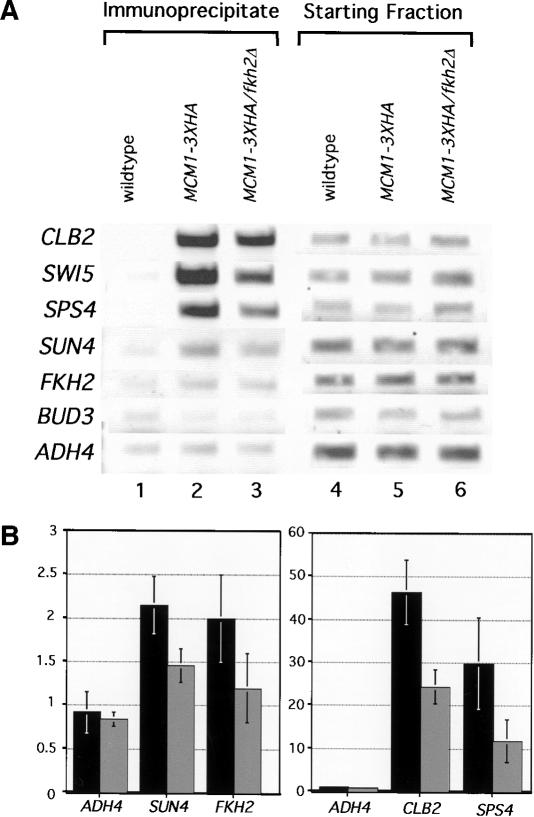
To test whether Mcm1p was present at Fkh-controlled promoters in vivo that lacked Mcm1p-binding sites, Mcm1p–3xHA occupancy was determined for several of the promoters examined above by use of α-HA-directed chromatin immunoprecipitation (Fig. 6). As expected, Mcm1p–3xHA was present at both the CLB2 and SWI5 promoters that contain Mcm1p-binding sites. Significantly, however, Mcm1p–3xHA was also present at the FKH2, SPS4, and SUN4 promoters that contain weak or no predicted Mcm1p sites (Fig. 6B). Thus, Mcm1p was present at promoters that bound Fkh2p, regardless of whether they contained Mcm1p-binding sites, suggesting that Fkh2p–Mcm1p interactions recruited Mcm1p to these promoters.
Figure 6.
Fkh2p stabilized Mcm1p–DNA interactions at Fkh-controlled promoters in vivo. (A) Occupancy of a variety of promoters by Mcm1p–3xHA (lanes 2,3) was measured in vivo by performing chromatin immunoprecipitation experiments with an α-HA antibody as described in Figure 4. Three isogenic strains were analyzed and the relative levels of the relevant promoter fragments present in the immunoprecipitate (lanes 1–3) or prior to immunoprecipitation (starting fraction, lanes 4–6) are shown. The three isogenic strains were wild type (CFY145) transformed with pRS316 [wild type (FKH1 FKH2) lanes 1,4], mcm1Δ∷HIS3 (CFY1006) carrying a plasmid with MCM1–3xHA (Ycp lac33 MCM1–3xHA) (lanes 2,5, MCM1–3xHA), or mcm1Δ∷HIS3 fkh2Δ∷HISG (CFY1055) carrying the MCM1–3xHA plasmid (lanes 3,6, MCM1–3xHA fkh2Δ). (B) Quantitative chromatin immunoprecipitations were performed as described in Figure 4. The quantitative data presented in the histograms were obtained using two isogenic strains that each contained the plasmid expressing MCM1–3xHA and either a wild-type chromosomal FKH2 gene (MCM1–3xHA; black bars) or a deletion of FKH2 (MCM1–3xHA fkh2Δ; gray bars). The values were normalized relative to chromatin immunoprecipitation performed in the wild-type strain (CFY145) as described in Figure 4. The low values for Mcm1p binding to the FKH2 and SUN4 promoters caused variation in the absolute values obtained in individual experiments and, hence, large error values. However, these error values overestimate the actual variation observed, because although the absolute values changed between experiments, the trend was the same.. For example, enrichment of the FKH2 and SUN4 promoters by Mcm1p–3xHA was always lower in the fkh2Δ strain compared with the FKH2 strain.
If interactions with Fkh2p play a role in stabilizing Mcm1p at Fkh-controlled promoters in vivo, then deletion of FKH2 should reduce occupancy of these promoters by Mcm1p. Therefore, we compared Mcm1p–3xHA at several promoters in strains that contained wild-type FKH2 or a deletion of FKH2 (fkh2Δ) (Fig. 6). Significantly, deletion of FKH2 reduced Mcm1p-occupancy of Fkh-controlled promoters regardless of whether these promoters possessed a Mcm1p-binding site. Strains containing a deletion of FKH2 expressed similar levels of Mcm1p compared with a wild-type strain (data not shown). Therefore, Fkh2p was required for a stable Mcm1p–DNA interaction at Fkh-controlled promoters in vivo.
Fkh2p/Mcm1p limited Fkh1p-occupancy of CLB2-cluster promoters in vivo
At promoters that contained a Mcm1p-binding site, Fkh2p bound more efficiently than Fkh1p in vivo. At promoters that lacked a Mcm1p-binding site, Fkh1p bound more efficiently than Fkh2p in vivo (Fig. 4). Together, these data were consistent with the hypothesis that Fkh1p and Fkh2p/Mcm1p compete for promoter-occupancy at Fkh-controlled promoters. Therefore, we tested whether Fkh1p–3xHA occupancy of Fkh-controlled promoters was enhanced in a strain lacking FKH2 (Fig. 7A,B) and, conversely, whether Fkh2p–3xHA occupancy was enhanced in a strain lacking FKH1 (Fig. 7C). α-HA-directed chromatin immunoprecipitation experiments were performed in two isogenic FKH1–3xHA-containing strains that differed only at the FKH2 locus; one strain contained wild-type FKH2, whereas the other contained a fkh2Δ mutation (Fig. 7A,B). Significantly, at all promoters examined, regardless of whether they contained a Mcm1p-binding site, deletion of FKH2 enhanced promoter-occupancy by Fkh1p–3xHA. Similarly, chromatin immunoprecipitation experiments performed in two isogenic FKH2–3xHA-containing strains that differed only at the FKH1 locus indicated that the presence of Fkh1p limited the ability of Fkh2p to occupy several promoters in vivo. These data indicate clearly that the Fkh1p and Fkh2p proteins compete for occupancy of Fkh-controlled promoters in vivo.
Figure 7.
Fkh2p/Mcm1p and Fkh1p limit each other's occupancy of CLB2-cluster promoters in vivo. (A) Occupancy of a variety of promoters by Fkh1p–3xHA was measured in vivo by performing chromatin immunoprecipitation experiments with an α-HA antibody. Three isogenic strains were analyzed after immunoprecipitation (lanes 1–3) and prior to immunoprecipitation (lanes 4–6) for the presence of promoter fragments listed at left by use of PCR and the appropriate promoter-specific primers. The strains analyzed were isogenic except for the presence or absence of FKH1, FKH2, or a HA epitope tag: (FKH1 FKH2, wild type, CFY 145; FKH1–3xHA FKH2, CFY442; FKH1–3xHA fkh2Δ, CFY443). (B) Quantitative analysis of chromatin immunuprecipitations that were performed as in A and analyzed as described for Fig. 4. The quantitative data presented in the histograms were obtained using two isogenic strains that each contained a chromosomal copy of FKH1–3xHA and either a wild-type copy of FKH2 (FKH1–3xHA FKH2, CFY442, black bars) or a deletion of FKH2 (FKH1–3xHA fkh2Δ, CFY443, gray bars). The values were normalized relative to chromatin immunoprecipitations performed in the wild-type strain (CFY145) as described in Fig. 4. (C) Occupancy of a variety of promoters by Fkh2p–3xHA was measured in vivo by performing chromatin immunoprecipitation experiments with an α-HA antibody. Three isogenic strains were analyzed after immunoprecipitation (lanes 1–3) and prior to immunoprecipitation (lanes 4–6) for the presence of promoter fragments listed at left by use of PCR and the appropriate promoter-specific primers. The strains analyzed were isogenic except for the presence or absence of FKH1, FKH2, or a HA epitope tag: (FKH1 FKH2, wild type, CFY 145; FKH2–3xHA FKH1, CFY854; FKH2–3xHA fkh1Δ, CFY884).
Discussion
In this work, we addressed the biochemical mechanisms that determine relative promoter-occupancy by the Fkh1 and Fkh2 proteins in vivo. Our in vitro and in vivo data, together with data presented in our earlier study (Hollenhorst et al. 2000), support a model in which Fkh1p is in competition with a stable Fkh2p/Mcm1p complex for promoter-occupancy at Fkh-controlled promoters. We postulate that the levels and or binding/activity of Fkh1p are more limiting for promoter-occupancy, providing a simple explanation for how differentiation and cell cycle progression can be fine tuned through regulation of Fkh1p.
Cooperative interactions at Fkh-controlled promoters as a determinant of relative promoter-occupancy by Fkh1p and Fkh2p
Purified Fkh1p and Fkh2p bound their DNA targets efficiently in vitro and with similar specificity. Nevertheless, Fkh2p occupies most promoters within the CLB2 cluster to a greater extent than Fkh1p in vivo (Koranda et al. 2000; Kumar et al. 2000). One striking biochemical difference between Fkh1p and Fkh2p helped explain these promoter-occupancy differences. Specifically, Fkh2p, but not Fkh1p, showed cooperative binding with Mcm1p. Because the majority of CLB2-cluster promoters contain strong Mcm1p-binding sites, a Mcm1p/Fkh2 interaction could stabilize substantially Fkh2p's interaction with CLB2-cluster promoters. Thus, Mcm1p would be expected to lower the concentration of Fkh2p relative to Fkh1p required for binding in vivo. As the steady-state levels of Fkh1p and Fkh2p are similar in vivo (Hollenhorst et al. 2000), these data help explain why Fkh2p occupies promoters in this class more effectively than Fkh1p.
We note that Fkh2p, but not Fkh1p, exhibited cooperative binding on its own that was most obvious at the CLB2 promoter. This ability may be related to some feature of bona fide target promoters, as the SIR1 promoter that contained a high-affinity Fkh site was bound by Fkh1p but not Fkh2p in vitro and was not a bona fide target promoter in vivo. One feature of Fkh-controlled promoters that may have some role in stabilizing binding by Fkh2p in vivo is the presence of multiple Fkh-binding sites. However, it remains to be determined what role multiple Fkh-binding sites and Fkh2p cooperative binding play in Fkh-controlled gene expression in vivo.
Fkh1p more efficiently occupies a second class of Fkh-controlled promoters in vivo relative to Fkh2p. This class includes the BUD3 and the FKH2 promoters. A common feature of promoters in this class is that they lack an obvious Mcm1p-binding site. Although Mcm1p could enhance binding by Fkh2p to these promoters in vitro, the concentration of Mcm1p required for efficient Fkh2p binding was higher than at promoters that contain Mcm1p-binding sites, such as CLB2-cluster promoters. Thus, in vivo, Fkh1p would be expected to be more competitive for binding to promoters that lack Mcm1p-binding sites, consistent with our observations. It is probable that additional factors regulate Fkh1p binding to Fkh-controlled promoters in this class, as Fkh1p was not detected at the SIR1 or FKH1 promoters in vivo, even though each of these promoters contained a high-affinity Fkh-binding site.
A strategy for regulating Fkh-controlled gene expression
Our data lead us to postulate that changes in the levels of Fkh1p may have a more significant impact on relative promoter-occupancy and regulation of Fkh-controlled gene targets than changes in the levels of Fkh2p. In particular, if Fkh2p binds to its DNA targets as a complex with Mcm1p, the levels of Fkh2p may not be the most limiting factor for promoter-occupancy by Fkh2p under most physiological conditions. Rather, the availability of Mcm1p competent to interact with Fkh2p may be more limiting than Fkh2p levels. If promoter-occupancy by Fkh2p is not limiting in vivo, then one expectation is that transcription regulation by Fkh2p might occur primarily through recruitment of additional transcription regulators such as Ndd1p (Koranda et al. 2000). This expectation is consistent with the observations that Fkh2p-occupancy of CLB2-cluster promoters appears constant throughout the cell cycle (Koranda et al. 2000) and that high-copy expression of FKH2 has no obvious effect on the cell cycle or transcription (Hollenhorst et al. 2000). However, in contrast to Fkh2p, increasing the levels of Fkh1p by approximately fivefold in vivo has a significant effect on the cell cycle-regulated transcription of CLB2 and the cell cycle (Hollenhorst et al. 2000). Importantly, deletion of FKH1 has the opposite effect and leads to a slight enhancement of CLB2 mRNA levels through much of the cell cycle (Hollenhorst et al. 2000). These data indicate that one physiological role for Fkh1p may be to attenuate transcription of CLB2 by competing with Fkh2p for occupancy of the CLB2 promoter. Interestingly, the levels of FKH1 mRNA change significantly under a variety of different environmental conditions, whereas the levels of FKH2 mRNA do not (Chu et al. 1998; Causton et al. 2001), providing compelling circumstantial evidence that changes in Fkh1p levels are physiologically relevant to its role in gene expression.
SRF/forkhead interactions in mammalian tissue-specific transcription
Mcm1p is the yeast homolog of mammalian serum response factor (SRF; Hayes et al. 1988; Jarvis et al. 1989; Treisman and Ammerer 1992), a MADS-box transcription factor (Shore and Sharrocks 1995) that controls multiple genes required for cell proliferation and differentiation in a variety of mammalian tissues (Johansen and Prywes 1995). Recently, a forkhead transcription factor that binds to the promoters of smooth muscle-specific genes in combination with SRF has been identified (Hoggatt et al. 2000). An analysis of one of these promoters reveals an arrangement of SRF and forkhead DNA-binding sites remarkably similar to the arrangement of Mcm1p and forkhead-binding sites in CLB2-cluster promoters. Thus, SRF may bind cooperatively with a forkhead protein to these promoters that control genes required for the differentiation of smooth muscle. In addition, two different forkhead proteins, an activator and a repressor, have been implicated in the regulation of smooth muscle-specific gene transcription through the same Fkh-binding site (Hoggatt et al. 2000). Similar observations have been made for forkhead/winged helix control of other tissue-specific promoters (Sawaya and Luse 1994). Thus, the use of MADS-box/forkhead interactions and multiple forkhead proteins in the regulation of functionally related genes has been conserved to a significant degree between yeast and mammals
Materials and methods
Expression of purification of recombinant proteins
Baculoviruses expressing Fkh1p–6xHis, Fkh2p–6xHis, or Mcm1p–6xHis were produced by cloning the coding regions of each protein inframe with a C-terminal 6× histidine tag into pVL1392 (Pharmingen) and transfecting with linearized viral DNA (Baculogold) into insect Sf9 cells. For protein purification, ∼3 × 108 Sf9 cells were infected with the relevant virus. A nuclear extract was prepared and precipitated with ammmonium sulfate and resuspended in Buffer H (Bell et al. 1995). Fkh1p–6xHis and Fkh2p–6XHis were each applied to a SP-sepharose column pre-equilibrated in BufferH/0.2 M KCl. The columns were developed with a 0.2 to 0.75 linear KCl gradient and peak fractions of Fkh1p–6xHis and Fkh2p–6xHis were determined by SDS-PAGE and pooled. The pooled fractions of Fkh1p–6xHis, Fkh2p–6xHis, and an ammonium sulfate precipitate of a nuclear extract containing Mcm1p–6xHis were each equilibrated in Buffer N (50 mM Tris at pH 8, 0.5 M NaCl, 25 mM imidazole, 0.02% NP-40, 10% glycerol) and allowed to bind in batch overnight to ∼0.5 mL of charged Ni-sepharose. The resin was poured into a small column and the protein eluted in Buffer N/0.5 M imidazole. The fractions containing protein were transferred to Buffer H/0.15 M KCl by gel filtration. Protein concentrations were determined by comparison to a BSA standard using Coomassie staining of a SDS–polyacrylamide gel.
EMSAs
The 200-bp promoter fragments were obtained by PCR amplification of genomic DNA using the relevant PCR primers (Table 1) and subcloned into a pUC vector. To obtain radioactive probes for use in EMSAs, the relevant fragment was amplified from the appropriate subclone by performing standard PCR in the presence of [α-32P]dCTP. The amplified fragment was then purified from a polyacrylamide gel and quantified by liquid scintillation counting. Purified probes (1000 cpm), ∼45 pM final concentration, were incubated with an excess of the relevant purified protein as indicated in the figure legends in EMSA buffer (20 mM Hepes at pH 7.9, 50 mM KCl, 5 mM MgCl2, 3 mM DTT, 0.10 mg BSA, 0.25 mg/mL (final concentration) dI/dC, and 10% glycerol) for 15 min at room temperature followed by 15 min on ice. The samples were analyzed by electrophoresis through a 4.5% polyacrylamide gel at 200V at 4 °C for 2 h. The gels were dried and quantified using a PhosphorImager.
Table 1.
Primers used in this study
| Locus
|
Primer pair
|
|
|---|---|---|
| EMSA | ||
| ALK1 | GGTTTCCCTTCCCGTTATTATG | GGGTATGACTTGATAGTAATTC |
| ASE1 | CTGCCTATAAACAATAAGTCTG | CAGTGAAAAAACCAGGTATTCGAC |
| BUD3 | CCTAAGCTATAAATTAGGTC | GCAGGTAGCAGCAGTAGAATGC |
| BUD4 | CGAGAAACGCCCCCCTTTGTCCC | CCAACGAAGTTTAGTGAATGGAG |
| CDC5 | CCGGGTAGGGCATTATGTAGC | GAGTAACTGTTCAAGATATGCC |
| CDC20 | GCGGTTCTTTGTTAGCTTTCC | GGTGATAAATTCTTTGCCTGC |
| CLB2 | GCCAAAAAGACAGATTTTATTCCAAATGCGG | GCTATTTCCTCGAGGGACAGGCACC |
| FKH1 | CACTAGGGCTTGAAGAGCGTAGG | GCCGGTATTCATCTAAACCTCTTACC |
| FKH2 | CGCTCGCCCTGTGCCATTTGG | GCCTTTTGTTCCTTAGTATGATCTTG |
| HAT1 | GGTGAAGCTTGAAGCTCGC | CGTAACAGCCAACTTCGAAAC |
| HST3 | GTATTTCCCCTTTGGCACATTCG | GCAGCACCAGCATCATAATCC |
| LYS2 | GAAACACAGCGTGCAGTTTCTTATTTCG | GCACGAACATAAATCTCACCTATTTCGCC |
| SIR1 | CCGCCCACAAACACAAAGG | GAGGAACAATTCTATTAAATTCA |
| SWI5 | CGATGCCTTTTTGTCTTCCGTGCC | GGATGAAATTTTCACTTTCGTACTTTAACC |
| YJL051w | CACTGTTTAAAAGTTATGGCGAGAACG | CCACCTCTTCAAAGGATAATGCG |
| YLR084c | GATTTACATAGCAACAATGAAAATCCG | CGCGAAAAAATGGGAAGAATGGC |
| ChIP | ||
| ACT1 | CGGTATTGTCACCAACTGGGACGATATGG | GCAGCGGTTTGCATTTCTTGTTCGAAGTCC |
| ADH4 | GGCTACTAACGGTGGGGAAATCGGAGAC | GCACAGGCATCGGTGATTGGGTTAGAGGC |
| BUD3 | GCAGGTAGCAGCAGTAGAATGC | CCGTTCATCTCATAGTGC |
| BUD4 | CGAGAAACGCCCCCCTTTGTCCC | GGTAGACATCTTAGAAGGTGATCG |
| CDC20 | GGTGATAAATTCTTTGCCTGC | CGTTAGTTGAACTTGAATTCCG |
| CLB2 | GCCAAAAGACAGATTTTATTCCAAATGCGG | CGCTTTTCAGAAGTATCAATTCG |
| FKH1 | CGATTACTGGTTGTCTTCAATGCAAGG | CCAAGCCTTTCTTTGCTAATTGAACGC |
| FKH2 | GCAACATTTTGATGGCCTCTTCTTCC | GCGAGGGAATCGGGAAGATTGGC |
| HAT1 | CGTAACAGCCAACTTCGAAAC | GAAAATGCACCTTCTCCAACTC |
| SIR1 | CCGCCCACAAACACAAAGG | GCAATAACAGATAAAGATAAGG |
| SPS4 | CTGACATTGAAAAGGCAAAGCAACTG | GCAACGCGTCTGACGCTGCTGC |
| SUN4 | GGTTACCCGACATATATGCTGG | CGGCGAAAAAGGAGTGTAGAG |
| SWI5 | CGATGCCTTTTTGTCTTCCGTGCC | GGGCCAATGTTATTTCTGTCTTAAAGG |
Yeast strains
The yeast strains used in this study were constructed by use of standard yeast molecular genetic techniques (Guthrie and Fink 1991) as described previously (Hollenhorst et al. 2000). The plasmid containing MCM1–3xHA (Ycp lac33 MCM1–HA) was provided by Elizabeth Grayhack (Kuo et al. 1997).
Chromatin immunoprecipitation and analysis of immunoprecipitated DNA
Chromatin immunoprecipitations were performed as described (Strahl-Bolsinger et al. 1997) except that the standard cross-linking time was 15 min. Quantitative chromatin immunoprecipitations were performed by including 2 μCi of α-[32P]dCTP in each PCR reaction. PCR was directed by the relevant primer pairs listed in Table 1. Radiolabeled products were separated by gel electrophoresis through a 4.5% polyacrylamide gel and quantified using a PhosphorImager. Fold-enrichments of a promoter in these experiments are reported as values that have been normalized by dividing the amount of promoter fragment obtained from a chromatin immunoprecipitation of an epitope-tagged strain by the amount obtained from a wild-type (untagged) strain.
Acknowledgments
We thank Elizabeth Grayhack for her generosity in providing several MCM1 plasmids, and Kelly Gardner and Madeleine DeBeer for protocols. We also thank Michael Sheets, Jaerek Marszalek, and members of the Fox and Sheets laboratories for advice and constructive criticism, and Ulrika Muller for significant help in performing the experiment shown in Figure 4C. This work was supported by a grant from the National Institutes of Health to C.A.F. and a grant to the University of Wisconsin Medical School under the Howard Hughes Medical Institute Research Resources Program for Medical Schools. C.A.F. is a recipient of a Burroughs Wellcome Fund Career Award in the Biomedical Sciences.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL cfox@facstaff.wisc.edu; FAX (608) 262-5253.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.906201.
References
- Althoefer H, Schleiffer A, Wassmann K, Nordheim A, Ammerer G. Mcm1 is required to coordinate G2-specific transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:5917–5928. doi: 10.1128/mcb.15.11.5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati B, Land H. Myc–Max–Mad: A transcription factor network controlling cell cycle progression, differentiation and death. Curr Opin Genet Dev. 1994;4:102–108. doi: 10.1016/0959-437x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- Bell SP, Mitchell J, Leber J, Kobayashi R, Stillman B. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell. 1995;83:563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- Breeden LL. Cyclin transcription: Timing is everything. Curr Biol. 2000;10:586–588. doi: 10.1016/s0960-9822(00)00634-5. [DOI] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Futcher B. Cyclins and the wiring of the yeast cell cycle. Yeast. 1996;12:1635–1646. doi: 10.1002/(SICI)1097-0061(199612)12:16%3C1635::AID-YEA83%3E3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. San Diego, CA: Academic Press; 1991. [Google Scholar]
- Hayes TE, Sengupta P, Cochran BH. The human c-fos serum response factor and the yeast factors GRM/PRTF have related DNA binding specificities. Genes & Dev. 1988;2:1713–1722. doi: 10.1101/gad.2.12b.1713. [DOI] [PubMed] [Google Scholar]
- Hoggatt AM, Kriegel AM, Smith AF, Herring BP. Hepatocyte nuclear factor-3 homologue 1 (HFH-1) represses transcription of smooth muscle-specific genes. J Biol Chem. 2000;275:31162–31170. doi: 10.1074/jbc.M005595200. [DOI] [PubMed] [Google Scholar]
- Hollenhorst PC, Bose ME, Mielke MR, Muller U, Fox CA. Forkhead genes in transcriptional silencing, cell morphology and the cell cycle: Overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae. Genetics. 2000;154:1533–1548s. doi: 10.1093/genetics/154.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hromas R, Costa R. The hepatocyte nuclear factor-3/forkhead transcription regulatory family in development, inflammation, and neoplasia. Crit Rev Oncol Hematol. 1995;20:129–140. doi: 10.1016/1040-8428(94)00151-i. [DOI] [PubMed] [Google Scholar]
- Jarvis EE, Clark KL, Sprague GFJ. The yeast transcription activator PRTF, a homolog of the mammalian serum response factor, is encoded by the MCM1 gene. Genes & Dev. 1989;3:936–945. doi: 10.1101/gad.3.7.936. [DOI] [PubMed] [Google Scholar]
- Johansen PE, Prywes R. Serum response factor: Transcriptional regulation of genes induced by growth factors and differentiation. Biochim Biophys Acta. 1995;1242:1–10. doi: 10.1016/0304-419x(94)00014-s. [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes & Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- Kaufmann E, Knochel W. Five years on the wings of fork head. Mech Dev. 1996;57:3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- Kaufmann E, Muller D, Knochel W. DNA recognition site analysis of Xenopus winged helix proteins. J Mol Biol. 1995;248:239–254. doi: 10.1016/s0022-2836(95)80047-6. [DOI] [PubMed] [Google Scholar]
- Koranda A, Schleiffer A, Endler L, Ammerer G. Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature. 2000;406:94–98. doi: 10.1038/35017589. [DOI] [PubMed] [Google Scholar]
- Kron SJ, Styles CA, Fink GR. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Reynolds DM, Shevchenko A, Shevchenko A, Goldstone SD, Dalton S. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr Biol. 2000;10:896–906. doi: 10.1016/s0960-9822(00)00618-7. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Nadeau ET, Grayhack EJ. Multiple phosphorylated forms of the Saccharomyces cerevisiae Mcm1 protein include an isoform induced in response to high salt concentrations. Mol Cell Biol. 1997;17:819–832. doi: 10.1128/mcb.17.2.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E, Clark KL, Burley SK, Darnell JE., Jr Hepatocyte nuclear factor 3/fork head or “winged helix” proteins: A family of transcription factors of diverse biologic function. Proc Natl Acad Sci. 1993;90:10421–10423. doi: 10.1073/pnas.90.22.10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levrero M, De Laurenzi V, Costanzo A, Gong J, Wang JY, Melino G. The p53/p63/p73 family of transcription factors: Overlapping and distinct functions. J Cell Sci. 2000;113:1661–1670. doi: 10.1242/jcs.113.10.1661. [DOI] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: Regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydall D, Ammerer G, Nasmyth K. A new role for MCM1 in yeast: Cell cycle regulation of SWI5 transcription. Genes & Dev. 1991;5:2405–2419. doi: 10.1101/gad.5.12b.2405. [DOI] [PubMed] [Google Scholar]
- Maher M, Cong F, Kindelberger D, Nasmyth K, Dalton S. Cell-cycle regulated trasncription of the CLB2 gene is dependent on Mcm1 and a ternary complex factor. Mol Cell Biol. 1995;15:3129–3137. doi: 10.1128/mcb.15.6.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema RH, Kops GJ, Bos JL, Borgering BM. AFX-like forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–787. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Nakamura N, Ramaswamy S, Vazquez F, Signoretti S, Loda M, Dellers WR. Forkhead transcription factors are critical effectors of cell death and cell cycle arrest downstream of PTEN. Mol Cell Biol. 2000;20:8969–8982. doi: 10.1128/mcb.20.23.8969-8982.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overdier DG, Porcella A, Costa RH. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol Cell Biol. 1994;14:2755–2766. doi: 10.1128/mcb.14.4.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pic A, Lim FL, Ross SJ, Veal EA, Johnson AL, Sultan MR, West AG, Johnston LH, Sharrocks AD, Morgan BA. The forkhead protein Fkh2 is a component of the yeast cell-cycle transcription factor SFF. EMBO J. 2000;19:3750–3761. doi: 10.1093/emboj/19.14.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya PL, Luse DS. Two members of the HNF-3 family have opposite effects on a lung transcriptional element; HNF-3a stimulates and HNF-3b inhibits activity of region I from the Clara cell secretory protein (CCSP) promoter. J Biol Chem. 1994;269:22211–22216. [PubMed] [Google Scholar]
- Shore P, Sharrocks D. The MADS-box family of transcription factors. Eur J Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K. Comprehensive identification of cell-cycle regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. Sir2 and Sir4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Kirito K, Kashii Y, Uchida M, Watanabe T, Endo H, Endoh T, Sawada KK, Ozawa K, Komatsu N. Forkhead family transcription factor FKHRL1 is expressed in human megakaryocytes and regulates cell cycling as a downstream molecule of TPO signaling. J Biol Chem. 2001;276:15082–15089. doi: 10.1074/jbc.M007958200. [DOI] [PubMed] [Google Scholar]
- Treisman R, Ammerer G. The SRF and MCM1 transcription factors. Curr Opin Genet Dev. 1992;2:221–226. doi: 10.1016/s0959-437x(05)80277-1. [DOI] [PubMed] [Google Scholar]
- Yang Q, Bassel-Duby R, Williams RS. Transient expression of a winged-helix protein, MNF-β, during myogenesis. Mol Cell Biol. 1997;17:5236–5243. doi: 10.1128/mcb.17.9.5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Spellman PT, Volpe T, Brown PO, Botstein D, Davis TN, Futcher B. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature. 2000;406:90–94. doi: 10.1038/35017581. [DOI] [PubMed] [Google Scholar]