Abstract
The 22q11.2 deletion syndrome (22q11DS) is associated with an increased prevalence (20–30%) of schizophrenia. Therefore, it is likely that one or more genes within the 22q11.2 region are causally related to schizophrenia. Recently, a significant association with schizophrenia in the general population was reported for three SNPs in phosphatidyl-inositol-4-kinase-catalytic-α (PIK4CA), a gene located in the 22q11.2 region. In the current study, we tested the hypothesis that the same PIK4CA risk-alleles would be associated with schizophrenia in individuals with 22q11DS. Our analysis of the PIK4CA genotypes in a sample of 79 adults with typical 22q11.2 deletions, comparing those with schizophrenia to those without, revealed a significant association. Our findings represent an independent replication of the previously reported PIK4CA association with schizophrenia in the general population. Second, the results of this study indicate that variation at PIK4CA may be a relevant factor influencing the risk of schizophrenia in individuals with 22q11DS.
Keywords: schizophrenia, chromosome 22q11.2 deletion syndrome, PIK4CA, susceptibility gene
In 22q11.2DS patients, schizophrenia is far more common than in the general population (20%–30% vs. 1%) [Bassett and Chow, 2008]. Conversely, 22q11.2 deletions are found in 0.75% (95% Confidence Interval: 0.5–1.2%) of patients with schizophrenia, which is approximately 30 times more frequently than in the general population [Bassett and Chow, 2008; Hoogendoorn et al., 2008]. These findings strongly suggest that the 22q11.2 region harbors one or more schizophrenia-susceptibility genes. Nevertheless, approximately 70–80% of 22q11.2DS patients do not develop schizophrenia. This suggests the influence of other factors, including genetic variation of the remaining alleles within the 22q11.2 deletion region.
Recently, we reported a significant association of three SNPs within the phosphatidyl-inositol-4-kinase-catalytic-α gene (PIK4CA) with schizophrenia in the general population [Jungerius et al., 2007]. PIK4CA is located within the distal half of the region that is typically deleted in 22q11.2DS. We hypothesized that the same PIK4CA SNPs could contribute to the risk of schizophrenia in individuals with 22q11.2DS. We examined the distribution of these PIK4CA SNPs in adults with 22q11.2DS, comparing those with schizophrenia (“cases”) to those without (“controls”).
Adults with 22q11.2DS, followed at the Clinical Genetics Research Program, Centre for Addiction and Mental Health (CAMH) at the University of Toronto, Canada, were studied. Participants provided written informed consent, and the study was approved by the Research Ethics Boards of the University of Toronto, CAMH, and University Health Network.
Eighty-four subjects of Caucasian descent were included and assessed by experienced psychiatrists for lifetime DSM-IV psychiatric diagnoses using standard methods, as previously described [Bassett et al., 2007]. In order to exclude a stratification error related to intellectual level, we used IQ results available for 77 participants [Bassett et al., 2007].
Approximately 13% of 22q11.2 deletions differ from the most commonly found 3 Mb deletion [Shaikh et al., 2000]. In the majority of this subgroup a smaller proximal deletion involves a region that does not contain PIK4CA. Therefore, we used multiplex ligation-dependent probe amplification (MLPA) [Jalali et al., 2008] to verify the involvement of PIK4CA in the 22q11.2 deletions. Of the initial 84 subjects, seven (8.3%) were found to have smaller or atypical 22q11.2 deletions, five of which did not include PIK4CA. The latter subjects were therefore excluded, leaving 79 subjects confirmed to be hemizygous at the PIK4CA gene for genotyping studies.
We performed genotyping of previously reported PIK4CA SNPs (rs165793, rs2072513 and rs165862) using an allele specific PCR method (KASPar assay, performed by KBiosciences, Hoddesdon, Hertfordshire, UK). In one sample, genotyping of rs2072513 did not generate reliable results.
Thirty-two patients were diagnosed with schizophrenia or schizoaffective disorder (“cases”; 15 males, 17 females, mean age 40.0 years, SD 8.7) while in 47 participants psychotic illness was excluded (“controls”; 22 males, 25 females, mean age 30.0 years, SD 9.4).
We compared allele and haplotype frequencies between cases and controls using chi-square or Fisher’s Exact tests. Sex distribution and mean IQ were compared between cases and controls using chi-square and Student’s t-test, respectively (see Table I). Given the modest size of this sample, no other SNPs were genotyped and no other phenotypes were included in the analysis to minimize multiple testing issues.
TABLE I.
Demographic Characteristics and Genotype Results in 22q11.2DS Cases and Controls
| 22q11.2DS schizophrenia (cases) | 22q11.2DS (controls) | p (Bonferroni corrected p)d | Odds ratio (95% confidence interval) | |
|---|---|---|---|---|
| Sample size (n) | n = 32 | n = 47 | ||
| Gender (M/Fa) | 0.88 | 0.88 | 0.995 | |
| Intelligence (mean FSIQ ± SD) | 69.0 ± 10.1 | 73.1 ± 10.1 | 0.081 | |
| rs165793b (G, A) | 32, 0 | 36, 11 | 0.002 (0.006) | 9.47 (1.16–77.56) |
| rs2072513c (C, T) | 15, 17 | 14, 32 | 0.140 (0.42) | 2.02 (0.79–5.14) |
| rs165862 (G, T) | 22, 10 | 22, 25 | 0.054 (0.162) | 2.50 (0.98–6.41) |
| Haplotypeb (TTA, other) | 0, 32 | 10, 36 | 0.004 | 8.61 (1.04–71.10) |
M/F is male to female ratio.
In both instances the odds ratios were calculated after artificially attributing one call to the genotype or haplotype with zero (respectively rs165793-G and the TTA haplotype) in the cases group.
In one control sample, genotyping of rs2072513 did not generate reliable results.
Correction of n = 3 single allele comparisons.
Cases and controls did not differ with regard to sex distribution (P = 0.995) or mean IQ; (cases: 69.0, SD 10.1, controls: 73.1, SD 10.1, P = 0.081). The intronic SNP rs165793 was significantly associated with schizophrenia; the G-allele was found in all 32 cases and in 36/47 controls (77%), P = 0.002. The G-allele of rs165862 was found in 22/32 (69%) cases versus 22/47 (47%) controls (P = 0.054). The C-allele of rs2072513 was found in 15/32 (47%) cases versus 14/46 (30%) controls (P = 0.140). Closer examination showed that the previously reported protective TTA haplotype (rs2072513, rs165862, rs165793) [Jungerius et al., 2007] was found in none of the 32 cases and in 10/46 (22%) controls (P = 0.004). The GCG haplotype showed an association with schizophrenia susceptibility, [cases: 15/32 (47%), controls: 13/46 (28%)], although this effect did not reach statistical significance in this sample (P = 0.092). The mean IQ of 66 rs165793 G-allele carriers (71.5, SD 10.8) was not significantly different from that of 11 A-allele carriers (70.9, SD 6.5, P = 0.868).
Calculation of the odds ratio (OR) for the most significantly associated risk allele (rs165793-G) is hampered by the fact that the contingency table of the allele distribution includes an empty cell. When artificially attributing one rs165793-A call in the cases group (cases n = 32, rs165793-G: n = 31, rs165793-A: n = 1), the resulting estimation of the OR is 9.47 (95% Confidence Interval: 1.16–77.56). Similarly, for the TTA haplotype, after attributing one TTA call in the cases group (cases n = 32, TTA: n = 1, other haplotypes: n = 31) the resulting OR is 8.61 (95% Confidence Interval: 1.04–71.10).
Genotypes of the five samples with 22q11.2 deletions that did not include PIK4CA were used as a quality control. Consistent with the shorter deletions identified by MLPA, heterozygosity at any of the three PIK4CA SNPs occurred exclusively in these samples. Of these five subjects, only one was diagnosed with schizophrenia; (PIK4CA rs165793 genotype: GG).
In this study we report a significant association of a common variant at PIK4CA with schizophrenia in adults with 22q11.2DS. We identified the same protective haplotype (TTA) as in our previous study [Jungerius et al., 2007] to be significantly more prevalent in controls. Also similar to the Jungerius et al. [2007] results (cases: 87.1%, controls: 79%, P = 1.25 × 10−5, OR = 1.80 95% Confidence Interval: 1.38–2.34), the rs165793-G-allele was found to be significantly more prevalent in the 22q11.2DS schizophrenia group.
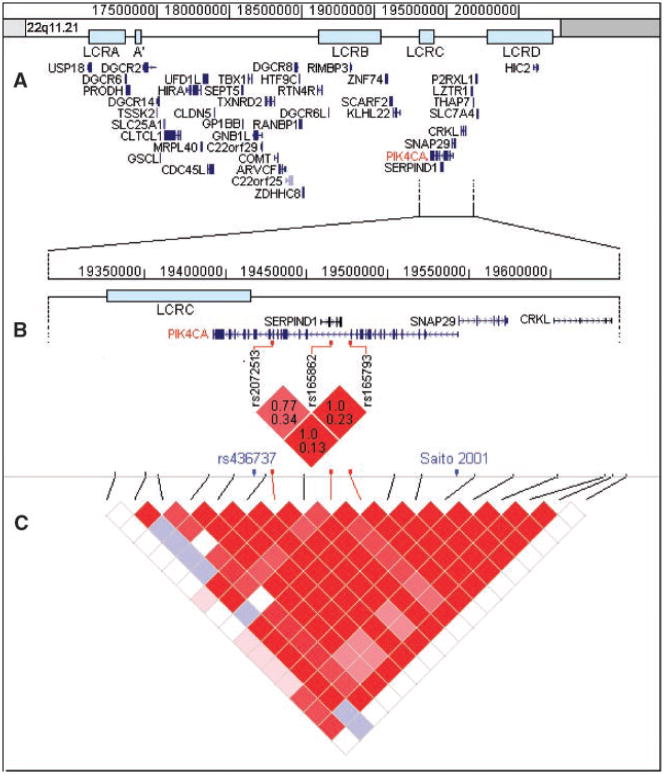
Although the OR of the TTA haplotype (8.61) is slightly less than the OR of the rs165793-G-allele (9.47) this difference is not significant. Therefore, it is difficult to draw firm conclusions with regard to the question of whether the effect of the rs165793-G-allele alone is greater than that of the effect of the TTA haplotype. As can be seen in Figure 1, there is linkage disequilibrium (LD) between the three genotyped markers (Fig. 1B). However, although substantial, the LD is far from complete with r-squared values ranging between 0.13 and 0.34. Therefore the significant association of the TTA-haplotype identified in the current study and in the study by Jungerius et al. cannot be fully explained by a “tagging effect” of the rs165793-A-allele with regard to the other two markers. In addition, the distribution of the GCG haplotype indicated an association with schizophrenia susceptibility, although this effect did not reach statistical significance in this sample (P = 0.092). Notably, Jungerius et al. found this haplotype in 29.5% of control subjects, similar to the rate found in the current study (28%).
FIG. 1.
Upper panel A: Genes located in the 22q11.2 region (source: RefSeq genes, UCSC genome browser March 2006 assembly; http://www.genome.ucsc.edu). The majority of 22q11.2 deletions occur between low copy repeats (LCRs) A and D. Middle panel B: Enlargement of the region around the three genotyped markers of the current study with LD plot. In each square the upper number is D′, the lower number is r-squared. In blue font: the only non-synonymous coding SNP with average heterozygosity >0.10 (rs436737) in this haploblock, and the SNP previously reported to be associated with schizophrenia [Saito et al., 2001; Wonodi et al., 2005] are denoted. Lower panel C: LD plot of the region around PIK4CA. Red vertical lines denote the three SNPs used in the current study. The LD plots are obtained from markers genotyped in CEU panel, analyzed with Haploview [Barrett et al., 2005].
LD in this region expands distally towards the SNAP29 gene (see Fig. 1C). Within this LD-block there is only one common non-synonymous coding SNP (rs436737, average heterozygosity 0.375). Other non-synonymous coding SNPs in this region are rare (i.e., average heterozygosity <0.1). Interestingly, an association with schizophrenia was reported in two studies for a polymorphism located in a region which is both the first intron of PIK4CA and the SNAP29 promotor region [Saito et al., 2001; Wonodi et al., 2005], (see Fig. 1B). Possibly, in these studies, as well as in the current study, the same signal is detected because of the LD in this region.
The findings of the current study represent an independent replication of our previous study with similar frequencies of PIK4CA alleles in this special sample of 22q11.2DS patients as in general population samples [Jungerius et al., 2007]. Further, the absence of effect of the rs165793 risk allele on IQ in the current study suggests an influence on brain function that is related to risk of schizophrenia in 22q11.2DS rather than to intellectual level. However, in order to better understand the neurobiological processes affected by PIK4CA, future studies using larger samples are needed to investigate whether PIK4CA is associated with basic cognitive processes that have been related to schizophrenia such as measures of executive functioning, verbal fluency or working memory.
PIK4CA is a catalytic enzyme in the phosphatidylinositol (PI) pathway, involved in the regulation of signal transduction, synaptic transmission and possibly of cell shape of neurons or oligodendrocytes [Jungerius et al., 2007]. Interestingly, PIK4CA is expressed in the gray matter, with higher expression in fetal than adult brain [Nakagawa et al., 1996], consistent with a neurodevelopmental pathogenesis of schizophrenia [Bassett et al., 2001].
In conclusion, 22q11.2DS patients who carry the rs165793-G-allele in their single copy of the PIK4CA gene are at increased risk of developing schizophrenia. The rather robust effect size of the reported association suggests that variation in PIK4CA affects the likelihood of expression of schizophrenia in 22q11.2DS, although it is likely that other schizophrenia risk genes reside within the 22q11.2 region. The findings of this study support the potential power of this population to examine risk factors for schizophrenia and associated phenotypes [Bassett and Chow, 2008]. Indeed, the results provide additional evidence that variation at PIK4A may be involved as one of the multiple genetic factors that contribute to the risk of schizophrenia in the general population.
Acknowledgments
The authors thank the patients and their family members for their participation, staff and students at the Clinical Genetics Research Program, and colleagues at the Toronto Congenital Cardiac Centre for Adults, Hospital for Sick Children and Centre for Addiction and Mental Health and many others for referring patients. This research was supported by grants from the W. Garfield Weston Foundation, Canadian Institutes of Health Research (MOP-79518), a NARSAD Distinguished Investigator Award (ASB) and Canada Research Chair in Schizophrenia Genetics (ASB). J.A.S. Vorstman M.D., Ph.D. was supported by a 2006 NARSAD Young Investigator Award, funded by Stephen and Constance Lieber.
References
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bassett AS, Chow EW. Schizophrenia 22q11.2 deletion syndrome. Curr Psychiatry Rep. 2008;10:148–157. doi: 10.1007/s11920-008-0026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AS, Chow EW, O’Neill S, Brzustowicz LM. Genetic insights into the neurodevelopmental hypothesis of schizophrenia. Schizophr Bull. 2001;27:417–430. doi: 10.1093/oxfordjournals.schbul.a006884. [DOI] [PubMed] [Google Scholar]
- Bassett AS, Caluseriu O, Weksberg R, Young DA, Chow EW. Catechol-O-methyl transferase and expression of schizophrenia in 73 adults with 22q11 deletion syndrome. Biol Psychiatry. 2007;61:1135–1140. doi: 10.1016/j.biopsych.2006.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogendoorn ML, Vorstman JA, Jalali GR, Selten JP, Sinke RJ, Emanuel BS, Kahn RS. Prevalence of 22q11.2 deletions in 311 Dutch patients with schizophrenia. Schizophr Res. 2008;98:84–88. doi: 10.1016/j.schres.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalali GR, Vorstman JA, Errami A, Vijzelaar R, Biegel J, Shaikh T, Emanuel BS. Detailed analysis of 22q11.2 with a high density MLPA probe set. Hum Mutat. 2008;29:433–440. doi: 10.1002/humu.20640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungerius BJ, Hoogendoorn ML, Bakker SC, Van’t SR, Bardoel AF, Ophoff RA, Wijmenga C, Kahn RS, Sinke RJ. An association screen of myelin-related genes implicates the chromosome 22q11 PIK4CA gene in schizophrenia. Mol Psychiatry. 2007 Sep 25; doi: 10.1038/sj.mp.4002080. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Goto K, Kondo H. Cloning, expression, and localization of 230-kDa phosphatidylinositol 4-kinase. J Biol Chem. 1996;271:12088–12094. doi: 10.1074/jbc.271.20.12088. [DOI] [PubMed] [Google Scholar]
- Saito T, Guan F, Papolos DF, Rajouria N, Fann CS, Lachman HM. Polymorphism in SNAP29 gene promoter region associated with schizophrenia. Mol Psychiatry. 2001;6:193–201. doi: 10.1038/sj.mp.4000825. [DOI] [PubMed] [Google Scholar]
- Shaikh TH, Kurahashi H, Saitta SC, O’Hare AM, Hu P, Roe BA, Driscoll DA, McDonald-McGinn DM, Zackai EH, Budarf ML, Emanuel BS. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: Genomic organization and deletion endpoint analysis. Hum Mol Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- Wonodi I, Hong LE, Avila MT, Buchanan RW, Carpenter WT, Jr, Stine OC, Mitchell BD, Thaker GK. Association between polymorphism of the SNAP29 gene promoter region and schizophrenia. Schizophr Res. 2005;78:339–341. doi: 10.1016/j.schres.2005.03.023. [DOI] [PubMed] [Google Scholar]