Abstract
Interaction of cell surface glycoproteins with endogenous lectins on the cell surface regulates formation and maintenance of plasma membrane domains, clusters signaling complexes, and controls the residency time of glycoproteins on the plasma membrane. Galectin-9 is a soluble, secreted lectin that binds to glycoprotein receptors to form galectin–glycoprotein lattices on the cell surface. Whereas galectin-9 binding to specific glycoprotein receptors induces death of CD4 Th1 cells, CD4 Th2 cells are resistant to galectin-9 death due to alternative glycosylation. On Th2 cells, galectin-9 binds cell surface protein disulfide isomerase (PDI), increasing retention of PDI on the cell surface and altering the redox status at the plasma membrane. Cell surface PDI regulates integrin function on platelets and also enhances susceptibility of T cells to infection with HIV. We find that galectin-9 binding to PDI on Th2 cells results in increased cell migration through extracellular matrix via β3 integrins, identifying a unique mechanism to regulate T-cell migration. In addition, galectin-9 binding to PDI on T cells potentiates infection with HIV. We identify a mechanism for regulating cell surface redox status via a galectin–glycoprotein lattice, to regulate distinct T-cell functions.
Control of the cell surface redox environment regulates essential functions by altering disulfide bonds in cell surface proteins (1, 2). Though disulfide bonds can be essential for protein stability or conformation, disulfide bonds in some cell surface proteins can undergo reversible reduction, so that the presence or absence of disulfide bonds can act as a switch to turn on and off specific protein activities (1, 2). Though factors that control the cell surface redox environment are not well understood, the thiol content of cell surface proteins is dynamically regulated; for example, activated B and T lymphocytes have increased cell surface thiols compared with resting cells, with the greatest increase seen on CD4 T cells (3).
Protein disulfide isomerases (PDIs) are a family of soluble oxidoreductases that act in the endoplasmic reticulum to promote disulfide bond formation and efficient folding of nascent proteins (4). Specific PDIs have also been identified at the surface of lymphocytes, platelets, endothelial cells, hepatocytes, and cancer cells (4–6). Cell surface PDI (primarily P4HB) can catalyze reduction of disulfide bonds in cell surface proteins. For example, PDI reduction of disulfide bonds in β-integrins regulates adhesion and migration of platelets and tumor cells (4–7), and PDI has been proposed to alter the conformation of viral fusion proteins and their cell surface receptors to facilitate viral entry into target cells (5, 8, 9). PDI can directly interact with protein substrates, as PDI associates with β3 integrin on the surface of platelets (7, 10), and PDI forms a complex with HIV gp120, CD4, and CXCR4 on the surface of T cells (5, 9, 11–13). However, it is not known how PDI is retained on the cell surface to dynamically modify thiols in cell surface proteins.
Galectins, a family of mammalian lectins, control numerous biological functions, including cell proliferation and death, adhesion and migration, and interaction of host cells with microbial pathogens, by binding to glycan ligands on specific glycoprotein or glycolipid receptors (14–16). Galectin-9, expressed by T cells, eosinophils, endothelial cells, dendritic cells, and macrophages (14, 15), can kill T cells and thymocytes (17). Galectin-9 kills CD4 Th1 cells but spares CD4 Th2 cells (18); one mechanism for resistance of CD4 Th2 cells to galectin-9 is the abundance of α2,6-linked sialic acids on the surface of Th2 cells, which blocks galectin-9 binding to glycan receptors required for cell death (17). Th2 cells have also been proposed to be resistant to galectin-9 because these cells lack the cell surface receptor Tim-3 (18); however, as Tim-3− T cells are susceptible to galectin-9 cell death (17), we identified additional T-cell surface receptors for galectin-9.
We identified PDI as a unique T-cell surface receptor for galectin-9. Galectin-9 binding to murine Th2 cells increased PDI abundance at the cell surface, as well as the abundance of cell surface thiols. This effect enhanced β3 integrin-mediated migration of murine Th2 cells through extracellular matrix; the galectin-9–mediated increase in cell surface PDI also enhanced human T-cell infection by HIV. Many galectins retain cell surface glycoproteins at the plasma membrane via formation of galectin–glycoprotein lattices (16, 19, 20). Our present work demonstrates that galectin-9 can regulate the T-cell surface redox environment, identifies PDI and β3 integrin (CD61) as markers of Th2 cells, and describes unique roles for galectin-9 and PDI in regulating T-cell migration and HIV infection.
Results
Galectin-9 Is a Ligand for T-Cell Surface PDI.
We found that galectin-9 triggered death of T cells lacking Tim-3 (17), implicating additional T-cell glycoprotein receptors for galectin-9. We confirmed that galectin-9 bound to T cells in a lactose inhibitable manner (Fig. 1A and Fig. S1A). We isolated galectin-9 binding glycoproteins from Tim-3− CEM T cells by galectin-9 affinity chomatography of solubilized membranes, eluting bound glycoproteins with lactose (Fig. 1B). Mass spectrometry of eluted glycoproteins identified nine proteins as galectin-9 receptors; surprisingly, none of these were known galectin receptors, such as CD45 or CD44 (21–23). However, three proteins that bound galectin-9 are members of the PDI family: PDI (P4HB), PDIA3 (ERP57, GRP58), and PDIA6 (ERP5).
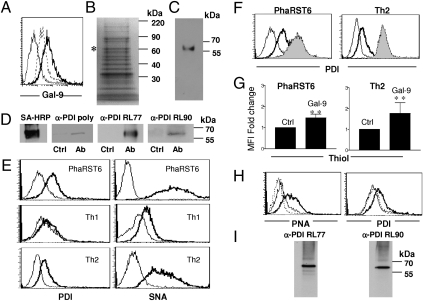
Fig. 1.
Galectin-9 binds to T-cell surface PDI. (A) Galectin-9 binds T cells. PhaRST6 cells were treated with PBS (thin line), 0.1 μM biotin–galectin-9 (thick line), or 0.1 μM biotin–galectin-9 plus 100 mM lactose (dotted line), and bound galectin-9 detected with 0.1 μg streptavidin-PE. (B) Isolation of T-cell glycoproteins that bind galectin-9. T-cell membrane proteins bound to a galectin-9 affinity column were eluted with lactose and stained with SYPRO Ruby. The band (*) was determined to be PDI (P4HB) by mass spectrometry. (C) Reactivity with anti-PDI mAb RL77 confirmed PDI in eluate from galectin-9 affinity column. (D) Cell surface PDI coprecipitates with galectin-9. After plasma membrane biotinylation and addition of galectin-9, PhaRST6 cell extracts were treated with IgG control (Ctrl) or anti–galectin-9 (Ab) and bound material precipitated with protein G sepharose. Precipitates were probed with streptavidin-HRP, polyclonal anti-PDI, or anti-PDI mAbs RL77 or RL90. (E) PDI is abundantly expressed on PhaRST6 and murine Th2 cells, but not Th1 cells; thick line, anti-PDI; thin line, isotype control. α2,6-linked sialic acid, detected with SNA, is abundant on PhaRST6 and murine Th2 cells, compared with Th1 cells; thick line, biotinylated SNA; thin line, biotinylated BSA. (F) Galectin-9 binding increases abundance of cell surface PDI. 0.1 μM galectin-9 was added to PhaRST6 and murine Th2 cells and cell surface PDI detected by flow cytometry: thin line, IgG control; bold line, anti-PDI on control cells; gray solid, anti-PDI on galectin-9 treated cells. (G) 0.1 μM galectin-9 increased abundance of free thiols, detected with Alexa Fluor 488 C5 maleimide on PhaRST6 and murine Th2 cells; results are mean ± SE of five experiments for PhaRST6 cells and three experiments for Th2 cells, each in triplicate, shown as fold-change in mean fluorescence intensity (MFI) compared with control. **P < 0.0025. (H) Galectin-9–mediated increase in cell surface PDI requires O-glycans. (Left) Jurkat E6-1 cells were treated without (thick line) or with (thin line) 2 mM benzyl-α-GalNAc for 72 h and phenotyped with biotinylated PNA (dotted line, biotinylated BSA) to demonstrate loss of cell surface O-glycans. (Right) Treatment with benzyl-α-GalNAc reduced galectin-9 (0.1 μM)–mediated increase in cell surface PDI (dotted line, no galectin-9; thin line, galectin-9 on benzyl-α-GalNAc–treated cells; thick line, galectin-9 on control cells). (I) PNA reactivity indicates O-glycans on PDI. Biotinylated PNA was added to Jurkat T-cell lysates, bound glycoproteins precipitated, and probed with anti-PDI mAbs RL77 and RL90.
Because PDI can be found at the plasma membrane, where it regulates cell surface redox status (1, 2, 4), we focused on this receptor. We confirmed that PDI was present in eluate from galectin-9 affinity matrix by immunoblotting (Fig. 1C). To confirm that PDI is a T-cell surface receptor for galectin-9, we biotinylated the surface of PhaRST6 cells that express α2,6-linked sialic acid on cell surface glycans, and are thus resistant to galectin-9 death (17), before addition of recombinant galectin-9. We immunoprecipitated galectin-9 from cell lysates and detected biotinylated PDI in the precipitate (Fig. 1D), and confirmed this with Th2 cells (Fig. S1A). We did not detect biotinylation of intracellular actin in PhaRST6 cells (Fig. S1B), confirming that labeled PDI was at the cell surface.
We examined cell surface PDI expression on PhaRST6 cells and primary murine CD4 Th1 and Th2 cells. There was significant expression of PDI on PhaRST6 and Th2 cells, but minimal PDI expression on Th1 cells (Fig. 1E). Th2 cells and PhaRST6 cells also express α2,6-linked sialic acids on cell surface glycans (17, 24); this glycan structure is detected by the plant lectin Sambuccus nigra agglutinin (SNA; Fig. 1E). Thus, Th2 cells can be distinguished from Th1 cells by two features, α2,6-sialylation detected with SNA (24) and cell surface PDI; Th2 cells bear specific glycans that prevent galectin-9–induced death, and express cell surface PDI that can bind galectin-9 in a carbohydrate-dependent manner.
Galectin-9 Retains Cell Surface PDI.
Soluble galectins bind glycoprotein receptors on the cell surface to retain receptors at the plasma membrane (16, 19, 20). The addition of exogenous recombinant galectin-9 to PhaRST6 cells and to primary murine Th2 cells resulted in increased abundance of PDI, indicating that galectin-9 specifically retained PDI at the T-cell surface (Fig. 1F); galectin-1 and galectin-3 addition had no effect on abundance of cell surface PDI (Fig. S1C). We also detected an increase in cell surface thiols on PhaRST6 and Th2 cells treated with galectin-9 (Fig. 1G), demonstrating that increased cell surface PDI resulted in increased thioreductase activity on cell surface substrates. PDI abundance at the cell surface was increased by exogenous galectin-9 in a time- and dose-dependent manner, with increased cell surface PDI detected within 15 min and maximal by 2 h (Fig. S1D). The increase in cell surface PDI was inhibited by 100 mM lactose, confirming that galectin-9 binding to PDI is carbohydrate dependent (Fig. S1E). The increase in cell surface PDI after galectin-9 binding was not due to increased synthesis of PDI mRNA (Fig. S1F), indicating that retention of existing PDI, rather than increased de novo synthesis, was responsible for increased cell surface PDI. Galectin-9 could bind to either N- or O-linked glycans on cell surface glycoproteins; as the amino acid sequence of PDI lacks canonical N-glycosylation sites, we asked if galectin-9 could bind O-glycans on PDI. The galectin-9–mediated PDI increase at the cell surface required O-glycans, as the increase was abolished when cells were treated with benzyl-α-GalNAc to block O-glycan elongation (Fig. 1H). Moreover, the presence of O-glycans on PDI was determined by lectin precipitation with peanut agglutinin (PNA), which recognizes core 1 O-glycans (Fig. 1I).
Galectin-9 Enhanced T-Cell Migration Is Dependent on PDI Thioreductase Activity.
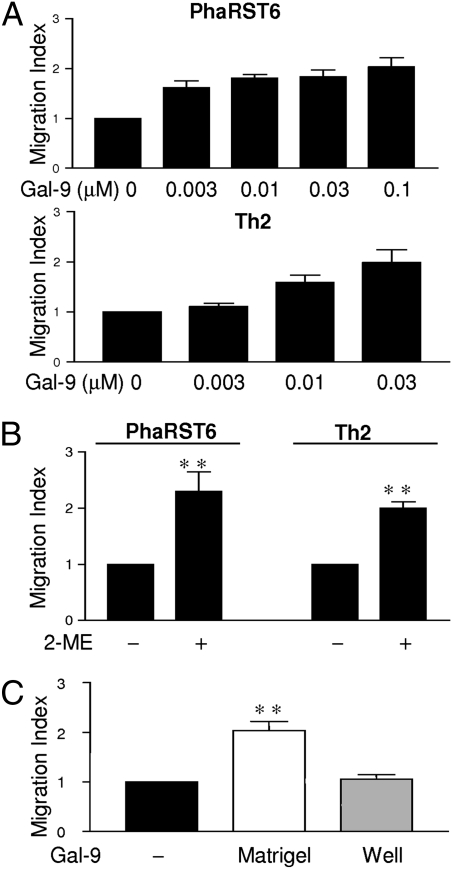
Regulation of exofacial redox status mediates critical cellular events, including leukocyte adhesion and migration (1, 2). As both PhaRST6 T cells and Th2 cells are resistant to galectin-9 death, we asked if galectin-9 could regulate the migration of these cells that express abundant cell surface PDI, using a Matrigel migration assay (25). The addition of galectin-9 significantly enhanced migration of PhaRST6 cells and primary Th2 cells (Fig. 2A); as expected, when Th1 cells were used, most cells died at the surface of the galectin-9–coated Matrigel. Galectin-9–enhanced migration was phenocopied by replacing galectin-9 with 2-mercaptoethanol in Matrigel, indicating that altering cell surface redox status was sufficient to increase migration (Fig. 2B). Importantly, though galectin-9 is chemotactic for eosinophils (26), we observed no increase in T-cell migration if galectin-9 was only added to the lower chamber (Fig. 2C).
Fig. 2.
Galectin-9 enhances Th2 and PhaRST6 T-cell migration. (A) Migration of PhaRST6 and Th2 through Matrigel was enhanced by galectin-9 in a dose-dependent fashion. (B) Increased T-cell migration was phenocopied by inclusion of 2-ME (6 mM) in Matrigel. (C) Migration of PhaRST6 cells through Matrigel was only enhanced when galectin-9 was added directly to Matrigel, but not when 0.1 μM galectin-9 was added to the bottom well of the migration chamber. Results are mean ± SE of three experiments, each in triplicate. **P < 0.025.
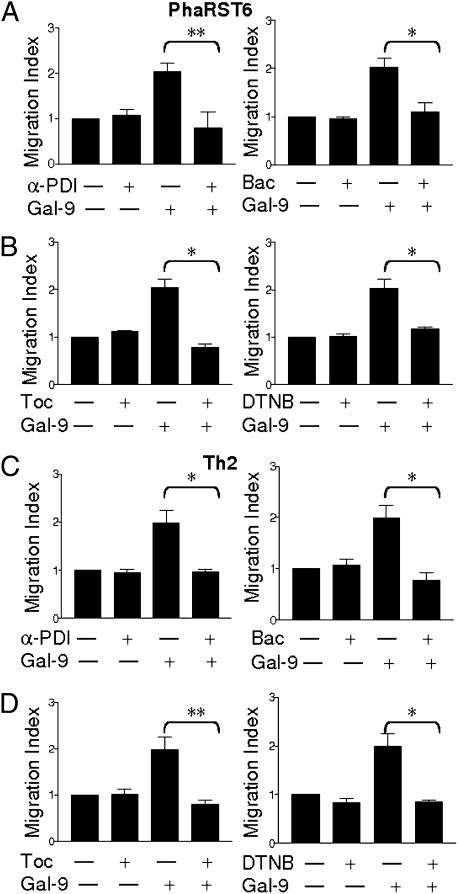
Increased migration of PhaRST6 and primary Th2 cells was PDI dependent, as the effect was reversed by inclusion of anti-PDI antibody in the matrix and by bacitracin, an inhibitor of PDI enzymatic activity (5, 7, 27) (Fig. 3 A and C). Cell-impermeable thiol scavengers tocinoic acid and 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) also reversed the galectin-9–mediated increase in PharST6 and Th2 cell migration (Fig. 3 B and D). Our data demonstrate that galectin-9 directly enhances T-cell migration through matrix by binding to cell surface PDI to retain PDI and thus increase PDI disulfide reductase activity at the plasma membrane.
Fig. 3.
Increased T-cell migration mediated by galectin-9 is PDI dependent. Galectin-9 (0.1 μM) enhanced migration of PhaRST6 cells (A and B) and Th2 cells (C and D) through Matrigel was abrogated by 1:10,000 anti-PDI (α-PDI), 3 mM bacitracin (Bac), and thiol scavengers 0.03 mM tocinoic acid (Toc) and 0.01 mM DTNB. Inhibitors reduced migration in a dose-dependent manner; indicated doses are shown. Results are mean ± SE of three experiments, each in triplicate. *P < 0.01, **P < 0.0025.
PDI-Associated CD61 Promotes T-Cell Migration Enhanced by Galectin-9.
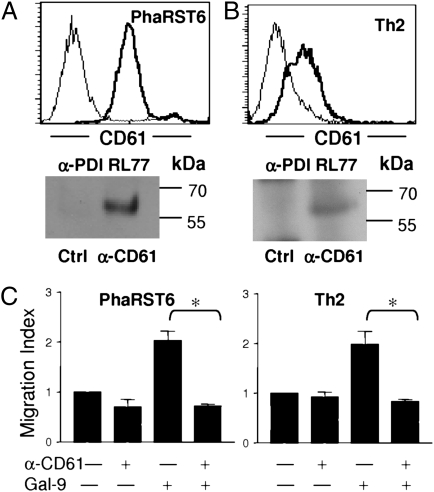
PDI regulates the function of several integrins, including β1, β2, and β3, by reducing disulfide bonds to convert integrins into a high-affinity conformation (7, 28, 29). β3 integrins regulate platelet adhesion; β3 is also known as CD61, and on platelets, PDI reduces disulfide bonds in CD61 to promote binding to ligands (7, 28). CD61 expression has also been reported on specific T-cell subsets (30–34). We confirmed that PhaRST6 and primary Th2 cells express cell surface CD61 (Fig. 4 A and B), and that PDI associates with CD61, as PDI specifically coimmunoprecipitated with CD61 from membrane extracts of PhaRST6 and Th2 cells (Fig. 4 A and B). Antibody to CD61 reversed galectin-9 enhancement of PhaRST6 and Th2 cell migration through Matrigel (Fig. 4C). We did not identify CD61 as a T-cell receptor for galectin-9 in our affinity purification (Fig. 1); thus, we propose that galectin-9 enhances T-cell migration by retaining cell surface PDI, where PDI associates with CD61 to enhance migration.
Fig. 4.
Galectin-9 enhancement of T-cell migration involves CD61. (A Upper) CD61 (thick line) is expressed on PhaRST6 cells; isotype control (thin line). (Lower) PDI associates with CD61 on PhaRST6 cells; T-cell membrane extracts were immunoprecipitated with IgG control (Ctrl) or anti-CD61 mAb HMbeta3.1 (α-CD61), and precipitate probed with anti-PDI mAb RL77. (B Upper) CD61 (thick line) is expressed on human Th2 cells; isotype control (thin line). (Lower) PDI associates with CD61 on human Th2 cells; T-cell membrane extracts were immunoprecipitated with IgG control (Ctl) or anti-CD61 mAb Y2/51 (α-CD61), and precipitate probed with anti-PDI mAb RL77. (C) Galectin-9 (0.1 μM) enhancement of T-cell migration was abrogated by anti-CD61 mAb; a representative dose (1:10,000 for PhaRST6 cells, 1:30,000 for Th2 cells) is shown. Results are mean ± SE of three experiments, each in triplicate. *P < 0.01.
Galectin-9 Enhances HIV-1 Infection of T Cells in a PDI-Dependent Manner.
PDI enzymatic activity as a cell surface disulfide reductase is also known to promote HIV-1 infection of human T cells (5, 9, 11–13). We asked if galectin-9 would enhance HIV-1 entry into target cells in a PDI-dependent manner. Galectin-9 increased abundance of PDI on Jurkat T cells in a dose- and time-dependent manner (Fig. 5 A and B). We infected Jurkat cells with high or low viral inocula in the presence or absence of galectin-9, and determined viral infectivity. Galectin-9 increased viral infectivity by two- to threefold at high viral inoculum and by four- to sixfold at low viral inoculum; enhancement was PDI dependent, as addition of bacitracin reversed the galectin-9 effect (Fig. 5C). Similar results were obtained with CEM T cells (Fig. S2). Galectin-1 was used as a positive control, because galectin-1 is known to enhance HIV infectivity by directly increasing viral attachment to cell surface receptors (35). Significantly, galectin-1 enhancement was maximal by 15 min, in contrast to the 2 h required for comparable galectin-9 enhancement (Fig. 5D), underscoring the qualitatively different modes of enhancement of galectin-9 and galectin-1.
Fig. 5.
Galectin-9 enhancement of HIV-1 infection is PDI dependent. (A) PDI is expressed on Jurkat cells, and PDI is increased by addition of galectin-9 in a dose-dependent manner. Cell surface staining with IgG control (light gray filled) or anti-PDI without (dark gray filled) or with 0.01 μM (dotted line), 0.03 μM (thin line), or 0.1 μM (bold line) galectin-9 for 2 h. (B) PDI abundance on Jurkat cells was increased by 0.1 μM galectin-9 in a time-dependent manner; mean fluorescence intensity (MFI) ± SD from triplicate samples is shown. (C) Jurkat cells were infected with NL4-3 (CXCR4-tropic HIV-1 strain) at high (20 ng p24) or low (5 ng p24) viral input for 2 h. Galectin-9 (0.1 μM) with or without 3 mM bacitracin (Bac) or galectin-1 (20 μM) was added; after 2 h, cells were washed. At 48 h postinfection, the number of infected cells was quantified by intracellular viral p24 antigen staining detected by flow cytometry. Data from one of three experiments is shown. (D) Galectin-9 enhancement of HIV infection differs from galectin-1. Infection was performed as in C, except that viral entry was quantified by qPCR of early RT products at 6–8 h postinfection. Early RT products were normalized to β-actin levels for each condition, and the number of early RT products in the absence of galectin-9 was set at 1. Galectin-9 enhancement of HIV infection increases over 2 h, the same timeframe in which cell surface PDI increase is seen (B). Galectin-1 enhancement of HIV infection is optimal at 15 min. Results are shown as fold enhancement over HIV-1 alone. (E) Galectin-9 enhancement of HIV infection requires PDI activity. Infection of Jurkat cells was performed as in D using 5 ng (p24 equivalent) of viral inoculum. Results are shown as fold enhancement over HIV-1 alone. Striped bar, no galectin-9; solid bars, 0.1 μM galectin-9; B, 3 mM bacitracin; P, anti-PDI mAB RL77 1:3000; D, 0.01 mM DTNB; T, 0.03 mM tocinoic acid; C, DMSO as vehicle control. Results are mean ± SE of three to five replicate experiments, each done in duplicate.
To confirm that galectin-9 enhanced HIV infection at the level of virus entry, HIV infectivity was measured by quantitative PCR (qPCR) for early reverse transcriptase (RT) products 6–8 h postinfection. Viral entry was enhanced by the addition of galectin-9 in a PDI-dependent manner, because enhancement was reversed by PDI inhibitors and thiol scavengers (Fig. 5E); this enhancement was even more marked than that measured by p24 staining (Fig. 5C). This finding is consistent with the known role of PDI in catalyzing disulfide bond rearrangements in the HIV envelope glycoprotein that are required for productive membrane fusion and viral entry (5, 9, 11, 12). Thus, galectin-9–mediated stabilization of PDI activity at the cell surface enhances HIV infection, an effect that may have implications for in vivo HIV spread and pathogenesis (36).
Discussion
We report a unique function for galectin-9 in regulating the cell surface redox environment to influence both T-cell migration and infection by HIV. The pleiotropic effects of galectins have been noted in a number of systems; whereas galectins affect many cellular processes, different effects result from a specific galectin binding to a specific set of glycoprotein receptors that directly regulate specific events, such as TCR or growth factor receptor signaling (16, 19, 20). Importantly, though galectin-9 can be detected intracellularly in many cell types, galectin-9 is secreted and binds back to the surface of T cells and endothelial cells in a carbohydrate-dependent manner (37, 38) (Fig. S1G). Thus, galectin-9 binding to PDI at the plasma membrane may influence many T-cell events regulated by the redox status of cell surface glycoproteins that associate with PDI, such as CD61 or ADAM17 (7, 28, 39). Additionally, lymphocyte cell surface-associated PDI has a well-documented role in catalyzing disulfide bond rearrangements in HIV envelope glycoproteins (gp120/41) that promote virus-cell membrane fusion and viral entry (13). Indeed, PDI inhibitors are being studied as a potential new class of host-targeted anti-HIV therapeutics (40). Though some PDI substrates, such as CD61 and HIV gp120, have been identified, the full repertoire of cell surface proteins that may be modified by PDI is not known. This work identifies PDI as a cell surface receptor for galectin-9, and reveals the biological relevance of galectin-9/PDI interactions by demonstrating that galectin-9 binding can increase endogenous PDI activity to enhance integrin-mediated migration and augment HIV infection of T cells.
The different effects of galectin-9 on Th2 vs. Th1 cells are striking. Galectin-9 kills Th1 cells (18), but regulates migration of Th2 cells; the responses of Th2 vs. Th1 cells can be explained by differential expression of glycoprotein receptors as well as specific glycans. We found that N-glycans are required for galectin-9–mediated cell death (17), and that blocking N-glycan termini with α2,6-linked sialic acid, a glycan structure that is abundant on Th2 cells (Fig. 1E) (24), abrogated galectin-9 T-cell death (17). On PDI, galectin-9 appears to bind to O-glycans (Fig. 1 H and I); PDI has no canonical N-glycosylation sites, and, though the O-glycosylation status of PDI has not been previously described, reactivity with the lectin PNA indicates that PDI bears O-glycans. In addition, blocking O-glycan elongation with benzyl-α-GalNAc abolished the galectin-9–mediated increase in cell surface PDI. In addition to α2,6-linked sialic acid, both PDI and CD61, a PDI substrate, are also abundant on Th2 cells (Figs. 1E and 4B). Thus, specific glycosylation of specific glycoprotein receptors that differ in expression between Th2 and Th1 cells accounts for the different effects of galectin-9 on these two populations.
Galectin-9 is highly expressed by leukocytes, fibroblasts, and endothelial cells in inflamed tissues (14, 15, 27, 38). Increased galectin-9 expression in inflamed tissues may dynamically regulate the redox environment on the T-cell surface in vivo and affect T-cell function. Importantly, the amount of galectin-9 required for these effects may be relatively modest, as we observed effects on T-cell migrations at nanomolar concentrations of galectin-9 (Fig. 2); the potency of galectin-9 may result from the ability of this tandem repeat galectin to form multivalent oligomers (41). That galectin-9 promotes Th2 cell migration while killing Th1 cells suggests that galectin-9 expression will be of particular significance in disease mediated by Th2-type immune responses, such as asthma (23); galectin-9 expression is increased in lungs of mice in a model of allergic airway disease, and galectin-9 is also a chemokine for eosinophils (27, 42). Suppressing local expression of galectin-9 may be an approach to mitigate inflammation and tissue damage in asthma. Similarly, reducing local expression of galectin-9 in genital mucosa may reduce infection of T cells with HIV. The mechanism by which we propose that this effect occurs, i.e., creation of a galectin–glycoprotein lattice to retain PDI on the plasma membrane, has been demonstrated in a number of other cell types, with specific galectins, including galectin-9, retaining specific glycoprotein receptors (16, 19, 20, 43, 44). Identification of PDI as a T-cell glycoprotein receptor for galectin-9 reveals a unique mechanism for galectin lattices to regulate a variety of critical T-cell functions by controlling redox status at the cell surface.
Materials and Methods
Cells and Reagents.
CEM, Jurkat E6-1, and PhaR2.1 cells stably transfected with ST6Gal I-expressing vector (PhaRST6) were maintained as in Bi et al. (17). Recombinant galectin-1 and galectin-9M (medium length linker) were prepared as described (17, 45). See SI Materials and Methods for reagents and suppliers.
Identification of Galectin-9 Binding Proteins by Affinity Chromatography.
Galectin-9-binding proteins from CEM T cells were isolated and identified as in Stillman et al. (22) with minor modifications (SI Materials and Methods).
Immunoblotting and Immunoprecipitation.
A 10-μg elute from galectin-9 affinity matrix was separated by SDS/PAGE, transferred to nitrocellulose, and probed with PDI mAb. For immunoprecipitation with galectin-9, lysates of biotinylated PhaRST6 cells were used as described in SI Materials and Methods. For lectin precipitation, 107 Jurkat E6-1 cells were solubilized, and 50 μg of biotinylated peanut agglutinin was added with streptavidin agarose beads overnight at 4 °C. Precipitates were probed with anti-PDI mAb RL77 and RL90. For CD61 precipitation, PhaRST6 cells and human Th2 cells were precipitated with 2 μL of hamster anti-mouse CD61 mAb or mouse anti-human CD61 mAb, respectively, or relevant IgG control, and precipitates were probed with anti-PDI mAb RL77.
Detection of Cell Surface Galectin-9.
A total of 106 PhaRST6 cells were blocked with 1% biotinylated BSA in PBS at 4 °C for 30 min, washed, and labeled with 0.1 μM biotinylated galectin-9 with or without 100 mM lactose, followed by 0.1 μg streptavidin-PE. To detect endogenous cell surface galectin-9, cells were labeled with 0.2 μg of anti–galectin-9 with or without 100 mM lactose or with irrelevant IgG control at 4 °C for 1 h, followed by 0.1 μg PE-conjugated goat anti-mouse IgG. Cells were analyzed on a FACScan (BD) using CellQuest software.
Mouse CD4 T-Cell Isolation, Polarization, and T-Cell Phenotyping.
Murine C57BL/6 or human CD4+ T cells were Th1 or Th2 polarized (46–48) and phenotyped (SI Materials and Methods).
T-Cell Migration Through Matrigel.
Cell migration assays were performed as described (25) with the indicated amount of galectin-9 added to rehydrated Matrigel for 1 h at room temperature (SI Materials and Methods). Antibodies (anti-PDI, CD61), bacitracin, thiol scavengers (tocinoic acid, DTNB), or 2-ME were loaded at indicated concentrations to the upper chamber 30 min before cell loading. Bacitracin was boiled 10 min before use (49). To test galectin-9 as a chemokine, 0.1 μM galectin-9 was added to the lower chamber.
HIV-1 Infection.
Jurkat E6-1 or CEM T cells were incubated with indicated concentrations of galectin-9 in the absence or presence of anti-PDI, bacitracin, tocinoic acid, or DTNB for 15 min. High (20 ng) or low (5 ng) inocula of HIV-1 (IIIB) were spinoculated (770 × g) on cells for 2 h at 37 °C. Viral infectivity was determined by intracellular staining for viral p24 antigen 48 h postinfection, or by quantitative PCR for early viral transcripts 8 h postinfection. For intracellular p24 antigen detection, infected cells were fixed with 2% paraformaldehyde, permeabilized with methanol and 0.1% Nonidet P-40, and p24 antigen detected with KC57-FITC (1:1,000). Quantitative PCR was used to detect early RT products in the R/U5 region of the LTR as described (50). Endogenous β-globin levels were measured to normalize R/U5 values.
Supplementary Material
Acknowledgments
We thank Lesley Earl, Mabel Pang, Xin Qiao, and Qingmei Jia for discussion, and Vanessa Minassian, Linsey Jacobs, and Jennifer Thu Vu for technical assistance. Support was provided by National Institutes of Health Grants R21HL102989 (to L.G.B.), R01AI060694 (to L.G.B. and B.L.), R21AI092218 (to B.L.), and T32AR053463 (to P.W.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017954108/-/DCSupplemental.
References
- 1.Jordan PA, Gibbins JM. Extracellular disulfide exchange and the regulation of cellular function. Antioxid Redox Signal. 2006;8:312–324. doi: 10.1089/ars.2006.8.312. [DOI] [PubMed] [Google Scholar]
- 2.Hogg PJ. Disulfide bonds as switches for protein function. Trends Biochem Sci. 2003;28:210–214. doi: 10.1016/S0968-0004(03)00057-4. [DOI] [PubMed] [Google Scholar]
- 3.Lawrence DA, Song R, Weber P. Surface thiols of human lymphocytes and their changes after in vitro and in vivo activation. J Leukoc Biol. 1996;60:611–618. doi: 10.1002/jlb.60.5.611. [DOI] [PubMed] [Google Scholar]
- 4.Turano C, Coppari S, Altieri F, Ferraro A. Proteins of the PDI family: Unpredicted non-ER locations and functions. J Cell Physiol. 2002;193:154–163. doi: 10.1002/jcp.10172. [DOI] [PubMed] [Google Scholar]
- 5.Fenouillet E, Barbouche R, Courageot J, Miquelis R. The catalytic activity of protein disulfide isomerase is involved in human immunodeficiency virus envelope-mediated membrane fusion after CD4 cell binding. J Infect Dis. 2001;183:744–752. doi: 10.1086/318823. [DOI] [PubMed] [Google Scholar]
- 6.Goplen D, et al. Protein disulfide isomerase expression is related to the invasive properties of malignant glioma. Cancer Res. 2006;66:9895–9902. doi: 10.1158/0008-5472.CAN-05-4589. [DOI] [PubMed] [Google Scholar]
- 7.Lahav J, et al. Sustained integrin ligation involves extracellular free sulfhydryls and enzymatically catalyzed disulfide exchange. Blood. 2002;100:2472–2478. doi: 10.1182/blood-2001-12-0339. [DOI] [PubMed] [Google Scholar]
- 8.Jain S, McGinnes LW, Morrison TG. Thiol/disulfide exchange is required for membrane fusion directed by the Newcastle disease virus fusion protein. J Virol. 2007;81:2328–2339. doi: 10.1128/JVI.01940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenouillet E, Barbouche R, Jones IM. Cell entry by enveloped viruses: Redox considerations for HIV and SARS-coronavirus. Antioxid Redox Signal. 2007;9:1009–1034. doi: 10.1089/ars.2007.1639. [DOI] [PubMed] [Google Scholar]
- 10.Manickam N, Sun X, Li M, Gazitt Y, Essex DW. Protein disulphide isomerase in platelet function. Br J Haematol. 2008;140:223–229. doi: 10.1111/j.1365-2141.2007.06898.x. [DOI] [PubMed] [Google Scholar]
- 11.Markovic I, et al. Thiol/disulfide exchange is a prerequisite for CXCR4-tropic HIV-1 envelope-mediated T-cell fusion during viral entry. Blood. 2004;103:1586–1594. doi: 10.1182/blood-2003-05-1390. [DOI] [PubMed] [Google Scholar]
- 12.Gallina A, et al. Inhibitors of protein-disulfide isomerase prevent cleavage of disulfide bonds in receptor-bound glycoprotein 120 and prevent HIV-1 entry. J Biol Chem. 2002;277:50579–50588. doi: 10.1074/jbc.M204547200. [DOI] [PubMed] [Google Scholar]
- 13.Papandréou MJ, et al. Mapping of domains on HIV envelope protein mediating association with calnexin and protein-disulfide isomerase. J Biol Chem. 2010;285:13788–13796. doi: 10.1074/jbc.M109.066670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu F-T, Rabinovich GA. Galectins: Regulators of acute and chronic inflammation. Ann N Y Acad Sci. 2010;1183:158–182. doi: 10.1111/j.1749-6632.2009.05131.x. [DOI] [PubMed] [Google Scholar]
- 15.Vasta GR. Roles of galectins in infection. Nat Rev Microbiol. 2009;7:424–438. doi: 10.1038/nrmicro2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garner OB, Baum LG. Galectin-glycan lattices regulate cell-surface glycoprotein organization and signalling. Biochem Soc Trans. 2008;36:1472–1477. doi: 10.1042/BST0361472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bi S, Earl LA, Jacobs L, Baum LG. Structural features of galectin-9 and galectin-1 that determine distinct T cell death pathways. J Biol Chem. 2008;283:12248–12258. doi: 10.1074/jbc.M800523200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu C, et al. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol. 2005;6:1245–1252. doi: 10.1038/ni1271. [DOI] [PubMed] [Google Scholar]
- 19.Lau KS, et al. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 20.Grigorian A, Torossian S, Demetriou M. T-cell growth, cell surface organization, and the galectin-glycoprotein lattice. Immunol Rev. 2009;230:232–246. doi: 10.1111/j.1600-065X.2009.00796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Earl LA, Bi S, Baum LG. N- and O-glycans modulate galectin-1 binding, CD45 signaling, and T cell death. J Biol Chem. 2010;285:2232–2244. doi: 10.1074/jbc.M109.066191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stillman BN, et al. Galectin-3 and galectin-1 bind distinct cell surface glycoprotein receptors to induce T cell death. J Immunol. 2006;176:778–789. doi: 10.4049/jimmunol.176.2.778. [DOI] [PubMed] [Google Scholar]
- 23.Katoh S, et al. Galectin-9 inhibits CD44-hyaluronan interaction and suppresses a murine model of allergic asthma. Am J Respir Crit Care Med. 2007;176:27–35. doi: 10.1164/rccm.200608-1243OC. [DOI] [PubMed] [Google Scholar]
- 24.Toscano MA, et al. Differential glycosylation of TH1, TH2 and TH-17 effector cells selectively regulates susceptibility to cell death. Nat Immunol. 2007;8:825–834. doi: 10.1038/ni1482. [DOI] [PubMed] [Google Scholar]
- 25.He J, Baum LG. Endothelial cell expression of galectin-1 induced by prostate cancer cells inhibits T-cell transendothelial migration. Lab Invest. 2006;86:578–590. doi: 10.1038/labinvest.3700420. [DOI] [PubMed] [Google Scholar]
- 26.Hirashima M, et al. Galectin-9 in physiological and pathological conditions. Glycoconj J. 2004;19:593–600. doi: 10.1023/B:GLYC.0000014090.63206.2f. [DOI] [PubMed] [Google Scholar]
- 27.Mandel R, Ryser HJ-P, Ghani F, Wu M, Peak D. Inhibition of a reductive function of the plasma membrane by bacitracin and antibodies against protein disulfide-isomerase. Proc Natl Acad Sci USA. 1993;90:4112–4116. doi: 10.1073/pnas.90.9.4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lahav J, Gofer-Dadosh N, Luboshitz J, Hess O, Shaklai M. Protein disulfide isomerase mediates integrin-dependent adhesion. FEBS Lett. 2000;475:89–92. doi: 10.1016/s0014-5793(00)01630-6. [DOI] [PubMed] [Google Scholar]
- 29.Swiatkowska M, et al. Ero1alpha is expressed on blood platelets in association with protein-disulfide isomerase and contributes to redox-controlled remodeling of alphaIIbbeta3. J Biol Chem. 2010;285:29874–29883. doi: 10.1074/jbc.M109.092486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy A, Liu X, Pahan K. Myelin basic protein-primed T cells induce neurotrophins in glial cells via alphavbeta3 [corrected] integrin. J Biol Chem. 2007;282:32222–32232. doi: 10.1074/jbc.M702899200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lacy-Hulbert A, et al. Beta 3 integrins regulate lymphocyte migration and cytokine responses in heart transplant rejection. Am J Transplant. 2007;7:1080–1090. doi: 10.1111/j.1600-6143.2007.01757.x. [DOI] [PubMed] [Google Scholar]
- 32.Gerber DJ, Pereira P, Huang SY, Pelletier C, Tonegawa S. Expression of alpha v and beta 3 integrin chains on murine lymphocytes. Proc Natl Acad Sci USA. 1996;93:14698–14703. doi: 10.1073/pnas.93.25.14698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wierzbicki P, et al. Beta 3 integrin expression on T cells from renal allograft recipients. Transplant Proc. 2006;38:338–339. doi: 10.1016/j.transproceed.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 34.Doucey MA, et al. The beta1 and beta3 integrins promote T cell receptor-mediated cytotoxic T lymphocyte activation. J Biol Chem. 2003;278:26983–26991. doi: 10.1074/jbc.M302709200. [DOI] [PubMed] [Google Scholar]
- 35.Ouellet M, et al. Galectin-1 acts as a soluble host factor that promotes HIV-1 infectivity through stabilization of virus attachment to host cells. J Immunol. 2005;174:4120–4126. doi: 10.4049/jimmunol.174.7.4120. [DOI] [PubMed] [Google Scholar]
- 36.Matthias LJ, Azimi I, Tabrett CA, Hogg PJ. Reduced monomeric CD4 is the preferred receptor for HIV. J Biol Chem. 2010;285:40793–40799. doi: 10.1074/jbc.M110.190579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chabot S, et al. Regulation of galectin-9 expression and release in Jurkat T cell line cells. Glycobiology. 2002;12:111–118. doi: 10.1093/glycob/12.2.111. [DOI] [PubMed] [Google Scholar]
- 38.Imaizumi T, et al. Interferon-gamma stimulates the expression of galectin-9 in cultured human endothelial cells. J Leukoc Biol. 2002;72:486–491. [PubMed] [Google Scholar]
- 39.Willems SH, et al. Thiol isomerases negatively regulate the cellular shedding activity of ADAM17. Biochem J. 2010;428:439–450. doi: 10.1042/BJ20100179. [DOI] [PubMed] [Google Scholar]
- 40.Gowthaman U, Jayakanthan M, Sundar D. Molecular docking studies of dithionitrobenzoic acid and its related compounds to protein disulfide isomerase: Computational screening of inhibitors to HIV-1 entry. BMC Bioinformatics. 2008;9(Suppl 12):S14. doi: 10.1186/1471-2105-9-S12-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Earl LA, Bi S, Baum LG. Galectin multimerization and lattice formation are regulated by linker region structure. Glycobiology. 2011;21:6–12. doi: 10.1093/glycob/cwq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sziksz E, et al. Galectin-9 in allergic airway inflammation and hyper-responsiveness in mice. Int Arch Allergy Immunol. 2010;151:308–317. doi: 10.1159/000250439. [DOI] [PubMed] [Google Scholar]
- 43.Ohtsubo K, et al. Dietary and genetic control of glucose transporter 2 glycosylation promotes insulin secretion in suppressing diabetes. Cell. 2005;123:1307–1321. doi: 10.1016/j.cell.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 44.Mishra R, Grzybek M, Niki T, Hirashima M, Simons K. Galectin-9 trafficking regulates apical-basal polarity in Madin-Darby canine kidney epithelial cells. Proc Natl Acad Sci USA. 2010;107:17633–17638. doi: 10.1073/pnas.1012424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pace KE, Hahn HP, Baum LG. Preparation of recombinant human galectin-1 and use in T-cell death assays. Methods Enzymol. 2003;363:499–518. doi: 10.1016/S0076-6879(03)01075-9. [DOI] [PubMed] [Google Scholar]
- 46.Thornton AM. In: Current Protocols in Immunology. Coligan JE, et al., editors. Hoboken, NJ: Wiley; 2003. pp. 3.5A.1–3.5A.11. [Google Scholar]
- 47.Fitch FW, Gajewski TF, Hu-Li J. In: Current Protocols in Immunology. Coligan JE, et al., editors. Hoboken, NJ: Wiley; 2006. pp. 3.13.1–3.13.15. [DOI] [PubMed] [Google Scholar]
- 48.Moonis M, Lee B, Bailer RT, Luo Q, Montaner LJ. CCR5 and CXCR4 expression correlated with X4 and R5 HIV-1 infection yet not sustained replication in Th1 and Th2 cells. AIDS. 2001;15:1941–1949. doi: 10.1097/00002030-200110190-00005. [DOI] [PubMed] [Google Scholar]
- 49.Rogelj S, Reiter KJ, Kesner L, Li M, Essex D. Enzyme destruction by a protease contaminant in bacitracin. Biochem Biophys Res Commun. 2000;273:829–832. doi: 10.1006/bbrc.2000.3029. [DOI] [PubMed] [Google Scholar]
- 50.Hong PW, Nguyen S, Young S, Su SV, Lee B. Identification of the optimal DC-SIGN binding site on human immunodeficiency virus type 1 gp120. J Virol. 2007;81:8325–8336. doi: 10.1128/JVI.01765-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.