Abstract
The histone variant H3.3 and the canonical histone H3.1, which differ in only 4- to 5-aa positions, are coexpressed in complex multicellular eukaryotes from fly to human and plant. H3.3 is mainly associated with active chromatin by replacing H3.1 through chaperones such as histone regulator A, death domain associated protein DAXX, thalassemia/mental retardation syndrome X-linked homolog ATRX, or proto-oncogene protein DEK and plays important roles in the germline, epigenetic memory, and reprogramming. However, the signals within H3.3 that serve as a guide for its dynamic deposition or depletion in plant chromatin are not clear. Here, we show that Arabidopsis histone H3.3 differs from H3.1 by 4-aa sites: amino acids 31, 41, 87, and 90. Although histone H3.1 is highly enriched in chromocenters, H3.3 is present in nucleolar foci in addition to being diffusely distributed in the nucleoplasm. We have evaluated the function of the 4 aa that differ between H3.1 and H3.3. We show that amino acid residue 87, and to some extent residue 90, of Arabidopsis histone H3.3 are critical for its deposition into rDNA arrays. When RNA polymerase I-directed nucleolar transcription is inhibited, wild type H3.3, but not H3.3 containing mutations at residues 31 and 41, is depleted from the rDNA arrays. Together, our results are consistent with a model in which amino acids 87 and 90 in the core domain of H3.3 guide nucleosome assembly, whereas amino acids 31 and 41 in the N-terminal tail of Arabidopsis H3.3 guide nucleosome disassembly in nucleolar rDNA.
Keywords: H3 dynamics, histone code, nucleolar dominance, chromatin remodeling
The fundamental repeat unit of packaging of eukaryotic genomic DNA is predominantly the nucleosome where DNA is wrapped around a histone octamer that contains two molecules of each core histone: H2A, H2B, H3, and H4. The four core histones share the common histone folding domain (HFD), which is composed of three α-helices (α1, α2, and α3) separated by two loops (L1 and L2) (1). The N- and C-terminal tails stretch out of the nucleosome core and are subject to diverse posttranslational modifications (PTMs) including methylation, acetylation, phosphorylation, ubiquitination, and poly-ADP ribosylation (2, 3). In addition, the incorporation of histone variants into nucleosomes has been proposed to provide another mechanism for modifying chromatin structure (4).
In addition to the centromere-specific histone 3 variant (CenH3), which can be deposited into the centromeric nucleosomes in tetrameric (5, 6) or hexameric states (7), the histone H3 family includes another universal histone H3 variant H3.3 (4) or H3.2 in alfalfa, a plant histone H3.3 (8). In most animals, histone variant H3.3 differs by 4-aa residues from H3.1, which are residue 31 in the N-terminal tail and residues 87, 89, and 90 in the HFD near the beginning of α2 (1, 9). In contrast to the replication-coupled deposition of H3.1 during S-phase mediated by chromatin-assembly factor 1 (CAF1) (4), H3.3 in eukaryotes incorporates into nucleosomes mostly in a replication-independent manner (9) through the histone chaperones, including the histone regulator A (HIRA), ATRX, death-associated protein DAXX, and DEK (10–17). H3.3 is deposited primarily in promoters, gene regulatory sites, and regions with PTMs that are associated with transcribed genes (18). H3.3 has been shown to play roles in epigenetic reprogramming (19) and memory (20). When the H3.3 gene was knocked out in Drosophila melanogaster, flies exhibited sterility, and the transcriptional defects were able to be rescued by overexpression of H3.1, which shows that H3.3 is not necessary for somatic development (21, 22). In addition, H3.3 mutants display meiotic defects in chromosome condensation in spermatocytes, which is dependent on residues 87, 89, and/or 90 but not on methylation of H3.3K4 or phosphorylation of Ser31, suggesting a role of H3.3 in the germline (22).
H3.3 can be displaced dynamically by H3.1 and reloaded at the site of transcription in D. melanogaster (23). The nucleosome turnover might have functional significance in epigenome maintenance, gene regulation, and DNA replication (24). However, the signals within H3.3 that serve as a guide for its dynamic deposition or depletion in plant chromatin are not clear. Here, we have evaluated the function of 4 aa that differ between Arabidopsis histone H3.1 and H3.3, and we have identified the specific signals within H3.3 that serve as guides for its dynamic deposition into or depletion from the H3.3-containing nucleosomes.
Results and Discussion
Arabidopsis Histone H3.3 Is Highly Enriched at Nucleolar rDNA Foci in Addition to Being Diffusely Distributed in the Nucleoplasm.
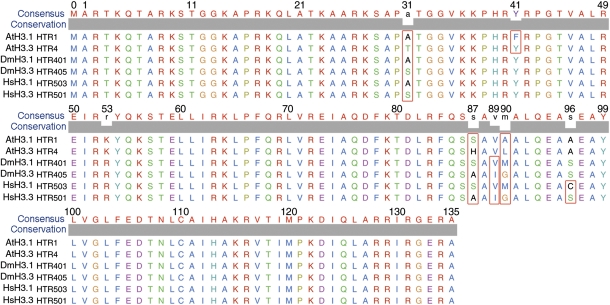
The Arabidopsis genome encodes six H3.3 or H3.3-like genes, including histone three related 4(HTR4, At4g40030), HTR5(At4g40040), HTR6(At1g13370), HTR8(At5g10980), HTR14(At1g75600), and the male gamete-specific HTR10/AtMGH3(At1g19890) (25). To investigate the regions in the plant histone H3.3 variant that are responsible for guiding its incorporation into and its depletion from nucleosomes, we first examined the sequence alignment of canonical Arabidopsis histone H3.1(HTR1) and variant H3.3(HTR4) proteins and compared them with corresponding sequences from other organisms (Fig. 1). The difference between histone H3.1 and H3.3 is defined by 4-aa residues at the sites 31 (Ala vs. Ser), 87 (Ser vs. Ala), 89 (Val vs. Ile), and 90 (Met vs. Gly) in Drosophila (26), and an additional site at 96 (Cys vs. Ser) is included in humans (27, 28). The difference of 4-aa residues between Arabidopsis histone H3.1/HTR1 and H3.3/HTR4 was observed at the sites 31 (Ala vs. Thr), 41 (Phe vs. Tyr), 87 (Ser vs. His), and 90 (Ala vs. Leu); they lack the amino acid change at residue 89 observed in other organisms, but contain an additional amino acid difference at residue 41. This plant-specific feature is conserved in both dicot and monocot plant species such as rice and maize (Figs. S1–S3) (19).The amino acid residues 87 and 90 are located in the α2-helix of the H3 core domain and residues 31 and 41 in the H3 N-terminal tail (1). The plant-specific Tyr41 is the site that passes through the minor groove of the DNA double helix around the nucleosome (1), and thus, it may play a role in the plant-specific regulation of genes by affecting the interaction between DNA and the histone core particle.
Fig. 1.
Multiple alignments of the typical histone H3.1 and H3.3 proteins from Arabidopsis thaliana (At), Drosophila melanogaster (Dm), and Homo sapiens (Hs). The different amino acid residues between H3.1 and H3.3 at the sites 31, 41, 87, 89, and 90 are highlighted by red rectangular boxes. The consensus amino acids and conservation level are shown at the top of the aligned sequence.
To study the subcellular organization of the Arabidopsis histone H3.1 and H3.3 variants, genomic DNA fragments of the H3.1/HTR1 and H3.3/HTR4 genes, including the coding region and their endogenous upstream regulatory sequences, were amplified and fused with green fluorescent protein (GFP). Transgenic Arabidopsis plants expressing these fusion proteins grow normally without any altered phenotypes. As expected, these histone proteins localize in the nucleus in vivo. At the subnuclear level, H3.1/HTR1-GFP was observed to be enriched at DAPI-dense heterochromatic chromocenters (19, 29), and it was also observed to be a diffuse signal in the nucleoplasm with little or no signal in the nucleolus (Fig. 2 A–C). In contrast, H3.3/HTR4-GFP was observed to be highly enriched at nucleolar rDNA foci, and it was also observed to be a diffuse signal in the nucleoplasm without obvious chromocenter labeling (Fig. 2 D–F). Arabidopsis contains various cell types with different endopolyploidy levels, nuclear sizes, and shapes (30). Interestingly, the nucleolar foci of H3.3-GFP are normally two in number in the 2C cell types such as guard or small-leaf epidermal cells with round nuclei, and the two foci likely represent a pair of nucleolar organizer regions (NORs) (Fig. S4A). In contrast, the number of nucleolar foci of H3.3-GFP ranges from 4 to ∼16 in the endoreduplicated cell types (30, 31) such as larger root or leaf epidermal cells with elongated nuclei (Fig. 2D and Fig. S4B); this is likely because of the endoreduplication of NORs in these cell nuclei.
Fig. 2.
Distinct localization patterns between the plant histone H3.1/HTR1 and H3.3/HTR4. (A) H3.1/HTR1-GFP in a nucleus of a leaf epidermal cell. (B) DAPI staining of the nucleus; dense chromocenters are resolved (pseudocolored red). (C) Overlay of A and B. H3.1/HTR1-GFP colocalizes with DAPI-dense chromocenters in addition to diffuse signal in the nucleoplasm. (D) H3.3/HTR4-GFP in the nucleus of a leaf epidermal cell. (E) DAPI staining of the nucleus (pseudocolored red). (F) Overlay of D and E. H3.3/HTR4-GFP is enriched in nucleolar foci in addition to a diffuse signal in the nucleoplasm. (Scale bar: 10 μm.)
Amino Acid Residue 87, Histidine, of Arabidopsis Histone H3.3 Is More Critical than Residue 90, Leucine, for Its Deposition into rDNA Arrays.
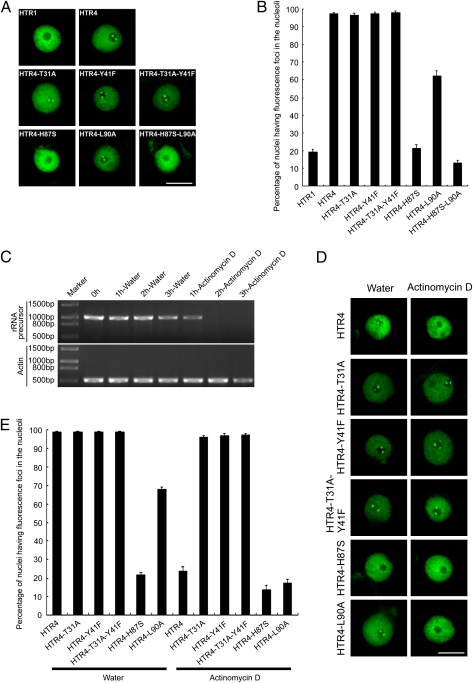
Because histone H3.3 is deposited at sites of active transcription, one would expect disruption and replacement of the relevant nucleosomes (24, 32, 33). A higher turnover rate was observed in Drosophila (23, 24) and alfalfa histone H3.2, a homolog of H3.3 (34), compared with histone H3.1. Because plant H3.3 differs from H3.1 in only 4-aa residues and it exhibits a distinct deposition pattern in rDNA arrays, we used nucleoli as an in vivo assay system to investigate how these 4-aa residues contribute to the deposition of histone H3.3 at nucleolar rDNA. We transiently expressed WT H3.1/HTR1-GFP, H3.3/HTR4-GFP, and various forms of site-mutated H3.3/HTR4-GFP fusions independently in tobacco leaf epidermal cells. Similar to the results observed in transgenic Arabidopsis cell nuclei, WT HTR4/H3.3-GFP was observed to be highly enriched in nucleolar foci in addition to a diffuse signal in the nucleoplasm of tobacco leaf cell nuclei, whereas HTR1/H3.1-GFP exhibited weak to no signal in the nucleoli (Fig. 3 A and B). Chromocenters were not obvious in tobacco nuclei expressing HTR1/H3.1-GFP, possibly because of differences in chromatin organization between tobacco and Arabidopsis. A series of mutated forms of H3.3/HTR4 were generated by altering individual or combinatorial amino acid residues in the core and N-terminal domains of HTR4/H3.3 at sites 31, 41, 87, and 90 to their corresponding residue in HTR1/H3.1, respectively. These mutated forms of H3.3/HTR4 were fused to GFP and transiently expressed in tobacco leaf cells, and the percentage of nuclei having fluorescent foci in the nucleoli was scored using a deconvolution fluorescence microscopy system (Fig. 3 A and B). When threonine 31 of HTR4/H3.3 is mutated to its counterpart residue alanine of HTR1/H3.1 (or HTR4-T31A) and fused to GFP, HTR4-T31A-GFP foci were observed in nucleoli at a similar level to the WT H3.3 (Fig. 3 A and B). Similarly, when tyrosine 41 of HTR4/H3.3 is mutated to its counterpart residue phenylalanine of HTR1/H3.1 (or HTR4-Y41F) or both the amino acids 31 and 41 are mutated to generate HTR4-T31A-Y41F, the HTR4-Y41F-GFP and HTR4-T31A-Y41F-GFP foci in nucleoli were also largely not affected compared with WT HTR4/H3.3 (Fig. 3 A and B). This suggested that amino acid residues 31 and 41 in the N-terminal tail of plant H3.3 do not play a significant role in contributing to the deposition of H3.3 into nucleolar DNA. However, when histidine 87 of HTR4/H3.3 is mutated to its corresponding residue serine of HTR1/H3.1 to generate HTR4-H87S, the percentage of nuclei having HTR4-H87S-GFP foci in the nucleoli decreases dramatically to about 21% compared with that of WT H3.1 at about 19% (Fig. 3 A and B). When leucine 90 of HTR4/H3.3 is mutated to its corresponding residue alanine of HTR1(H3.1) or HTR4-L90A and fused to GFP, the percentage of nuclei having HTR4-L90A-GFP foci in the nucleoli decreases to a level of ∼62% (Fig. 3 A and B). When both histidine 87 and leucine 90 of histone HTR4/H3.3 are mutated to HTR4-H87S-L90A, the nucleolar signal of HTR4-H87S-L90A-GFP looks like WT H3.1-GFP (Fig. 3 A and B). These results suggest that histidine 87 of plant histone HTR4/H3.3 plays a more critical role than leucine 90 of HTR4/H3.3 in defining its deposition to nucleoli, although both histidine 87 and leucine 90 in the core domain of plant histone H3.3 are important for nucleolar deposition of HTR4/H3.3.
Fig. 3.
Roles of the amino acids residues 31, 41, 87, and 90 of plant histone H3.3/HTR4 in its deposition to or depletion from rDNA. (A) WT H3.1/HTR1 and H3.3/HTR4 and sites-mutated forms of HTR4 including HTR4-T31A, HTR4-Y41F, HTR4-T31A-Y41F, HTR4-H87S, HTR4-L90A, and HTR4-H87S-L90A were fused to GFP and transiently expressed in tobacco leaf cells; representative images are shown. (B) Quantitative analysis of these GFP-tagged proteins (A); the percentages of nuclei having fluorescent foci in the nucleoli were scored for these fusions. Error bars indicate SDs (n = 9). (C) Transcription of tobacco rRNA precursor and an actin gene was examined by RT-PCR and the amplified fragments were analyzed in an ethidium bromide-stained gel. The leaf discs were treated with actinomycin D every 1 h up to 3 h. Samples incubated in water were used as controls. (D) WT H3.3/HTR4 and sites-mutated forms of HTR4 including HTR4-T31A, HTR4-Y41F, HTR4-T31A-Y41F, HTR4-H87S, and HTR4-L90A were fused to GFP and transiently expressed in tobacco leaf cells. The leaf discs were treated with actinomycin D; representative images are shown. Samples incubated in water were used as controls. (E) Quantitative analysis of these GFP-tagged proteins (D); the percentages of nuclei having fluorescent foci in the nucleoli were scored for these fusions. Error bars indicate SDs. For HTR4, HTR4-T31A, HTR4-Y41F, and HTR4-T31A-Y41F treated with water and actinomycin D, n = 7. For HTR4-H87S and HTR4-L90A treated in water, n = 9. For HTR4-H87S and HTR4-L90A treated in actinomycin D, n = 10. (Scale bars: 10 μm.)
Amino Acids 31 and 41 in the N-Terminal Tail of H3.3 Guide Nucleosome Disassembly from Nucleolar rDNA.
Next, we were interested in investigating the behavior of histone H3.3 when transcription is shut down. To this end, we monitored the signal of H3.3 in plant nucleoli when RNA polymerase I (Pol I) -directed transcription in nucleoli is inhibited by a low concentration of actinomycin D (35). The inhibition of RNA Pol I-directed transcription was investigated by reverse transcription polymerase chain reactions (RT-PCR) of ribosomal RNA precursor using a forward primer at the 5′ end of 5.8S rRNA and a reverse primer in the region of 26S rRNA. A predominant inhibition of rRNA transcription was observed when the tobacco leaf discs were treated with actinomycin D at 0.04 μg/mL for 2 h, which was indicated by failure to amplify the rRNA precursor. In contrast, controls exhibited clear bands of the expected size of the ribosomal precursor (Fig. 3C). Under these conditions of actinomycin D treatment, the mRNA of the introduced GFP-tagged histone H3.3 gene (Fig. S5A) and the tagged protein signal intensity in the nuclei (Fig. S5B) were maintained at a nearly constant level, indicating that, as expected, the expression of H3.3-GFP is not affected by incubation of leaf discs with this concentration of actinomycin D for 2 h. When WT HTR4/H3.3-GFP is transiently expressed in cells that are subsequently treated with actinomycin D, the HTR4/H3.3-GFP signal decreases from nucleoli gradually but not from the nucleoplasm (Fig. S5C). In 2 h, the H3.3-GFP signal in the nucleoli is largely depleted (Fig. 3 D and E). Under these conditions, the nucleoli are intact, which was indicated by the localization of AtNucleolin-YFP and AtFibrillarin 2 (AtFib2) -YFP, two typical nucleolar markers (Fig. S6). In control cells treated with water, no change in the HTR4/H3.3-GFP signal, nucleoli, and nucleoplasm was observed (Fig. 3 D and E). Together, these results indicate that the nucleolar chromatin containing H3.3 is transcriptionally active. However, HTR4-T31A-GFP, HTR4-Y41F-GFP, and HTR4-T31A-Y41F-GFP foci still remain in nucleoli in a similar pattern and at a similar level to the WT HTR4/H3.3-GFP when cells are treated with actinomycin D (Fig. 3 D and E). In contrast, on actinomycin D treatment, the percentages of cells having HTR4-H87S-GFP and HTR4-L90A-GFP foci in the nucleoli decrease from ∼21% and ∼62% (Fig. 3 A and B) to ∼14% and ∼17%, respectively (Fig. 3 D and E), suggesting that only amino acid residues 31 and 41 in the N-terminal tail of plant H3.3 play a role in defining the release of HTR4 from nucleolar nucleosomes when RNA Pol I-directed transcription is inhibited.
Conclusions
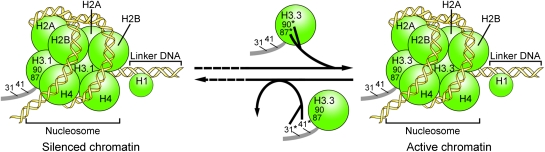
In summary, we have characterized the distinct localization of plant histones H3.1 and H3.3 and investigated how the amino acid residues 31, 41, 87, and 90 of plant histone H3.3, which distinguish it from H3.1, contribute to the deposition onto or depletion from nucleolar chromatin in an RNA Pol I transcription-dependent manner. Our results support a model (Fig. 4) in which amino acid residues 87 and 90 in the core domain of H3.3 guide its deposition into nucleolar chromatin when the nucleolus is transcriptionally active, whereas amino acid residues 31 and 41 in the N-terminal tail of H3.3 guide its depletion from rDNA arrays when transcription is silenced. The observation that histone H3.3 is depleted from rDNA arrays on silencing of the rDNA genes in interphase nuclei raises the possibility that histone H3.3 replacement by histione H3.1 can occur outside of S-phase. It is known that histone H3.1 is deposited into chromatin predominantly in a DNA replication-dependent manner during S-phase (4). However, some Arabidopsis histone H3.1 genes were also observed to be highly expressed outside of S-phase in addition to their DNA replication-coupled expression (25), raising the possibility that histone H3.1 may, under certain circumstances, also be deposited into chromatin in interphase nuclei in a DNA replication-independent manner in addition to its bulk incorporation into replicating chromatin. Intriguingly, two Arabidopsis histone H3.3 genes were found to express in S-phase in addition to their constitutive expression (25). Additional studies are needed to evaluate the potential incorporation of histone H3.3 into replicating chromatin and its role in epigenetic memory of active genes (20).
Fig. 4.
A model for the dynamics of plant H3.3/HTR4 in nucleoli. The amino acid residues 87 and 90 in the core domain of H3.3 guide its deposition to active nucleolar chromatin, whereas the amino acid residues 31 and 41 in the N-terminal tail of H3.3 guide its depletion from the rDNA arrays when transcription is silenced. Dashed lines mean that the switches between active chromatin and silenced chromatin likely include multiple steps in addition to the loading and unloading of histone H3.3.
In mammalian cells, Ser-31 is found to be phosphorylated (36); therefore, it is of future interest to investigate whether the amino acid residue Thr31 and the plant-specific Tyr41 are subject to PTMs that influence nucleosome disassembly. Interestingly, no defects during sexual reproduction were detected in a null mutant of At-hira (At3g44530) (19), the plant homolog of HIRA, which plays a role in DNA replication-independent assembly of histone H3.3-containing nucleosomes (4). In addition, a survey of plant homologs of animal H3 chaperones suggests that a different mechanism might be involved in the removal of parental H3 from the zygote nucleus (19). Therefore, it is of intense interest to identify and study plant chaperones that exhibit an association with the four amino acid residues 31, 41, 87, and 90 in histone H3.3 and mediate the assembly and disassembly of the related nucleosomes.
Materials and Methods
Multiple Alignment Analysis of Histone H3 Proteins.
Protein sequences of A. thaliana (At) histones H3.1/HTR1 and H3.3/HTR4, D. melanogaster (Dm) histones H3.1/HTR401 and H3.3/HTR405, and Homo sapiens (Hs) histones H3.1/HTR503 and H3.3/HTR501 were downloaded from http://www.chromdb.org/. Multiple alignment of the protein sequences was performed and exported using University of California at San Francisco Chimera (http://www.cgl.ucsf.edu/chimera) (37).
Constructs.
The constructs used in this study are described in SI Materials and Methods.
Plant Growth and Transformation.
The constructs described above were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation (38). A. thaliana plants (ecotype Columbia) were transformed by the floral dip method (39). Plants were grown in a growth chamber under 16-h light and 8-h dark conditions at 23 °C.
Transient Expression, Drug Treatment, and DAPI Staining.
Transient expression was performed using tobacco infiltration (38). The drug treatment and DAPI staining are described in SI Materials and Methods.
Microscopy and Quantitative Analysis of the Fluorescent Foci in the Nuclei.
Image stacks of nuclei were acquired at room temperature with a DeltaVision PersonalDV system (Applied Precision) using an Olympus UPLANAPO water-immersion objective lens (60×/1.20 numerical aperature) (38). The settings of the microscope are described in SI Materials and Methods. The fluorescent foci in the nuclei were scored using 7–10 Tobacco leaf discs transformed by GFP-tagged WT H3.1 or H3.3 or by various mutated forms at residues 31, 41, 87, and 90 with or without actinomycin D treatment. For each disk, 100–300 nuclei were scored.
Reverse Transcription Polymerase Chain Reaction (RT-PCR) Analysis.
RT-PCR analysis for the transcription of tobacco rRNA precursor is described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the Fang and Spector laboratories for insightful discussions, The Arabidopsis Information Resource (TAIR) for providing cDNAs of HTR1 and HTR4, and Steven Henikoff for critical reading of the manuscript. D.S. is supported by National Institutes of Health/National Institute of General Medical Sciences Grant 42694-22. Financial support was provided by National Natural Science Foundation of China Grant 91019006 (to Y.F.) and Chinese Academy of Sciences Grant KSCX2-YW-N-099 (to Y.F.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017882108/-/DCSupplemental.
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talbert PB, Henikoff S. Histone variants—ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 5.Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y. Tetrameric organization of vertebrate centromeric nucleosomes. Proc Natl Acad Sci USA. 2010;107:20317–20322. doi: 10.1073/pnas.1009563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 8.Waterborg JH. Multiplicity of histone h3 variants in wheat, barley, rice, and maize. Plant Physiol. 1991;96:453–458. doi: 10.1104/pp.96.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 10.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 11.Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta. 2004;1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 12.Goldberg AD, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci USA. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong LH, et al. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–360. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drané P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sawatsubashi S, et al. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev. 2010;24:159–170. doi: 10.1101/gad.1857410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phelps-Durr TL, Thomas J, Vahab P, Timmermans MC. Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell. 2005;17:2886–2898. doi: 10.1105/tpc.105.035477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 19.Ingouff M, et al. Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr Biol. 2010;20:2137–2143. doi: 10.1016/j.cub.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 20.Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- 21.Hödl M, Basler K. Transcription in the absence of histone H3.3. Curr Biol. 2009;19:1221–1226. doi: 10.1016/j.cub.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 22.Sakai A, Schwartz BE, Goldstein S, Ahmad K. Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr Biol. 2009;19:1816–1820. doi: 10.1016/j.cub.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz BE, Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 2005;19:804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okada T, Endo M, Singh MB, Bhalla PL. Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J. 2005;44:557–568. doi: 10.1111/j.1365-313X.2005.02554.x. [DOI] [PubMed] [Google Scholar]
- 26.Ahmad K, Henikoff S. Histone H3 variants specify modes of chromatin assembly. Proc Natl Acad Sci USA. 2002;99(Suppl 4):16477–16484. doi: 10.1073/pnas.172403699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gendler K, Paulsen T, Napoli C. ChromDB: The Chromatin Database. Nucleic Acids Res. 2008;36:298–302. doi: 10.1093/nar/gkm768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elsaesser SJ, Goldberg AD, Allis CD. New functions for an old variant: No substitute for histone H3.3. Curr Opin Genet Dev. 2010;20:110–117. doi: 10.1016/j.gde.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I. Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc Natl Acad Sci USA. 2002;99:14584–14589. doi: 10.1073/pnas.212325299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melaragno JE, Mehrotra B, Coleman AW. Relationship between Endopolyploidy and Cell Size in Epidermal Tissue of Arabidopsis. Plant Cell. 1993;5:1661–1668. doi: 10.1105/tpc.5.11.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang Y, Hearn S, Spector DL. Tissue-specific expression and dynamic organization of SR splicing factors in Arabidopsis. Mol Biol Cell. 2004;15:2664–2673. doi: 10.1091/mbc.E04-02-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wirbelauer C, Bell O, Schübeler D. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 2005;19:1761–1766. doi: 10.1101/gad.347705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ooi SL, Priess JR, Henikoff S. Histone H3.3 variant dynamics in the germline of Caenorhabditis elegans. PLoS Genet. 2006;2:e97. doi: 10.1371/journal.pgen.0020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waterborg JH. Dynamic methylation of alfalfa histone H3. J Biol Chem. 1993;268:4918–4921. [PubMed] [Google Scholar]
- 35.Perry RP. Selective effects of actinomycin D on the intracellular distribution of RNA synthesis in tissue culture cells. Exp Cell Res. 1963;29:400–406. [Google Scholar]
- 36.Hake SB, et al. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc Natl Acad Sci USA. 2005;102:6344–6349. doi: 10.1073/pnas.0502413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 38.Fang Y, Spector DL. Live cell imaging of plants. Cold Spring Harb Protoc. 2010;2:pdb.top68. doi: 10.1101/pdb.top68. [DOI] [PubMed] [Google Scholar]
- 39.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.