Abstract
Female mammals produce milk to feed their newborn offspring before teeth develop and permit the consumption of solid food. Intestinal enterocytes dramatically alter their biochemical signature during the suckling-to-weaning transition. The transcriptional repressor Blimp1 is strongly expressed in immature enterocytes in utero, but these are gradually replaced by Blimp1− crypt-derived adult enterocytes. Here we used a conditional inactivation strategy to eliminate Blimp1 function in the developing intestinal epithelium. There was no noticeable effect on gross morphology or formation of mature cell types before birth. However, survival of mutant neonates was severely compromised. Transcriptional profiling experiments reveal global changes in gene expression patterns. Key components of the adult enterocyte biochemical signature were substantially and prematurely activated. In contrast, those required for processing maternal milk were markedly reduced. Thus, we conclude Blimp1 governs the developmental switch responsible for postnatal intestinal maturation.
Keywords: enterocyte maturation, gut development, nutrition, cell fate
The defining characteristic of placental mammals is the presence of highly specialized mammary glands that secrete milk. During the suckling phase, neonates rely entirely on a lactose-rich milk diet. Immature enterocytes are highly specialized for pinocytosis and processing of milk constituents. During the suckling-to-weaning transition, as teeth develop enabling consumption of solid foods high in sucrose and maltose, intestinal enzymatic pathways adapt appropriately to mediate efficient carbohydrate breakdown. The regulatory mechanisms responsible for controlling this developmental switch in enterocyte gene expression profiles remain unknown.
The mouse gut tube is initially comprised of a pseudostratified layer of definitive endoderm surrounded by splanchnic mesoderm. Starting at approximately embryonic day (E) 14.5, the mesenchyme begins to invaginate into the endoderm to form the finger-like villi that protrude into the luminal space. Formation of terminally differentiated cell types including absorptive enterocytes, mucus secreting goblet cells, and enteroendocrine cells proceeds from E14.5 onward. Subsequent growth of the epithelial sheet is driven by proliferative cells restricted to the intervillus pockets (1). Starting at birth, their derivatives migrate into the underlying mesenchyme to generate the intestinal crypts. In the adult intestine, stem cells at the base of the crypts proliferate continuously to replace epithelial cells being shed from the villus tips (2).
The transcriptional repressor Blimp1, encoded by the Prdm1 gene, contains an N-terminal PR/SET domain and five C-terminal C2H2 zinc fingers that mediate nuclear import and DNA binding (3). Originally identified as a silencer of β-IFN gene expression (4), and a master regulator of plasma cell differentiation (5), Blimp1 acts downstream of Bmp/Smad signals in the early embryo to silence the default somatic pathway and specify primordial germ cells (6, 7). Blimp1 also regulates development of the posterior forelimb, caudal pharyngeal arches, and sensory vibrissae (8), and governs gene expression profiles in T-cell subsets (9), the sebaceous gland (10), and skin epidermis (11).
Here we report that Blimp1 acts as a master regulator of intestinal maturation. Blimp1 is strongly expressed throughout the epithelium of the embryonic gut. Beginning at birth, expression becomes down-regulated in the subset of intervillus cells that give rise to the developing crypts. The crypt derived adult enterocytes that emerge at postnatal day (P) 12 and repopulate the entire epithelium lack Blimp1 expression. Conditional inactivation of Blimp1 results in globally misregulated gene expression patterns, and compromises small intestine (SI) tissue architecture, nutrient uptake, and survival of the neonate. In combination with transcriptional profiling, the present experiments demonstrate that Blimp1/Prdm1 orchestrates orderly and extensive reprogramming of the postnatal intestinal epithelium.
Results
Down-Regulated Blimp1 Expression in Crypt Progenitors Beginning at Birth.
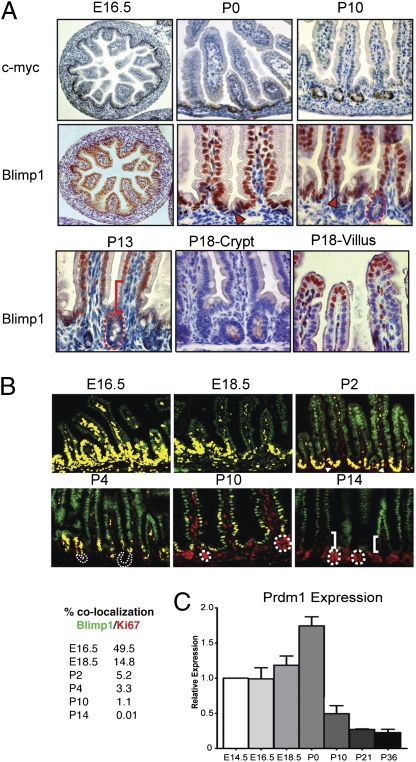
Blimp1 is strongly expressed throughout the embryonic gut endoderm (8, 12). Expression is maintained in the fetal SI throughout development (Fig. 1A), but, beginning at birth, the pattern dramatically changes. Expression is lost in a subset of c-Myc+ cells within the intervillus pockets. During the next 10 to 12 d, this population expands to form the mature crypts (Fig. 1A). In striking contrast, immature enterocytes retain Blimp1 expression (Fig. 1A). Slightly later Blimp1+ cells shift distally, and by P18 expression is strictly confined to the luminal villus tips (Fig. 1A). From postweaning onward, the intestinal epithelium lacks Blimp1 expression.
Fig. 1.
Dynamic Blimp1 expression patterns during intestinal maturation. (A) At E16.5, c-Myc+ cells are restricted to the intervillus pockets but become confined to developing crypts from P0 onward. Blimp1 is broadly expressed throughout the fetal endoderm. At P0, individual Blimp1− cells are detectable within the intervillus pockets (red arrow), and by P10 populate the entire maturing crypt (dashed red line). At approximately P13, Blimp1− cells enter the proximal villi, displacing immature Blimp1+ enterocytes (red bar). By P18, Blimp1 expression becomes restricted to the distal tip cells and endothelial cells in the lamina propria. (B) Quantification of Blimp1 (green, 605-nm Qdot) and Ki67 (red, 655-nm Qdot) coexpression via multi spectral imaging with yellow mask reveals decreasing percentages of double positives during villus morphogenesis and postnatal crypt formation. By P14, colocalization is no longer detectable (0.01%). (C) qPCR analysis confirms decreasing Prdm1 expression levels in total intestinal tissue (box, mean; bar, ± SEM; n > 10 for each data point).
Next, we compared cell proliferation (by Ki67 labeling) and Blimp1 expression during formation of the crypt villus axis (Fig. 1B). At E16.5, expression colocalizes to the intervillus pockets and proximal villus. Significant numbers of double-positive cells are retained at P2 in the proximal villi, but the percentage decreases during the next few days. A few double-positive cells persist at P10, but are subsequently lost. From P14 onward, Blimp1+ enterocytes are rapidly displaced as mature Blimp1− enterocytes emerge from the crypts and migrate up the villus axis. In contrast, Ki67+ cells are strictly confined to the crypts. To confirm that Blimp1− adult crypt stem cells derive from Blimp1+ fetal progenitors, Blimp1.Cre transgenic mice (6) were crossed to the R26R reporter strain (13) and stained for LacZ activity at P36. As expected, LacZ-positive crypts as well as labeled progeny were detectable along the entire villus axis (Fig. S1).
Conditional Inactivation Results in Postnatal Lethality.
We crossed the Villin.Cre transgenic strain that expresses Cre in yolk sac and gut endoderm from E12.5 onward (14) to Prdm1CA/CA animals (15) to selectively eliminate Blimp1 functions in the developing SI. As expected in Villin.Cre;Prdm1CA/CA embryos (referred to as Blimp1 mutants), expression in the intestinal epithelium was virtually undetectable by E18.5 (Fig. S2). Similarly, Western blot analysis at E18.5 confirms loss of Prdm1 expression (Fig. S2).
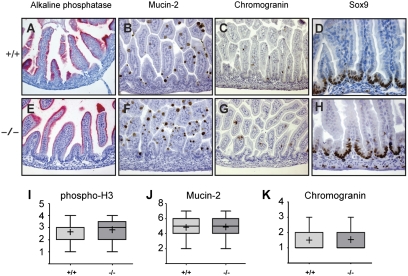
Blimp1 conditional inactivation loss has no noticeable effect on gross morphology or formation of mature cell types, including enterocytes and goblet and enteroendocrine cells, before birth (Fig. 2). Expression levels of Math1, Klf4, and Hes1 (16, 17, 18) were unaltered (Fig. S2). Sox9 expression required for postnatal Paneth cell formation (19) was unaffected (Fig. 2). Similarly, as judged by phosphohistone 3 and Ki67 staining and BrDu incorporation, the proliferative activity of intervillus pocket cells was unperturbed at E18.5 (Fig. 2 and Fig. S2). c-Myc is a well characterized Blimp1 target gene in B lymphocytes (20) and functions as a key downstream effector of Wnt signaling in the intestine (21). However, we found here that Blimp1 inactivation fails to influence c-Myc expression (Fig. S2).
Fig. 2.
Blimp1 inactivation fails to disturb intestinal morphogenesis before birth. (A–H) Conditional loss of Blimp1 has no noticeable effect on expression of mature markers of enterocyte (alkaline phosphatase), goblet (mucin-2), and enteroendocrine cells (chromogranin), or Sox9, a marker of Paneth cell progenitors. (I–K) More than 20 completely intact villi from proximal sections stained for phosphohistone 3, mucin 2, or chromogranin were counted (box, interquartile range; horizontal line, median; vertical line, minimum to maximum values excluding outliers; cross, mean).
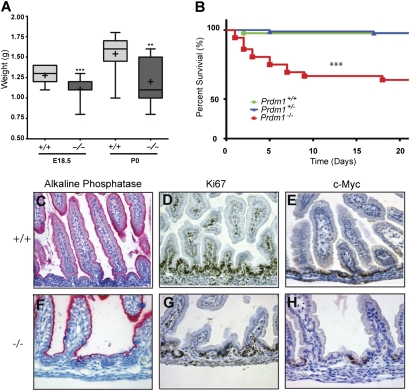
Mutant offspring were born at the predicted Mendelian frequencies (Table S1), and display normal suckling behavior. However, compared with control littermates, these pups are growth-retarded, and nearly half (P = 0.0001) died within 10 d of birth (Fig. 3 A and B). The SI tissue architecture was clearly disturbed (Fig. 3 C–H and Fig. S3). Villus density was reduced. Moreover, intervillous cells were highly vacuolated, retained only patchy Ki67/c-Myc expression, and strongly expressed the enterocyte marker alkaline phosphatase at the apical surface, a phenotype consistent with premature terminal differentiation. Interestingly, in Tcf4-deficient embryos, loss of Wnt-mediated proliferation similarly results in failure to maintain the intervillus stem cell compartment and causes cells to acquire a terminal columnar phenotype starting at E16.5 (22).
Fig. 3.
Loss of Blimp1 disrupts tissue architecture and causes postnatal lethality. (A) Blimp1 mutant offspring display significantly decreased body weights compared with control littermates. (B) Kaplan–Meier survival curve (n ≥ 35 for all genotypes). (C–H) Alkaline phosphatase, Ki67, and c-Myc staining at P3 reveals changed tissue architecture in mutant intestines. Mutants display reduced villus density and occupancy of intervillous regions by differentiated columnar epithelium. Intervillus regions show reduced proliferative potential as judged by Ki67 staining and fewer shallow crypt structures are evident. c-Myc expression is confined to the rare shallow crypts.
The SI of adult survivors invariably contains both WT and LacZ-marked deleted cells (Fig. S3). Thus, the incompletely penetrant phenotype is caused by a few exceptional WT cells that escape excision during embryogenesis and rapidly expand postnatally to support villus homeostasis. However, the initial weight loss observed at P0 persists into adulthood and is accompanied by decreased SI surface area (Fig. S3). Thus, conditional Blimp1 inactivation in the fetal gut epithelium has long-lasting effects on SI growth and development.
Global Changes in Gene Expression Patterns Revealed by Transcriptional Profiling.
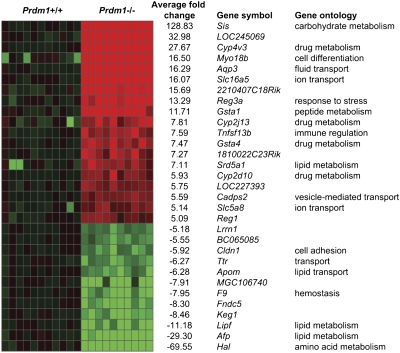
We used the Illumina array platform to compare WT versus mutant transcripts in SI tissue at late gestational (E18.5) stages. We identified 1,595 differentially expressed genes (Illumina DiffScore > 13, equivalent to P < 0.05) widely distributed throughout the genome. Of these, 664 were up- and 948 down-regulated. Functional annotation clustering analysis with DAVID 6.7 (23) demonstrated the highest enrichment scores for energy metabolism, vacuole function, and molecular transport categories (Fig. S4). Genes showing greater than fivefold changes (DiffScore > 100) are summarized in Fig. 4. Candidate misregulated genes were validated by quantitative PCR (qPCR) and scored for the presence of Blimp1 binding site motifs containing the core consensus sequence GAAAG (24) by using an affinity weighted matrix (Genomatix; Table S2).
Fig. 4.
Summary of misregulated genes identified in transcriptional profiling experiments. Transcripts with greater than fivefold changes in expression with Illumina DiffScores of more than 100 are shown. Gene Ontology identifies highest enrichment scores for categories associated with metabolic processes, vacuole function, and molecular transport. Fold changes in individual samples are normalized to mean Prdm1+/+ signal. The heat-map scale ranges from 10-fold reduced (green) to normal (black) and 10-fold increased (red).
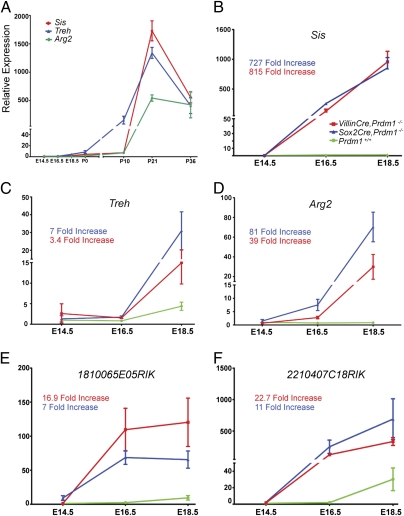
Numerous transcripts normally restricted to adult enterocytes were dramatically up-regulated. Up-regulated components of the adult biochemical signature include the carbohydrate processing enzymes sucrase isomaltase (Sis) and trehalase (Treh). Sis and Treh expression levels normally increase 10-fold between E14.5 and birth, and 1,500- and 2,000-fold, respectively, during the third week of postnatal life (Fig. 5A). Similarly, arginase 2 (Arg2), required for arginine breakdown in the adult intestine (25), normally increases 500-fold between P10 and weaning (Fig. 5A). The cytochrome p450 4v3 subunit, involved in fatty acid hydrolysis, three members of the glutathione transferase family (Gsta1, Gsta2, Gsta4), (26), as well as two RIKEN ESTs cloned from intestine libraries (27) normally expressed by adult enterocytes, were highly up-regulated (Table S2). Interestingly, the abrupt appearance of the adult biochemical signature that normally occurs between P10 and P21, just shortly before weaning (Fig. 5A), precisely mirrors the rapid disappearance of Blimp1-positive immature enterocytes. qPCR analysis of mutant fetal SI demonstrates these adult markers are ectopically activated starting at E14.5 of development, just before the onset of villus morphogenesis (Fig. 5 B–F).
Fig. 5.
Blimp1 regulates intestinal maturation during the suckling-to-weaning transition. (A) qPCR analysis of adult enterocyte markers reveals a dramatic shift in transcriptional profiles between P10 and P21. Mutant intestines obtained by crossing the conditional allele with Villin.Cre (red line) or the epiblast-specific Sox2.Cre (blue line) deleter strains acquire the adult biochemical signature at E16.5 as judged by dramatically up-regulated expression of Sis, (B), Treh (C), and Arg2 (D), and two RIKEN EST clones from an adult intestine library (E and F). (Symbol, mean; bar, ± SEM; n > 10 genotypes.)
In contrast we found many immature enterocyte markers were down-regulated (Fig. 4 and Table S2). For example, expression of the digestive enzyme neuraminidase (Neu1), responsible for the breakdown of sialic acid (28, 29), peaks near birth and decreases rapidly between P10 and weaning (P21; Fig. S5A). Milk is deficient in the essential amino acid arginine and de novo synthesis resulting from the breakdown of glutamine and proline in neonates, is in part mediated by arginosuccinate synthetase (Ass1) and arginosuccinate lyase (Asl) (30). Ass1 levels normally peak at E18.5 and then rapidly decrease (Fig. S5A). The ability of the neonatal SI to absorb intact macromolecules via pinocytosis depends on lysosomal enzymes, including N-acetylgalactosaminidase (Naga), β-gal (Glb1), and β-glucuronidase (GusB) (31). These transcripts are strongly expressed during the suckling period, but decrease before weaning (Fig. S5A). qPCR analysis confirmed markedly reduced expression levels in mutant enterocytes (Fig. S5 B–F). The reduced body weights of Blimp1 conditional mutants can be explained by decreased expression of pathway components required for milk processing, as well as uptake of nutrients and growth factors from the amniotic fluid.
Loss of Blimp1 Causes Fetal Intestinal Epithelium to Adopt an Adult Enterocyte Fate.
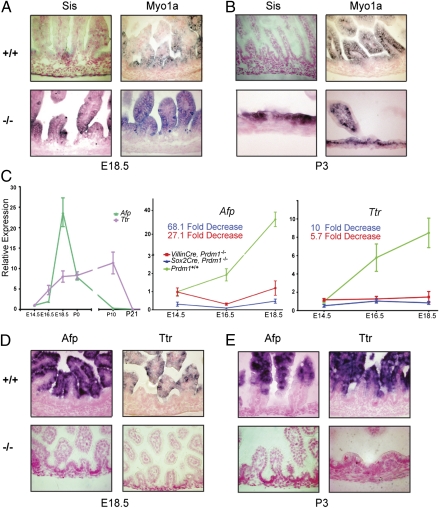
The brush border membrane protein Sis is initially expressed in crypt-derived mature enterocytes that emerge at P12 and start migrating distally to the villus tips (Fig. S6). In situ hybridization experiments confirmed that Sis is prematurely expressed in Blimp1-deficient villous epithelium from E16.5 onward (Fig. 6 A and B). Similarly, the brush border myosin Myo1A (32) is ectopically expressed throughout the mutant epithelium (Fig. 6 A and B). In contrast, Afp and Ttr, which are abundantly expressed in extraembryonic yolk sac endoderm and the developing gut epithelium (33, 34) (Fig. 6 C–E and Fig. S7), are markedly down-regulated (Figs. 4 and 6C). In WT embryos, expression increases between E14.5 and birth (Fig. 6C) and is restricted to the villus epithelium (Fig. 6D). These immature Afp/Ttr+ cells are gradually displaced by adult enterocytes (Fig. 6E). We found that mutant villi are completely devoid of Afp/Ttr+ cells (Fig. 6 D and E), but, in contrast, expression in mutant yolk sac tissue is unaffected (Fig. S7). These findings strongly argue that the mutant SI epithelium converts to an adult enterocyte phenotype.
Fig. 6.
Blimp1 loss causes the fetal intestinal epithelium to adopt an adult enterocyte phenotype. (A and B) WT enterocytes weakly express Myo1A and lack Sis expression at E18.5 and P3, whereas mutants strongly express both Sis and Myo1A. (C) qPCR analysis of developmentally regulated Afp and Ttr expression levels. (D and E) At E18.5 and P3, in contrast to WT, mutant enterocytes completely lack Afp and Ttr expression.
Discussion
Classical biochemical studies identified the key enzymatic and transport pathways responsible for uptake and digestion of maternal milk by newborn mammals (31, 35). In mice, the immature enterocyte profile becomes down-regulated beginning at approximately 2 wk after birth and rapidly shifts to the adult program during the third week of life (Fig. 5A and Fig. S5A). Early work established that this developmental switch is mediated by intrinsic rather than environmental or dietary cues (36). An attractive idea is that immature fetal enterocytes might be replaced by a second population with a radically altered biochemical signature. Our experiments demonstrate that the master transcriptional regulator Blimp1 plays a fundamental role in the developing intestine controlling this critical shift in gene expression profiles.
Blimp1 is strongly expressed in the definitive endoderm that gives rise to the embryonic gut tube shortly after it emerges from the anterior primitive streak during gastrulation (7, 12). Expression is maintained in both proliferating and quiescent intestinal epithelial cells throughout fetal development. However, beginning at birth, a subset of mitotically active c-Myc+ cells in the intervillus pockets selectively down-regulates Blimp1 coincident with migration into the underlying mesenchyme to initiate crypt formation. Slightly later, starting at P12, mature adult Blimp1−/Sis+ enterocytes gradually displace the immature Blimp1+ population. Between birth and crypt maturation, residual actively proliferating Ki67+/Blimp1+ cells situated in the proximal villi maintain the immature population. The coordinately balanced activities of these distinctive cell populations ensures temporal overlap of immature and mature metabolic pathways, enabling the juvenile to simultaneously process both milk and solid food during the suckling-to-weaning transition.
In the developing forelimb bud, Blimp1 expression transiently marks progenitor cells that give rise to the zone of polarizing activity (8). This key signaling center is correctly specified in Blimp1 mutants but fails to expand, resulting in discrete loss of the posterior digits and ulna (8). Likewise, Blimp1 inactivation in the developing intestine may compromise the ability of intervillus cells to respond to localized cues required to maintain their undifferentiated status as crypt progenitors. Surprisingly, our analysis of misregulated genes revealed only a few transcription factors, cell cycle regulators, and components of growth factor signaling pathways. It is of course possible, if not likely, that postnatal intestinal maturation is also controlled in part by posttranscriptional regulatory mechanisms.
Transcriptional networks controlling gut development have been previously characterized. Conditional inactivation of Cdx2 in the early gut endoderm disrupts villus architecture and completely blocks enterocyte, goblet, and enteroendocrine cell terminal differentiation (37). The Cdx2 target Hnf4α normally activates Treh (38). Similarly, the Sis promoter region contains binding sites for Hnf1α, Gata4, Cdx2, and Cdx1 (39). These transcriptional activators are expressed in the embryonic and adult intestinal epithelium (40, 41, 42). However, Treh and Sis gene expression becomes activated only in mature crypt-derived enterocytes, suggesting that these promoter regions are probably inaccessible in immature Blimp1+ enterocytes.
Recent experiments suggest that Blimp1/Prdm1 transcriptional targets are cell type-specific (3). Here we identify several candidate direct targets, including Sis and the two RIKEN clones found to be dramatically up-regulated and containing Blimp1 binding motifs with the core consensus sequence GAAAG within promoter and distal regions. However, consistent with previous studies (43), we also observe numerous predicted Blimp1 binding sites present near down-regulated genes. Current models of transcriptional control have evolved far beyond simple on/off switches to encompass increasingly complex circuits governing gene expression. Indeed, recent reports demonstrate that Blimp1 binds to a distal site within intron 2 and represses its own expression via an autoregulatory feedback loop (11, 43). Genome-wide ChIP experiments identified nearly 300 Blimp1-occupied promoters in a human myeloma cell line (44). Similarly recent studies reveal dynamic changes in occupancy by the homeodomain protein CDX2 at hundreds to thousands of distinct sites cooperatively bound with other tissue-specific transcription factors in the adult intestine (45). Future experiments will investigate local and long-range regulatory interactions during Blimp1 target site selection in the developing intestine.
In contrast to previous studies of transcription factors required for allocation of terminally differentiated cell types, or those acting to control regional compartmentalization during digestive tract development, the present work demonstrates that the transcriptional repressor Blimp1/Prdm1 globally regulates gene expression profiles in the developing SI. Combinatorial protein partnerships with histone deacetylases (HDACs), the G9a methyltransferase, and LSD1, a lysine-specific demethylase, have all been implicated in silencing Blimp1 target genes during plasma cell terminal differentiation (3). We note with interest that expression of its epigenetic partners Hdac1 and Hdac2 is also down-regulated in the intestinal epithelium during later stages of development (46). It will be important to learn more about Blimp1 interactions with its epigenetic partners that cooperatively regulate global changes in chromatin architecture during postnatal reprogramming of intestinal enterocytes.
Materials and Methods
Animals.
Villin-Cre (14) and Prdm1BEH/+ (7) animals were intercrossed to generate heterozygous Villin-Cre, Prdm1BEH/+ males that were then mated with homozygous Prdm1CA/CA females (15) carrying the R26R allele (13) to generate WT (Prdm1CA/+), heterozygous (Villin-Cre, Prdm1CA/+, or Prdm1BEH/+) or null (Villin-Cre, Prdm1CA/BEH) offspring. Sox2.Cre, Prdm1BEH/+ males were crossed to homozygous Prdm1CA/CA females to generate Blimp1-null embryos (8). Blimp1.Cre transgenic animals (6) were used for fate mapping.
In Situ Hybridization, Histology, and Immunohistochemistry.
SI contents were removed by flushing with PBS solution and were dissected into proximal (duodenum), middle (jejunum), and distal (ileum) tissue segments and fixed in 4% paraformaldehyde overnight at 4 °C. Sections (5 μm) were prepared for immunohistochemistry. Only data from the duodenum are presented. Protocols and a full list of antibodies are presented in SI Materials and Methods. In situ hybridization on 8-μm paraffin sections was performed according to standard protocols (47) with DIG-labeled probes for Ttr, Afp, Sis, and Myo1A, counterstained with eosin and mounted. For H&E staining, tissue was fixed overnight in 4% PFA before processing (47).
Transcriptional Profiling and qPCR Validation.
Tissue between the base of the stomach and the top of the cecum was removed, freed of surrounding mesentery, flushed with ice-cold PBS solution, and stored in RNAlater (Sigma). RNA was extracted by using the RNeasy Mini kit (74104; Qiagen). Biotinylated cRNA (1,500 ng) from 11 Prdm1+/+ and 10 Prdm1−/− samples was hybridized to Illumina Mouse WG-6 mouse v2 Expression BeadChips. Samples were randomly loaded onto the six array positions within each BeadArray. BeadStation data were extracted and analyzed by using the Gene Expression Analysis Module V1.6.0 of GenomeStudio V2009.2 (Illumina). Differential probe expression was determined following rank-invariant normalization by using the Illumina custom Error model option with Benjamini and Hochberg false discovery rate. Probes with significantly different expression (difference score > 13, equivalent to P < 0.05) were analyzed by using DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/).
For qPCR analysis, mRNA was converted into cDNA by using the Superscript III enzyme and oligo dT primers (18080–051; Invitrogen). cDNA (10 μL) was amplified in a 15-μL fluorescein (FAM)-based PCR by using SensiMix master mix (QT705-05; Bioline). Cycling conditions included an initial denaturation step of 15 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and then 1 min at 60 °C. Probe and primer sets were designed by using the Universal probe library assay design center (Roche). Primer sequences are provided in Table S3. Expression of target genes was normalized to Actin cDNA and relative fold change compared with WT was calculated by using the δCt method as 2δ (ΔCtgene2 − ΔCtgene1). All reactions were performed by using technical triplicates.
Data Analysis.
Statistical analysis was performed by using the GraphPad Prism 5 statistic package. All data were checked for normality by using the D'Agostino and Pearson omnibus normality test and subsequently tested by using the Student t test or the Mann–Whitney nonparametric test.
Supplementary Material
Acknowledgments
We thank Cordelia Langford and Peter Ellis (Sanger Institute) for performing the arrays, Silvie Robine for Villin.Cre mice, and Gina Doody and Reuben Tooze for antibodies. The work was funded by grants from the Wellcome Trust. E.J.R. is a Wellcome Trust Principal Research Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition. Array data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE29658).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105852108/-/DCSupplemental.
References
- 1.Trier JS, Moxey PC. Morphogenesis of the small intestine during fetal development. Ciba Found Symp. 1979;70:3–29. doi: 10.1002/9780470720530.ch2. [DOI] [PubMed] [Google Scholar]
- 2.van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 3.Bikoff EK, Morgan MA, Robertson EJ. An expanding job description for Blimp-1/PRDM1. Curr Opin Genet Dev. 2009;19:379–385. doi: 10.1016/j.gde.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Keller AD, Maniatis T. Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 1991;5:868–879. doi: 10.1101/gad.5.5.868. [DOI] [PubMed] [Google Scholar]
- 5.Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 6.Ohinata Y, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 7.Vincent SD, et al. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development. 2005;132:1315–1325. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- 8.Robertson EJ, et al. Blimp1 regulates development of the posterior forelimb, caudal pharyngeal arches, heart and sensory vibrissae in mice. Development. 2007;134:4335–4345. doi: 10.1242/dev.012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 10.Horsley V, et al. Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland. Cell. 2006;126:597–609. doi: 10.1016/j.cell.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magnúsdóttir E, et al. Epidermal terminal differentiation depends on B lymphocyte-induced maturation protein-1. Proc Natl Acad Sci USA. 2007;104:14988–14993. doi: 10.1073/pnas.0707323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohinata Y, Sano M, Shigeta M, Yamanaka K, Saitou M. A comprehensive, non-invasive visualization of primordial germ cell development in mice by the Prdm1-mVenus and Dppa3-ECFP double transgenic reporter. Reproduction. 2008;136:503–514. doi: 10.1530/REP-08-0053. [DOI] [PubMed] [Google Scholar]
- 13.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 14.el Marjou F, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 15.Shapiro-Shelef M, et al. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 16.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 17.Katz JP, et al. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki K, et al. Hes1-deficient mice show precocious differentiation of Paneth cells in the small intestine. Biochem Biophys Res Commun. 2005;328:348–352. doi: 10.1016/j.bbrc.2004.12.174. [DOI] [PubMed] [Google Scholar]
- 19.Mori-Akiyama Y, et al. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology. 2007;133:539–546. doi: 10.1053/j.gastro.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 20.Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- 21.Sansom OJ, et al. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–679. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- 22.Korinek V, et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 23.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo TC, Calame KL. B lymphocyte-induced maturation protein (Blimp)-1, IFN regulatory factor (IRF)-1, and IRF-2 can bind to the same regulatory sites. J Immunol. 2004;173:5556–5563. doi: 10.4049/jimmunol.173.9.5556. [DOI] [PubMed] [Google Scholar]
- 25.Hurwitz R, Kretchmer N. Development of arginine-synthesizing enzymes in mouse intestine. Am J Physiol. 1986;251:G103–G110. doi: 10.1152/ajpgi.1986.251.1.G103. [DOI] [PubMed] [Google Scholar]
- 26.Patel HR, Hewer A, Hayes JD, Phillips DH, Campbell FC. Age-dependent change of metabolic capacity and genotoxic injury in rat intestine. Chem Biol Interact. 1998;113:27–37. doi: 10.1016/s0009-2797(98)00016-7. [DOI] [PubMed] [Google Scholar]
- 27.Kawai J, et al. RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- 28.Doell RG, Kretchmer N. Studies of small intestine during development. I. Distribution and activity of beta-galactosidase. Biochim Biophys Acta. 1962;62:353–362. doi: 10.1016/0006-3002(62)90097-5. [DOI] [PubMed] [Google Scholar]
- 29.Dickson JJ, Messer M. Intestinal neuraminidase activity of suckling rats and other mammals. Relationship to the sialic acid content of milk. Biochem J. 1978;170:407–413. doi: 10.1042/bj1700407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu G, Bazer FW, Tuo W, Flynn SP. Unusual abundance of arginine and ornithine in porcine allantoic fluid. Biol Reprod. 1996;54:1261–1265. doi: 10.1093/biolreprod/54.6.1261. [DOI] [PubMed] [Google Scholar]
- 31.Koldovský O, Herbst J. N-acetyl-beta-glucosaminidase in the small intestine and its changes during postnatal development of the rat. Biol Neonate. 1971;17:1–9. doi: 10.1159/000240296. [DOI] [PubMed] [Google Scholar]
- 32.Tyska MJ, Mooseker MS. A role for myosin-1A in the localization of a brush border disaccharidase. J Cell Biol. 2004;165:395–405. doi: 10.1083/jcb.200310031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrews GK, Dziadek M, Tamaoki T. Expression and methylation of the mouse alpha-fetoprotein gene in embryonic, adult, and neoplastic tissues. J Biol Chem. 1982;257:5148–5153. [PubMed] [Google Scholar]
- 34.Makover A, Soprano DR, Wyatt ML, Goodman DS. An in situ-hybridization study of the localization of retinol-binding protein and transthyretin messenger RNAs during fetal development in the rat. Differentiation. 1989;40:17–25. doi: 10.1111/j.1432-0436.1989.tb00809.x. [DOI] [PubMed] [Google Scholar]
- 35.Moog F, Denes AE, Powell PM. Disaccharidases in the small intestine of the mouse: normal development and influence of cortisone, actinomycin D, and cycloheximide. Dev Biol. 1973;35:143–159. doi: 10.1016/0012-1606(73)90012-2. [DOI] [PubMed] [Google Scholar]
- 36.Lebenthal E, Sunshine P, Kretchmer N. Effect of prolonged nursing on the activity of intestinal lactase in rats. Gastroenterology. 1973;64:1136–1141. [PubMed] [Google Scholar]
- 37.Gao N, White P, Kaestner KH. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev Cell. 2009;16:588–599. doi: 10.1016/j.devcel.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyd M, Bressendorff S, Møller J, Olsen J, Troelsen JT. Mapping of HNF4alpha target genes in intestinal epithelial cells. BMC Gastroenterol. 2009;9:68. doi: 10.1186/1471-230X-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boudreau F, et al. Hepatocyte nuclear factor-1 alpha, GATA-4, and caudal related homeodomain protein Cdx2 interact functionally to modulate intestinal gene transcription. Implication for the developmental regulation of the sucrase-isomaltase gene. J Biol Chem. 2002;277:31909–31917. doi: 10.1074/jbc.M204622200. [DOI] [PubMed] [Google Scholar]
- 40.Beck F. The role of Cdx genes in the mammalian gut. Gut. 2004;53:1394–1396. doi: 10.1136/gut.2003.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D'Angelo A, et al. Hepatocyte nuclear factor 1alpha and beta control terminal differentiation and cell fate commitment in the gut epithelium. Development. 2010;137:1573–1582. doi: 10.1242/dev.044420. [DOI] [PubMed] [Google Scholar]
- 42.Stegmann A, et al. Metabolome, transcriptome, and bioinformatic cis-element analyses point to HNF-4 as a central regulator of gene expression during enterocyte differentiation. Physiol Genomics. 2006;27:141–155. doi: 10.1152/physiolgenomics.00314.2005. [DOI] [PubMed] [Google Scholar]
- 43.Rutishauser RL, et al. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doody GM, et al. An extended set of PRDM1/BLIMP1 target genes links binding motif type to dynamic repression. Nucleic Acids Res. 2010;38:5336–5350. doi: 10.1093/nar/gkq268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verzi MP, et al. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev Cell. 2010;19:713–726. doi: 10.1016/j.devcel.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tou L, Liu Q, Shivdasani RA. Regulation of mammalian epithelial differentiation and intestine development by class I histone deacetylases. Mol Cell Biol. 2004;24:3132–3139. doi: 10.1128/MCB.24.8.3132-3139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating The Mouse Embryo: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.