Abstract
Cancer stem cells (CSCs) sustain tumor growth through their ability to self-renew and to generate differentiated progeny. These functions endow CSCs with the potential to initiate secondary tumors bearing characteristics similar to those of the parent. Recently the hair follicle stem cell marker CD34 was used to purify a CSC-like cell population from early skin tumors arising from treatment with 7,12-dimethylbenz[α]anthracene/12-o-tetradecanoylphorbol-13-acetate, which typically generates benign papillomas that occasionally progress to squamous cell carcinomas (SCCs). In the present study, we identify and characterize CSCs purified from malignant SCCs. We show that SCCs contain two highly tumorigenic CSC populations that differ in CD34 levels but are enriched for integrins and coexist at the SCC–stroma interface. Intriguingly, whether CD34lo or CD34hi, α6hiβ1hi populations can initiate secondary tumors by serial limit-dilution transplantation assays, but α6loβ1lo populations cannot. Moreover, secondary tumors generated from a single CSC of either subtype contain both CD34lo and CD34hi α6hiβ1hiCSCs, indicating their nonhierarchical organization. Genomic profiling and hierarchical cluster analysis show that these two CSC subtypes share a molecular signature distinct from either the CD34− epidermal or the CD34hi hair follicle stem cell signature. Although closely related, α6hiβ1hiCD34lo and α6hiβ1hiCD34hi CSCs differ in cell-cycle gene expression and proliferation characteristics. Indeed, proliferation and expansion of α6hiβ1hiCD34hi CSCs is sensitive to whether they can initiate a TGF-β receptor II–mediated response to counterbalance elevated focal adhesion kinase-mediated integrin signaling within the tumor. Overall, the coexistence and interconvertibility of CSCs with differing sensitivities to their microenvironment pose challenges and opportunities for SCC cancer therapies.
Keywords: cancer stem cell signature, epithelial–mesenchymal interactions, skin cancer
Cancers develop when cells acquire mutations in tumor-suppressor genes and proto-oncogenes that favor growth-promoting over growth-restricting processes, thereby unbalancing tissue homeostasis (1, 2). Indeed, cancer cells are generally proliferative, refractory to apoptotic cell death, and deficient in normal cellular differentiation. However, solid tumors such as cutaneous squamous cell carcinomas (SCCs) are not simply cancer cell clones but rather are complex structures composed of multiple cell types in unique microenvironments (3). How tumor architecture develops and how it is maintained over time is still poorly understood for most cancers. Integral to these issues is whether deregulated proto-oncogenes and tumor-suppressor genes affect all cancer cells equally or perform specific functions within distinct cellular compartments of the tumor. Of particular importance is how these mutations affect those cancer cells that ensure long-term growth and survival of the tumor.
Cancer stem cells (CSCs) sustain tumor growth through their ability to self-renew and differentiate into hierarchically organized cancerous tissue (4, 5). These functions endow CSCs with the potential to initiate secondary tumors bearing characteristics similar to those of the parent. In SCCs, actively proliferating cancer cells reside at the tumor–stroma interface and differentiate into nontumorigenic pearls in the tumor center (6). The use of 7,12-dimethylbenz[α]anthracene (DMBA) and 12-o-tetradecanoylphorbol-13 acetate (TPA) is a well-established chemical carcinogen treatment that leads primarily to papillomas in the skin. Recently, it was shown that, like hair follicles (HFs), tumors formed by this chemical regimen contain a small population of cells that express the cell-surface glycoprotein CD34, a marker expressed by a variety of adult SCs. In a 500–50,000 cell serial transplant assay, CD34+ cells purified from these tumors were shown to possess increased tumor-initiating ability compared with unfractionated tumor cells (7). The extent to which CD34 defines CSCs is currently unknown. Also poorly understood are how CSCs self-renew, how they differentiate into non–tumor-initiating progeny in cutaneous SCC, and how they compare with stem cells and progenitor cells in normal tissue. These questions are pivotal to address for the development of therapies.
The TGF-β pathway is commonly deregulated in human cancers, including SCCs, where TGF-β functions initially as a tumor suppressor but promotes metastasis in late-stage carcinogenesis (8, 9). TGF-β receptor II (TβRII) is an essential component of the TGF-β pathway, and its conditional ablation in skin epithelium (TβRIIKO) accelerates the development of aggressive SCCs upon exposure to the chemical carcinogen DMBA (9). Concomitant with TβRII loss in keratinocytes is the hyperactivation of integrins and the integrin signal transducer focal adhesion kinase (FAK) (9), features that promote cell proliferation, cell survival, and carcinogenesis (10–13). Indeed, integrins and FAK are commonly up-regulated and are critical for the development of mouse and human solid tumors, including SCCs (10–15). The potent effects of TGF-β/TβRII and integrin/FAK signaling on SCC formation are particularly intriguing, given that normal stem cells (SCs) of epidermis and HFs are responsive to TGF-β signaling and display elevated integrin levels relative to their committed progeny (16–19). These features provide an ideal platform for exploring the consequences of perturbing these pathways on the characteristics of SCC tumors and their associated CSCs.
Results
FAK Function Is Critical for SCC Tumor Susceptibility in TβRII-Deficient Mice.
Mice lacking FAK are more refractory to SCC formation, whereas those lacking TβRII show enhanced tumor susceptibility. To investigate whether FAK/integrin signaling is critical for the development of TβRIIKO SCCs, we generated mice whose skin epithelium was conditionally null for both TβRII and FAK (dKO). As were TβRIIKO (9) and FAKKO (20), dKO skins were asymptomatic. However, complete carcinogenesis by topical DMBA treatments (two times per week) induced cutaneous SCCs in all genotypes, with some SCCs well contained but invasive and others less differentiated and very invasive. SCC initiation appeared sooner in TβRIIKO mice and later in FAKKO mice than in their wild-type littermates. Interestingly, the accelerated tumor initiation in TβRIIKO mice was not seen in dKO mice, which developed SCCs at rates indistinguishable from those of wild-type littermates (Fig. 1A).
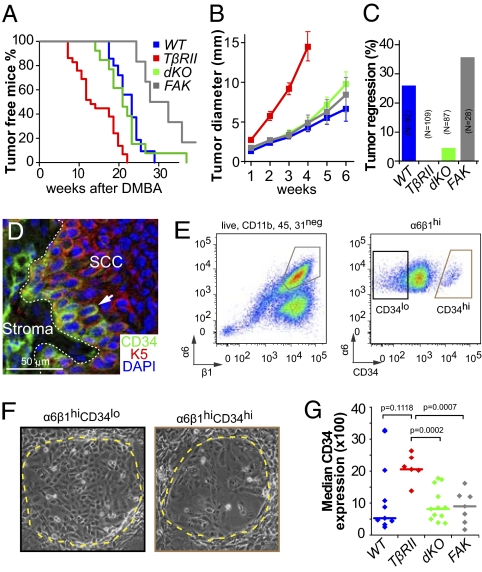
Fig. 1.
TβRII and FAK interact to control tumor initiation and growth and influence SCC composition. (A) Kaplan–Meier curves describing tumor-free survival after DMBA treatments of wild-type (blue), TβRIIKO (red), dKO (green), and FAKKO (gray) mice (P < 0.0001, Mantel–Cox log-rank test). Wild-type and dKO mice exhibit indistinguishable profiles (P = 0.271). Differences between TβRIIKO and dKO (P = 0.0004, Mantel–Cox log-rank test) and FAKKO and dKO (P = 0.0165, Mantel–Cox log-rank test) are statistically significant. (B) Tumor growth after initiation. TβRIIKO grow faster than other genotypes (n > 10; error bars indicate SEM). (C) Tumor regression after initiation depends upon TβRII function. (D) In SCCs, K5 is expressed in the undifferentiated cells at the tumor–stroma interface. Some K5+ cells express CD34. Although many CD34+ cells are in direct contact with the stroma, CD34+ cells also can be found at a distance from the tumor–stroma interface (arrow). (E) Representative flow cytometry profiles of cells isolated from primary SCCs and fractionated based on surface α6 (CD49f) and β1 (CD29) integrins after selecting live cells and eliminating CD11b+, CD45+, and CD31+ stromal cells. Both CD34lo and CD34hi α6β1hi cells (subfractionated into populations as in F) yielded large keratinocyte colonies (delineated by yellow dotted lines in representative phase contrast images) that could be passaged over the long term on fibroblast feeders. (G) Median CD34 expression in wild-type, TβRIIKO, dKO, and FAKKO SCCs reveals that the CD34hi population is expanded in TβRIIKO SCCs in a FAK-dependent manner. The means of median CD34 levels in the four SCC populations differ significantly (P = 0.02, ANOVA).
Once initiated, TβRIIKO SCCs grew faster than control SCCs (Fig. 1B). Additionally, although all SCCs executed a program resembling disorganized epidermal wound repair, TβRIIKO SCCs were the most poorly differentiated (Fig. S1). Such signs are typical of highly aggressive SCCs (11, 14).
FAK function appeared to be critical for the accelerated growth of TβRIIKO SCCs, because growth rates in dKO SCCs were comparable to those of FAKKO and control SCCs (Fig. 1B). Moreover, occasional newly developing tumors regressed in both control and FAKKO mice, suggesting either that they failed to generate CSCs for sustained long-term growth or that CSC self-renewal had been restricted by elevated suppressive activities within benign tumors (Fig. 1C) (2). By contrast, tumor regression was not observed in TβRIIKO mice and was rare in dKO animals. Together, these data suggest that TβRII/TGF-β and integrin/FAK signaling interact in controlling not only tumor initiation and growth but also the frequency with which benign tumors regress, persist, or progress to malignant SCCs.
Fractionating SCC Populations by Their Surface CD34 and Integrins and Functionally Testing Them for Self-Renewing Capacity in Vitro.
For the present study, we focused on tumors that progressed to SCCs in each of the four genetic backgrounds. Based on the notion that CSCs should reside within the relatively undifferentiated keratin 5(K5)/ keratin 14+ proliferative cells at the tumor–stroma interface (6), we posited that CSCs of SCCs should display abundant integrins. Indeed, all SCC cells located at the tumor–stroma interface expressed high levels of the hemidesmosomal α6 and β4 integrins and the focal adhesion marker β1 integrin, but only a fraction of these were CD34+ (Fig. 1D and Fig. S2). When coupled with the genotype-specific differences in SCC characteristics, these spatial differences in the intensity of CD34 and integrin staining at the tumor–stroma interface were suggestive of a heterogeneity that might be influenced by TβRII and/or FAK functions.
To place this heterogeneity in the context of proliferative potential, we fractionated these cancer cells from genotypically distinct primary SCCs by FACS. After eliminating stromal endothelial cells (CD31), lymphocytes (CD45), and macrophages (CD11b), we selected SCC keratinocytes based upon surface α6-integrin, β1-integrin, and CD34 levels (Fig. 1E). In contrast to α6hiβ1hi SCC cells, populations either α6loβ1hi or lacking integrins altogether failed to grow under our keratinocyte culture conditions. When α6hiβ1hi SCC cells were fractionated further into CD34hiα6hiβ1hi and CD34loα6hiβ1hi subpopulations, both cohorts formed colonies (Fig. 1F). Although colonies growing from CD34hiα6hiβ1hi SCC cells often appeared flatter and more differentiated, both cohorts could be passaged over the long term and independent of genotype.
The finding that SCC cells with long-term proliferative potential are uniformly enriched for integrins was consistent with transgenic studies linking integrin levels to SCC formation (21, 22). However, the CD34 variation was surprising. In seeking insights, we examined how the relative pool size of CD34hi cells compares among integrin-positive SCC populations of different genotypes. Interestingly, this pool was markedly expanded in TβRIIKO SCCs relative to dKO and FAKKO SCCs (Fig. 1G). Moreover, in control SCCs, in which the activities of TGF-β and FAK signaling are not defined, the pool was variable. These data further underscore the robust effects of cooperative action of TGF-β/TβRII and integrin/FAK signaling on SCC composition. Additionally, the expanded CD34hiα6hiβ1hi population within poorly differentiated TβRIIKO SCCs raises the possibility that their rapid tumor growth may be achieved by enhancing self-renewal and/or survival of CSCs while suppressing their differentiation. Moreover, because dKO SCCs are well differentiated and do not show an expansion of CD34hiα6hiβ1hi cells, this balance is predicated on FAK/integrin signaling.
Single-Cell Tumor-Initiating Functional Assays Reveal Two Interchangeable CSC Types in SCCs.
We hypothesized that, if our two integrin-rich SCC populations are truly CSCs, they should be able to initiate secondary and tertiary tumors and undergo self-renewal in the process. To test this hypothesis, we first transduced primary SCC cultures with retrovirus to express ubiquitously a triple-modality reporter (fluc-mrfp1-tk) (23). This reporter enabled us to mark the lineage permanently and to distinguish cancer parenchyma from stromal components. FACS-purified, transduced SCC cells were then suspended in Matrigel and injected intradermally into immunocompromised Nude mice to test their tumor-initiating potential (Fig. 2A). Approximately 10 d later, aberrant tumor growth was visible at the injection site, and tumors progressed to SCCs, as confirmed by histopathology.
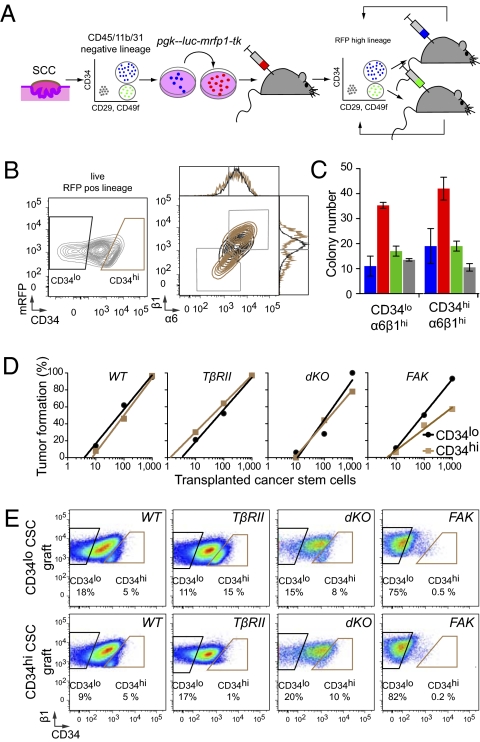
Fig. 2.
Cutaneous SCCs contain two interchangeable populations of CSCs that have tumor-initiating potential and are regulated by TβRII/TGF-β and FAK/integrin signaling. (A) Strategy to isolate CSCs. FACS-purified CD34hiCD29hiCD49fhiCD45−CD31−CD11b− cells are tagged with an RFP fluorescent reporter, and their long-term tumor-initiating potential is tested by serial orthotopic grafting experiments in immune-compromised Nude mice. (B) Representative flow cytometry profiles of RFP+ live cells isolated from secondary SCCs and separated based on their CD34 expression. Contour plots illustrate CD34lo (black) and CD34hi (brown) populations, which then were fractionated further based on α6(CD49f) and β1(CD29) integrin expression. Overlays illustrate that CD34hi subpopulations are either α6hiβ1hi or α6loβ1lo. (C and D) Colony formation (wild type, blue; TβRIIKO’ red; dKO,; green; FAKKO, gray; n > 2; error bars indicate SEM) and limit-dilution orthotopic transplantation experiments (n = 15–42 per data point; 576 grafts total) indicate that both α6β1hiCD34lo and α6β1hiCD34hi SCC cells form colonies in culture and tertiary SCCs in vivo, but their abilities depend upon TβRII and FAK function. (E) Flow cytometry profiles of tertiary SCCs illustrating the potential of CD34hi and CD34lo CSCs to generate CD34hi and CD34lo progeny interchangeably.
In orthotopic transplantation experiments, secondary SCCs displayed growth characteristics similar to those of their primary SCC origin and contained both CD34hi and CD34lo cell populations expressing both high and low levels of integrins (Fig. 2B). Additionally, as we had observed for the primary tumors, both high-integrin populations were effective in colony-forming assays, but neither low-integrin population was effective. Moreover, in these assays the levels of CD34 did not make a substantial difference (Fig. 2C). That said, it was notable that more colonies always formed from TβRIIKO than from FAKKO and dKO tumor-initiating cells.
These culture results were recapitulated faithfully in vivo. Thus, for all four genotypes, primary SCC cells with highest surface integrins exhibited the ability to initiate secondary tumors. However, the SCC-forming efficiencies of our purified CD34hiα6β1hi SCC cell populations varied considerably with genotype, showing the highest potential in TβRIIKO SCCs and the poorest in dKO and FAKKO SCCs (Fig. 2D). Interestingly, CD34hi and CD34lo cell populations did not appear to have a strict hierarchical relation or distinct differentiation potential, because serial transfer of either CD34hiα6β1hi or CD34loα6β1hi CSCs yielded progeny SCCs containing both CD34hi and CD34lo cell populations (Fig. 2E). Furthermore, CD34hi CSCs lost their CD34 expression and became indistinguishable from CD34lo CSCs when cultured in vitro, and cultures from either CSC type reestablished secondary tumors featuring both CD34hi and CD34lo CSCs when engrafted onto immunodeficient recipient mice (Fig. S3).
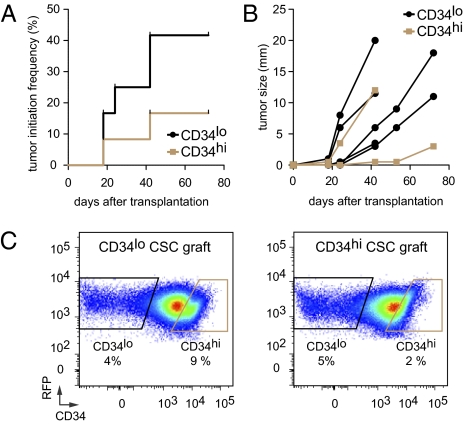
To document the interconvertibility, tumor-initiation, and differentiation potential of these two distinct types of putative CSCs, we performed single-cell sort and transplantation experiments. We focused on TβRIIKO SCCs that were the most aggressive of the four cell populations and showed the highest tumor-initiation efficiency in limit-dilution assays (Fig. 2D). Similar to our results in limit-dilution experiments, transplantation of single sorted CD34hiα6hiβ1hi and CD34loα6hiβ1hi CSCs from TβRIIKO SCC confirmed their high tumor-initiating potential (Fig. 3). We noted, however, that the tumor-initiation potential (Fig. 3A) and growth kinetics (Fig. 3B) were accelerated in secondary tumors that developed from single CD34loα6hiβ1hi CSCs compared with secondary tumors that developed from CD34hiα6hiβ1hi CSCs.
Fig. 3.
Tumor-initiation potential upon transplantation of single SCC CSCs. (A) Kaplan–Meier curve describing tumor-initiation frequency over time after transplantation of single CD34loα6hiβ1hi and CD34hiα6hiβ1hi cells into Nude mice. (n = 12, P = 0.185, Mantel–Cox log-rank test.) (B) Growth kinetics of tumors growing after transplantation of single CSCs. Although rates are variable, CD34loα6hiβ1hi CSCs have a tendency to initiate growth more rapidly than CD34hiα6hiβ1hi CSCs. (C) Flow cytometry profiles of tertiary SCCs. Note that single CD34hi and CD34lo CSCs have the potential to generate CD34hi and CD34lo progeny interchangeably within the tumors that they generate.
The ability of individual cells to generate SCCs improves by nearly three orders of magnitude the SCC tumor-initiating cell populations previously described (7) and unequivocally establishes these two cell populations as CSCs. Furthermore, the ability of CSCs to shift reversibly between CD34hi and CD34lo states indicates that neither CSC population is restricted in developmental potential. Rather, CSC interconversion appears to be sensitive to the microenvironment and CSCs’ ability to respond to it.
Transcriptional Profiling of CSCs from Four Different Genetic Backgrounds Identifies a CSC Molecular Signature for SCCs.
To understand further the relationship between these two CSC populations, we next addressed whether they might share a common transcriptional profile that distinguishes them from wild-type skin SCs. To this end, we carried out global gene-expression profiling of purified CD34hiα6hiβ1hi and CD34loα6hiβ1hi populations from two independent RFP-tagged SCC tumor samples for each genetic background. These 16 separate arrays then were contrasted with duplicate sets of CD34hiα6hiβ1hi HF-SCs and CD34loα6hiβ1hi epidermal SCs from 8-wk-old wild-type mice (17–19).
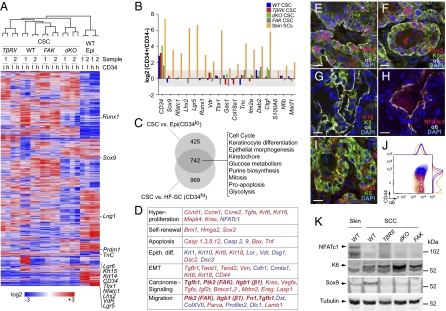
Corroborating our immunofluorescence data, overall CD34 levels were significantly lower in α6hiβ1hiCD34hi CSCs than in HF-SCs (Fig. 4 A and B). Surprisingly, however, many previously ascribed HF-SC markers, including leucine-rich repeat-containing G-protein coupled receptor 5 (Lgr5) (24), LIM homeobox 2 (Lhx2) (25), and nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 (Nfatc1) (26), also were weakly expressed or were absent in CSCs, as were established markers of epidermal and junctional zone/sebaceous gland SCs [e.g., leucine-rich repeats and immunoglobulin-like domains protein 1 (Lrig1) and Lgr6 (27, 28)]. In addition to their reduced expression, wild-type stem and progenitor markers showed no enrichment in CD34hiα6hiβ1hi compared with CD34loα6hiβ1hi CSCs (Fig. 4B). Although some HF-SC markers, including sex-determining region Y (SRY)-box 9 (Sox9) (29) and runt-related transcription factor 1 (Runx1) (30), were expressed at variable levels depending upon SCC background, they were still reduced relative to HF-SCs and were found at a distance from the core HF-SC marker cluster. Based upon hierarchical gene-cluster analyses, CSCs clustered together and were clearly more similar to each other than to either wild-type skin SC population (Fig. 4A).
Fig. 4.
Defining a cancer stem cell signature for SCCs. (A) Hierarchical cluster analysis of Affymetrix 430 2.0 array data indicates that, whether CD34+ or CD34−, SCC CSCs are more similar to each other than they are to wild-type SCs from either epidermis or HFs. Note that core markers of CD34+ HF-SCs, including Nfatc1, Lhx2, vitamin D receptor (VdR), and Lgr5, are strikingly reduced or absent in CSCs. Other adult SC markers [Lrig1, PR domain containing 1, with ZNF domain (Prdm1), and Sox9] are not uniformly elevated in CSCs. (B) Differential expression analyses indicate key distinctions between CSCs and wild-type SCs. Shown are fold changes (log2 scale) of previously described HF-SC markers between α6β1hiCD34hi and α6β1hiCD34lo cells of wild-type, TβRIIKO, dKO, and FAKKO SCCs and wild-type skin epithelium. Note that HF-SC marker genes other than CD34 are not differentially expressed in these two CSC populations. Bars indicate the average of two independent experiments. HF-SCs (α6hiβ1hiCD34hi) vs. epidermis (α6hiβ1hiCD34neg) serves as a positive control. (C) Venn diagram revealing a core CSC signature of 742 genes that are up-regulated by twofold or more (P < 0.05) in all CSCs irrespective of their genotype and whether compared with wild-type HF-SCs or epidermal progenitors. Functional hierarchical cluster analysis indicates enrichment for processes that are commonly deregulated in cancers. (D) Selected sets of skin genes that are elevated (red) or abrogated (blue) by more than twofold in CSCs compared with wild-type-SCs. (E–K) Immunofluorescence microscopy (E–I) and immunoblot analyses (K) validate the CSC signature and define the CSC niche at the tumor–stroma interface. Validation was performed on independent primary SCCs that developed in wild-type mice. (Scale bars: 20 μm.) FACS analysis in J shows that surface marker CD44, up-regulated in CSCs and localized at the tumor–stroma interface (I), is in the integrin-high population.
Because CSCs fell in a cluster distinct from normal skin SCs, we next sought to generate a CSC signature, i.e., genes up-regulated by twofold or more in CSCs, irrespective of genotype and CD34 status, relative to wild-type skin SCs (false-discovery rate, 0.05). Under these criteria, 742 genes formed the CSC signature (Fig. 4C and Dataset S1). Enriched in this signature were genes involved in cell cycle, mitosis, epithelial morphogenesis, hyperproliferation, apoptosis, and metabolism. They included many pathways commonly affected in carcinomas, including growth factor/signaling [Vegf-α; TGF-α; TGF-β1; MAPK4; breast cancer 1, early onset and breast cancer 2, early onset; v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog; integrin β1; protein tyrosine kinase 2 (FAK); and LIM and SH3 protein 1 (Lasp1)], self-renewal/hyperproliferation [BMI1 polycomb ring finger oncogene; high mobility group AT-hook 2 (HMGA2); Sox2], and epithelial-to-mesenchymal transitions [twist homolog 1 and twist homolog 2; vimentin; keratin 8/keratin 18; fibronectin] (Fig. 4 D–K). Conversely, genes down-regulated in CSCs relative to wild-type SCs included cell–cell adhesion genes [E-cadherin; α-catenin] (Datasets S2 and S3). Such differences are suggestive of cytoskeletal and adhesion remodeling within CSCs, and they correlated well with typifying features of the CSC microenvironment at the leading front of the tumor–stroma interface. Overall, differences between CSC and wild-type SC populations were confirmed for both up- and down-regulated genes (Fig. 4 E–K).
Differences Between CD34hi and CD34lo CSCs.
Although similarities trumped differences between CD34hi and CD34lo populations of integrin-rich CSCs, the differences proved to be interesting. By comparing their expression profiles in each SCC, we discovered that CD34hi CSCs of TβRIIKO SCCs were enriched for transcripts encoding cell-cycle and DNA-repair proteins, whereas CD34hi CSCs of other genetic backgrounds showed comparatively greater enrichment for extracellular matrix (ECM), migration, and antiapoptosis genes (Fig. 5A and Dataset S4).
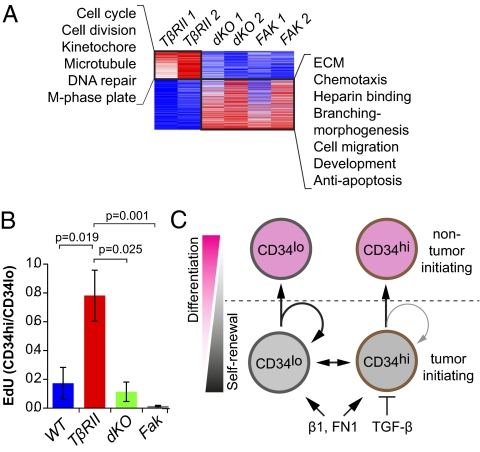
Fig. 5.
TGF-β/FAK signaling-dependent differences in proliferative activities of α6β1hiCD34hi CSCs. (A) Differential expression and functional hierarchical cluster analysis of α6β1hiCD34hi CSCs. Note that fast-cycling (TβRIIKO) α6β1hiCD34hi CSCs are enriched for cell-cycle and DNA repair transcripts, whereas slow-cycling (dKO, FAKKO) α6β1hiCD34hi CSCs display elevated transcripts for ECM, morphogenesis, migration, and antiapoptosis pathways. (B) Flow cytometry data indicate that EdU (6-h pulse) incorporates predominantly in CD34lo and not in CD34hi CSCs, unless TβRII is compromised and FAK function is sustained. Bar graphs indicate relative abundance of EdU+ cells in α6β1hiCD34hi CSCs over EdU+ cells in α6β1hiCD34lo CSCs. Note that loss of Tgfβ signaling in the presence of elevated FAK signaling leads to a specific enrichment in the proliferative activity within the α6β1hiCD34hi CSCs. (n > 5; P < 0.0001, ANOVA; error bars indicate SEM.) (C) Working model in which CSCs exist in interchangeable α6β1hiCD34hi and α6β1hiCD34lo states. Activated β1 integrin and its ligand fibronectin (FN1) promote self-renewal of both CSCs, whereas active TGF-β/TβRII signaling primarily influences α6β1hiCD34hi CSCs, restricting their self-renewal and expansion.
To pursue possible differences in the proliferation rates of the populations, we pulsed mice 6 h before euthanizing with the nucleotide analog 5-ethynyl-2′deoxyuridine (EdU). When FAKKO, dKO, or control SCCs were analyzed by FACS, EdU incorporation localized predominantly to CD34loα6hiβ1hi CSCs, and only few CD34hiα6hiβ1hi CSCs were EdU+ (Fig. 5B). By comparison, EdU incorporation rates were similar in CD34loα6hiβ1hi and CD34hiα6hiβ1hi CSCs and were significantly higher in the TβRIIKO CD34hiα6hiβ1hi CSCs than in dKO and FAKKO CD34hiα6hiβ1hi CSCs. Thus, within FAKKO and dKO CSCs, differences in CD34 appeared to be a reflection of differential proliferative activities, in a fashion similar to normal HF-SCs. In striking contrast, CD34hiα6hiβ1hi CSCs from TβRIIKO SCCs incorporated amounts of EdU similar to those in CD34loα6hiβ1hi CSCs.
Discussion
Our results provide compelling evidence that multiple CSC pools exist along the tumor–stroma interface in cutaneous SCCs. These CSC pools are interconvertible and lack a clear hierarchical organization as long as high levels of integrin expression are maintained. However, they differentiate irreversibly and lose their tumor-initiating potential as integrin expression is attenuated when CSCs depart from the tumor–stroma interface. Intriguingly, the distinct but coexisting CSC pools differ in their proliferative properties. As such, SCC CSCs’ behavior is similar to that of homeostatic skin where rapidly proliferating CD34lo epidermal SCs and slow-cycling CD34hi HF-SCs coexist and interconvert upon wounding or cell transplantation. Moreover, our studies of SCCs developed in chemically induced FAK- and/or TβRII-null skin epithelium revealed that CSC cycling activities within SCCs are influenced by their ability to respond to cues from their microenvironmental niche, which in this case is the tumor–stroma interface where TGF-β/TβRII and integrin/FAK signaling intersect. Finally, both transplantation and culture appear to reset the proliferative properties of these CSCs.
Three important findings came from our experiments. First was that our purification scheme further enriched for tumor-initiating SCC cells compared with a previously published strategy which purified on the basis of CD34 without integrins (7). Second, our findings suggest that high integrin expression is a more general marker for tumor-initiating CSCs within SCCs and that CD34 expression can distinguish between two specific subsets of tumor-initiating SCC cells which differ in cycling behavior. Finally, our data showed that TGF-β/TβRII and FAK/integrin signaling act in opposing fashion to control the self-renewal and tumor-initiation capabilities of CD34hiα6hiβ1hi cells, providing an explanation for the enhanced aggressiveness of TβRIIKO SCCs that retain and activate FAK/integrin function. Our identification of integrinhiCD34lo and integrinhiCD34hi tumor-initiating cells draws parallels between human and mouse SCCs. Both contain cancer cells with elevated integrin and FAK expression, whereas CD34-expressing cells have been found in mouse, but not in human, SCCs (7).
The ability of our CD34hiα6hiβ1hi and CD34loα6hiβ1hi populations of SCC cells to self-renew, be serially transferred over the long term, and initiate tumors at the single-cell level merited their definition as CSCs (4). In tumor-initiating assays, CD34loα6hiβ1hi populations were even more effective than CD34hiα6hiβ1hi populations at tumor initiation. That said, the percentage of CD34hiα6hiβ1hi cells within TβRIIKO SCCs was greater than in any other genetic background, and correspondingly, their overall efficiency in serial transplantation and tumor aggressiveness were greater also. This result underscores an important principle, namely that the ability of CSCs to respond to integrin signaling and suppress TGF-β responsiveness overrides the effects, if any, of CD34 levels in determining their long-term self-renewal and tumor-initiating characteristics. Given that SCCs lacking TβRII also were less differentiated than the SCCs formed on other genetic backgrounds, our data also imply that the ability to enhance signaling through FAK/integrin in the absence of active TGF-β signaling results in an impairment of the differentiation process. Our comparative studies of SCCs lacking TβRII alone versus those lacking both FAK and TβRII showed clearly that when FAK/integrin signaling was compromised, differentiation was restored in the TβRII-null SCCs.
Our results are intriguing in light of recent studies in melanoma, where tumor-initiating cells are abundant (31), marker genes are dynamically regulated (31, 32), and differences in cycling behaviors can be observed (32). However, despite the interconvertibility of our two CSC subtypes in SCCs, which like melanoma tumor-initiating cells, show differences in cycling behaviors, some cells within SCCs do show hierarchical relations: Low integrin-expressing SCC cells derive from high integrin-expressing cells, but they lack significant tumor-initiating potential and do not appear to be interconvertible (6).
By defining and analyzing CSCs from multiple SCCs of distinct genetic backgrounds, we have unearthed two populations that display heterogeneity across and within genotypes but differ primarily in their CD34 and cell-cycle gene expression. Enriching for CSCs, we achieved SCC-initiating studies with single CSCs. Such serial transfers enabled us to demonstrate that the two populations of CSCs are both tumorigenic and interconvertible and that their representation within SCCs is influenced markedly by their ability to respond to TβRII/TGF-β and integrin/FAK signaling at the tumor–stroma interface. The relative abilities of genetically distinct CSCs to escape inhibitory cues and exploit positive ones within their microenvironment provides a molecular understanding of the variations in SC numbers within different cancers. Our findings also place this flexible behavior of CSCs in the framework of how some cancers become faster growing and more aggressive (Fig. 5C). Our CSC signature provides insights into SCC behavior, draws interesting parallels between SCCs and other cancers, and offers a rich list of potential diagnostic and/or therapeutic targets for future investigations.
Methods
Comparative Pathology.
Tumor tissue was formalin fixed, paraffin embedded, sectioned, and stained with H&E. Histological analyses were performed by Suzana S. Couto (Memorial Sloan Kettering Cancer Center, New York). All tumors used for the present study were SCCs, which varied in their degree of severity.
Tumor Cell Preparation.
Tumors were dissected from mice and separated from normal skin, blood vessels, and connective tissue. Tumor tissue was minced and treated with 0.25% collagenase (C2670; Sigma) in HBSS (Gibco) for 60 min at 37 °C; 62.5 U/mL DNaseI (LS002138; Worthington) was added for the last 15 min of the collagenase treatment. The cell suspension was filtered with a 45-μm strainer. Retained cell clumps were dissociated further by treatment with 0.25% trypsin (Gibco) at 37 °C for 10 min and then were strained through a 45-μm mesh. The combined cell suspensions were diluted in wash buffer (PBS containing 2% chelexed FBS). Cells were pelleted at 300 × g for 10 min and then were resuspended in wash buffer, washed once, and incubated with surface antibodies for FACS.
Tumor Cell Transplantation.
Tumor cells were suspended in 50% Matrigel (356237; BD) in F medium at a concentration of 1, 10, 100, and 1000 cells/50 μL and injected s.c. into Nude mice. Tumor progression was documented photographically once every week from the time of inoculation to the experimental end point. Single-cell transplantation studies were performed by sorting a single cell per well in a 96-well plate containing 50% Matrigel (356237; BD) in F medium.
Cell-Cycle Analysis.
Cells were fixed in ice-cold 80% ethanol, and genomic DNA was labeled with 10 μg/mL propidium iodide in the presence of 250 μg/mL RNAseA in PBS. Cell-cycle analysis was performed by flow cytometry and analyzed using FlowJo software.
EdU Labeling and Detection.
Cell proliferation and S-phase entry in SCC were measured by injecting 50 μg/g EdU i.p. into mice 6 h before analysis. EdU incorporation was determined by flow cytometry using the Click-iT EdU Alexa Fluor 488 Cell Proliferation Assay Kit for flow cytometry (C35002; Invitrogen) following the manufacturer’s instructions.
Statistics.
Statistical and graphical data analyses were performed using Origin 7.5 (OriginLab) and Prism 5 (GraphPad) software.
Supplementary Material
Acknowledgments
We thank A. Levine and N. Stokes for their valuable technical support; G. Guasch for consultation and advice during the early phases of this project; B. Keyes for generously providing RNA samples for stem and progenitor cell microarray analyses; and S. Raghavan, A. Christiano, S. Beronja, B. Keyes, S. Williams, M. Rendl, and E. Gonzalez for discussions and comments on the manuscript. We are grateful for the help received from The Rockefeller University Flow Cytometry Center (which is supported by a grant from the Empire State Stem Cell fund through New York State Department of Health Contract C023046); the Bioimaging Resource Center; the Comparative Biology Center, (an American Association for Accreditation of Laboratory Animal Care facility) for their expert handling and care of the mice; Suzana S. Couto and the Memorial Sloan Kettering Comparative Pathology Core for pathology consultation and histological analyses and diagnoses of tumor tissues; and The Memorial Sloan Kettering Genomics Core Facility for RNA and microarray processing. This research also was supported by a grant from the Emerald Foundation (to E.F.) and by Grants R01-AR27883 (to E.F.) and 5K99-AR057260-02 (to M.S.) from the National Institutes of Health. E.F. is an investigator of The Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data discussed in this article have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE29328) (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29328).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107807108/-/DCSupplemental.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- 3.Egeblad M, Nakasone ES, Werb Z. Tumors as organs: Complex tissues that interface with the entire organism. Dev Cell. 2010;18:884–901. doi: 10.1016/j.devcel.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke MF, et al. Cancer stem cells—perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 5.Clevers H. The cancer stem cell: Premises, promises and challenges. Nat Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 6.Pierce GB, Wallace C. Differentiation of malignant to benign cells. Cancer Res. 1971;31:127–134. [PubMed] [Google Scholar]
- 7.Malanchi I, et al. Cutaneous cancer stem cell maintenance is dependent on β-catenin signalling. Nature. 2008;452:650–653. doi: 10.1038/nature06835. [DOI] [PubMed] [Google Scholar]
- 8.Bierie B, Moses HL. TGF-beta and cancer. Cytokine Growth Factor Rev. 2006;17:29–40. doi: 10.1016/j.cytogfr.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Guasch G, et al. Loss of TGFbeta signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell. 2007;12:313–327. doi: 10.1016/j.ccr.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLean GW, et al. The role of focal-adhesion kinase in cancer—a new therapeutic opportunity. Nat Rev Cancer. 2005;5:505–515. doi: 10.1038/nrc1647. [DOI] [PubMed] [Google Scholar]
- 11.Janes SM, Watt FM. New roles for integrins in squamous-cell carcinoma. Nat Rev Cancer. 2006;6:175–183. doi: 10.1038/nrc1817. [DOI] [PubMed] [Google Scholar]
- 12.McLean GW, et al. Specific deletion of focal adhesion kinase suppresses tumor formation and blocks malignant progression. Genes Dev. 2004;18:2998–3003. doi: 10.1101/gad.316304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitra SK, Schlaepfer DD. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr Opin Cell Biol. 2006;18:516–523. doi: 10.1016/j.ceb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 14.Mercurio AM. Invasive skin carcinoma—Ras and alpha6beta4 integrin lead the way. Cancer Cell. 2003;3:201–202. doi: 10.1016/s1535-6108(03)00049-7. [DOI] [PubMed] [Google Scholar]
- 15.Pylayeva Y, et al. Ras- and PI3K-dependent breast tumorigenesis in mice and humans requires focal adhesion kinase signaling. J Clin Invest. 2009;119:252–266. doi: 10.1172/JCI37160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones PH, Harper S, Watt FM. Stem cell patterning and fate in human epidermis. Cell. 1995;80:83–93. doi: 10.1016/0092-8674(95)90453-0. [DOI] [PubMed] [Google Scholar]
- 17.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Morris RJ, et al. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 20.Schober M, et al. Focal adhesion kinase modulates tension signaling to control actin and focal adhesion dynamics. J Cell Biol. 2007;176:667–680. doi: 10.1083/jcb.200608010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owens DM, Broad S, Yan X, Benitah SA, Watt FM. Suprabasal alpha 5 beta1 integrin expression stimulates formation of epidermal squamous cell carcinomas without disrupting TGFbeta signaling or inducing spindle cell tumors. Mol Carcinog. 2005;44:60–66. doi: 10.1002/mc.20118. [DOI] [PubMed] [Google Scholar]
- 22.Sugiyama M, Speight PM, Prime SS, Watt FM. Comparison of integrin expression and terminal differentiation capacity in cell lines derived from oral squamous cell carcinomas. Carcinogenesis. 1993;14:2171–2176. doi: 10.1093/carcin/14.10.2171. [DOI] [PubMed] [Google Scholar]
- 23.Ray P, Tsien R, Gambhir SS. Construction and validation of improved triple fusion reporter gene vectors for molecular imaging of living subjects. Cancer Res. 2007;67:3085–3093. doi: 10.1158/0008-5472.CAN-06-2402. [DOI] [PubMed] [Google Scholar]
- 24.Jaks V, et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 25.Rhee H, Polak L, Fuchs E. Lhx2 maintains stem cell character in hair follicles. Science. 2006;312:1946–1949. doi: 10.1126/science.1128004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horsley V, Aliprantis AO, Polak L, Glimcher LH, Fuchs E. NFATc1 balances quiescence and proliferation of skin stem cells. Cell. 2008;132:299–310. doi: 10.1016/j.cell.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watt FM, Jensen KB. Epidermal stem cell diversity and quiescence. EMBO Mol Med. 2009;1:260–267. doi: 10.1002/emmm.200900033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snippert HJ, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327:1385–1389. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- 29.Nowak JA, Polak L, Pasolli HA, Fuchs E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell. 2008;3:33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoi CS, et al. Runx1 directly promotes proliferation of hair follicle stem cells and epithelial tumor formation in mouse skin. Mol Cell Biol. 2010;30:2518–2536. doi: 10.1128/MCB.01308-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quintana E, et al. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roesch A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.