Abstract
An understanding of the balance between carbon and nitrogen assimilation in plants is key to future bioengineering for a range of applications. Metabolomic analysis of the model plant, Arabidopsis thaliana, using combined NMR-MS revealed the presence of two hemiterpenoid glycosides that accumulated in leaf tissue, to ~1% dry weight under repeated nitrate-deficient conditions. The formation of these isoprenoids was correlated with leaf nitrate concentrations that could also be assayed in the metabolomic data using a unique flavonoid–nitrate mass spectral adduct. Analysis of leaf and root tissue from plants grown in hydroponics with a variety of root stressors identified the conditions under which the isoprenoid pathway in leaves was diverted to the hemiterpenoids. These compounds were strongly induced by root wounding or oxidative stress and weakly induced by potassium deficiency. Other stresses such as cold, saline, and osmotic stress did not induce the compounds. Replacement of nitrate with ammonia failed to suppress the formation of the hemiterpenoids, indicating that nitrate sensing was a key factor. Feeding of intermediates was used to study aspects of 2-C-methyl-d-erythritol-4-phosphate pathway regulation leading to hemiterpenoid formation. The formation of the hemiterpenoids in leaves was strongly correlated with the induction of the phenylpropanoids scopolin and coniferin in roots of the same plants. These shunts of photosynthetic carbon flow are discussed in terms of overflow mechanisms that have some parallels with isoprene production in tree species.
For centuries mankind has relied on carbon, prehistorically fixed from the atmosphere by plants and algae, as a source of fuel and other materials. This is a dwindling resource and, to develop renewable sources of chemicals, knowledge of the biochemical pathways that link photosynthesis to the accumulation of useful materials will be paramount. Terpenoids (isoprenoids) are examples of carbon-rich products that occur prolifically across the plant kingdom. They are not only end products of chloroplast assimilation of photosynthetic carbon but also, by virtue of the phytyl side chains of chlorophyll, key elements of the machinery of photosynthesis. Some terpenoids are ubiquitous and are classed as primary metabolites (sterols, carotenoids, and phytyl side chains of chlorophyll); others are hormones (gibberellins, abscisic acid, brassinosteroids, and strigolactones). However, the majority of terpenoids, especially those that can accumulate to high levels in plants, are classed as secondary metabolites, although many have roles in chemical ecology. Manipulation of flux through the primary terpenoid pathways and the accumulation of terpenoids via the secondary pathways both have potential for exploitation.
Arabidopsis, the model plant, was, until recently, considered to be poorly endowed with terpenoids. For example, this species does not appear to contain oxidized sesqui- and diterpenes, compounds that are widespread in plants (1). There are some 30 terpene synthase genes in Arabidopsis (2) and recent analysis has revealed the presence of sesquiterpenes in flowers and monoterpenes in roots (1). Although the levels of terpenes are very low, the presence of the genes indicates that Arabidopsis remains a suitable model, especially for study of the central pathways of terpene biosynthesis.
Two pathways, producing the C5 units that form terpenes, coexist in plants; most primary and hormonal terpenoids are produced by the chloroplast-located 2-C-methyl-d-erythritol-4-phosphate (MEP) pathway (3) (Fig. S1). Notwithstanding the production of sterols and sesquiterpenes through the alternate mevalonate pathway, the MEP pathway is responsible for the majority of carbon flux into isoprenoids. The regulation of flux through the MEP pathway not only is crucial to plant growth and development but also has a role in the ability of the plant to react to changes in photosynthetic rate. Flux through the early part of the pathway varies with photosynthesis, by mechanisms involving gene and enzyme regulation in particular of 1-deoxy-d-xylulose-5-phosphate synthase (DXS) (4) and hydroxymethylbutenyl diphosphate reductase (HDR) (5). It is also known that plants need to balance N assimilation with photosynthesis-driven C metabolism (6). There is increasing evidence that flux through the MEP and phenylpropanoid (PP) pathways can be regulated by components of the nitrate (NO3−) assimilation pathway, particularly via the partitioning of phosphoenolpyruvate between cytosolic processes and chloroplastidic supply of precursors for these pathways (Fig. S1) (7). In this paper we report unique work on the metabolomics of Arabidopsis under root nutrient deprivation and other stresses and the induction of the biosynthesis of two hemiterpenoid glycosides at surprisingly high levels.
Results
Novel Metabolite Biomarkers in Leaves of Nutrient-Deprived Arabidopsis.
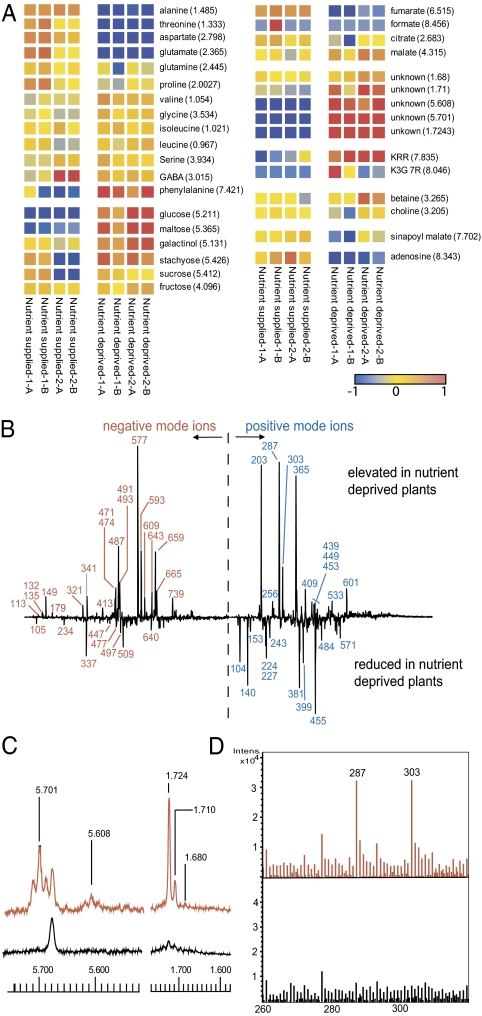
Combined [1H]-NMR and direct infusion electrospray ionization MS (DI-ESI-MS) is a technique that we have developed for rapid metabolite fingerprinting of plant extracts (8, 9). When applied to Arabidopsis tissue, the data obtained enable rapid assessment of concentrations and fluctuations of both primary and secondary metabolites. In this work, as a simple first experiment, we grew Arabidopsis in a hydroponic system with a full nutrient supply to the roots for 23 d to growth stage 1.12 (10) and then transferred the plants to water. Analysis of extracts of leaf tissue, harvested at 3 and 7 d after nutrient withdrawal, by parallel NMR and DI-ESI-MS revealed striking and repeatable differences in metabolite fingerprints between nutrient-deficient and nutrient-supplied plants (Fig. 1). Metabolite changes in the NMR spectra at 7 d are shown in the heatmap, Fig. 1A. Increases in carbohydrates, flavonoids (kaempferol glycosides), and in particular the amino acid phenylalanine (Phe) and decreases in sinapoyl malate and the amino acids Ala, Thr, Asp, and Glu were evident. However, the most striking feature of the NMR dataset was the presence of new signals for olefinic hydrogens at δ5.701 (major isomer) and 5.608 and aliphatic methyl groups at δ1.724 (major isomer), 1.710, and 1.680 in the nutrient-deprived plants. These signals are highlighted in Fig. 1C. These olefinic and methyl NMR signals were structurally linked to each other and also to signals at 4.18, 4.16, and 4.27 by 2D-NMR analysis (see next section). These data indicated that a small family of related compounds had been induced by the nutrient withdrawal and that there was associated reprogramming of metabolism involving Phe, flavonoids, and other primary metabolites.
Fig. 1.
Metabolite profiling of Arabidopsis leaf metabolites responding to root nutrient withdrawal. Data from 7-d deprivation are shown. Replicate codes 1 and 2 are biological and A and B are extraction replicates. (A) Heatmap of NMR data, derived by binning to 0.01 Hz followed by normalization of each bin to unit variance and mean centered scaling. (B) PC-1 loadings plots from PCA analysis of (−) and (+) ion DI-ESI-MS data. (C) Sections of NMR spectra depicting formation of unique metabolites in leaves of nutrient-deprived plants; red, nutrient deprived; black, nutrient supplied. (D) Sections of (+)DI-ESI-MS spectra depicting formation of unique metabolite with ions at m/z 287 and 303; red, nutrient deprived; black, nutrient supplied.
Positive and negative ion DI-ESI-MS fingerprints of the extracts were examined by principal component analysis (PCA). Biomarkers for nutrient deprivation, revealed in PC-1 loadings plots from that analysis, are depicted in Fig. 1B. The increase in flavonoid glycosides on nutrient depletion, seen in the NMR data, was confirmed in negative ion DI-ESI-MS by the increase in characteristic ions at m/z 577 (M-H; kaempferol 3,7-dirhamnoside, KRR) and 593 (M-H; kaempferol-3-rhamnoside-7-glycoside, KRG). Associated with the increase in these well-known flavonoids was a decrease in m/z 640, an ion shown to be related to KRR by MS-MS fragmentation (product ions at m/z 577, 431, and 285). However, the most striking feature of the DI-ESI-MS data was in the positive ion spectra where two significant ions at m/z 287 and 303 were observed only in starved plants. This was observed in both PCA loadings plots (Fig. 1B) and the spectra themselves (Fig. 1D).
Structure Determination of the Novel Nutrient Stress Biomarkers.
Further positive ion DI-ESI-MS data, collected at higher resolution, gave masses of 287.1076 and 303.0822 for the unique ions induced by nutrient deprivation. These masses are consistent (accuracy −11.6 ppm and −9.1 ppm, respectively) with a [M+Na]+ and [M+K]+ pair of adducts from a compound with the empirical formula C11H20O7. The possibility that these ions were [M+H]+ adducts of the flavonoid aglycones, kaempferol [C15H10O6, molecular weight (MW) 286] and quercetin (C15H10O7, MW 302) was ruled out as the masses of the measured ions were, respectively, 181 and 104 ppm removed from the calculated monoisotopic masses. Other computed formulas for the measured masses contained N, S, or very high numbers of unsaturations and these were not consistent with other data. 2D-NMR indicated that the discriminatory signals identified in the 1D-NMR could be assigned to at least two closely related glycosides of a C5-unsaturated diol. Chemical shift, connectivity, and coupling data (Fig. S2) indicated that the major compound could be (2E)-4-hydroxy-2-methyl-2-buten-1-yl-O-d-glucopyranoside [1] (Fig. 2). Further confirmation of the formula and presence of two isomers was obtained from GC-MS analysis of the plant material, derivatized by methoxyamination-trimethylsilylation (11). As shown in Fig. S3A, two unique peaks (ratio 4:1) with near identical mass spectra were present in the nutrient-starved samples. Molecular ions were absent but fragments at m/z 217, 204, 191, 147, and 129 were characteristic of trimethylsilylglycosides (12). The key fragment, m/z 157 (accurate masses 157.1048 and 157.1047 in the major and minor isomer, respectively) due to the aglycone, has the empirical formula C8H17OSi and corresponds to the structural fragment [(CH3)3SiOCH2CH = C(Me)CH2]− (calculated mass = 157.1049) that is consistent with the C5-enediol glycoside (hemiterpenoid glycoside, HTG) structure. 1D NOESY data of the major isomer indicated the trans (E) arrangement of the double bond and 2D-NMR spectroscopy placed the glycosidic linkage at the 1 position as shown in Fig. 2. The structures of this compound and the minor isomer were confirmed by synthesis, accomplished from (2E)-hydroxy-2-methylbut-2-enyl-4-acetate, an intermediate previously used in the synthesis of (2E)-4-hydroxy-3-methylbut-2-enyl diphosphate (HMBPP) (13). The synthesis (SI Materials and Methods) involved coupling of acetylbromoglucose to the 4-monoacetate, followed by deacetylation. Serendipitously, two isomers were produced that were identical (GC-MS and NMR) to the two HTGs observed in Arabidopsis. With synthetic material available comprehensive 2D-NMR investigations of both isomers were completed in isolation from contaminating plant carbohydrate. (SI Materials and Methods). This process led to the definitive conclusion that the natural compounds were the two regional isomers of the hemiterpenoid, (2E)-2-methylbut-2-en-1,4-diol glycoside, the major isomer being the 1-glycoside [1] and the minor isomer being the 4-glycoside [2] as shown in Fig. 2. 2-Methylbut-2-en-1,4-diol [3] was a by-product of the synthesis and was also available by deacetylation of the starting acetate. Small amounts of this compound were also detected in the NMR spectra (methyl group at δ1.68) of nutrient-deprived Arabidopsis extracts (Fig. S3B). 1H and 13C NMR data for all three synthetic products agreed well (Table S1) with previously reported occurrences of these and related compounds from other plant species (14–17).
Fig. 2.
Chemical structures.
Nitrate Deprivation Is a Key Inducer of HTGs.
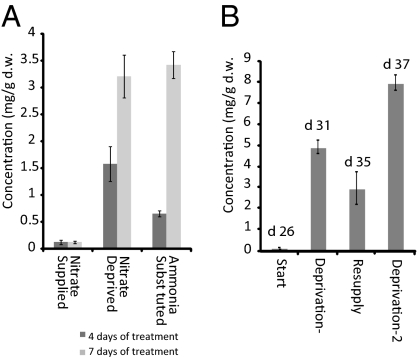
To define further the relationship between nutrient deprivation and the HTGs, the hydroponics system was used in experiments where individual nutrients were omitted from the full medium. Using metabolomic screening it was demonstrated that the prime inducer of the HTGs in leaves was root NO3− deprivation. When NH4+ was substituted for NO3− as a nitrogen source, the HTGs were still produced, indicating that NO3− sensing was the underlying mechanism behind the synthesis of HTGs (Fig. 3). Alternating periods of NO3− deprivation and resupply led to a stepwise accumulation of HTGs under periods of starvation, rising to ~1% dry weight. The HTGs did not appear to be substantially reassimilated on nitrate resupply. Other primary metabolites also changed during NO3− resupply. Ala, Thr, Asp, Asn, Gln, and Glu which, were all depleted under NO3− deprivation, recovered during the resupply period. Conversely, levels of metabolites that increased during NO3− deprivation (malate, Phe, sucrose, glucose, stachyose, and maltose) were seen to drop during resupply, to a level similar to that observed in NO3− supplied plants (Fig. S4).
Fig. 3.
Effect of nitrogen supply on HTG major isomer production. (A) Effect of NO3− deprivation and NH4+ ion replacement; (B) NO3− resupply experiment. Twenty-six-day-old plants were deprived of NO3− for 5 d, resupplied for 4 d, and deprived again for a further 2 d. Data are represented as mean ± SD.
Flavonoid Glycoside–Nitrate Adducts Formed in DI-ESI-MS Are Indicators of Tissue NO3− Levels.
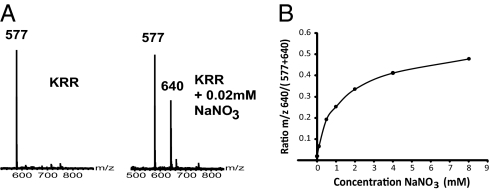
The m/z 640 ion routinely seen in untreated Arabidopsis leaf DI-ESI-MS negative ion spectra was absent in the NO3−-deprived plants (Fig. S5). From MS-MS experiments it was determined that this ion was a conjugate of the flavonoid glycoside, KRR, the first product ion being m/z 577 (KRR, M-H). Thereafter, fragmentation was identical to that of KRR (product ions at m/z 431 and 285). The accurate mass of the m/z 640 ion was 640.1541 kDa. This result is consistent (−9.8 ppm accuracy) with an empirical formula of C27H30O17N. The data indicated that m/z 640 arises from [KRR + NO3]− and that it is an in-source formed [+62] adduct. The absence of m/z 640 in the extracts from the NO3−-deprived tissue was consistent with lack of NO3− in the plant. The titer of the m/z 640 ion also serves as a second biomarker for NO3− starvation in Arabidopsis. Independent measurement of [NO3−] in samples of nutrient-deprived [3.6 ± 0.8 mg/g dry weight (d.w.) after 7 d] and nutrient-supplied plants (30–40 mg/g d.w.) (Table 1) provided support for this hypothesis. To confirm this, we isolated pure KRR from Arabidopsis by HPLC and collected DI-ESI-MS data from solutions of this compound spiked with 0.02 mM NaNO3 (Fig. 4A). We also added increasing concentrations of NaNO3 to solutions of the extracts of leaf tissue from NO3−-depleted plants. The effect of [NO3−] on the formation of m/z 640 is shown in Fig. 4B, demonstrating that the adduct intensity in the spectra can be correlated to [NO3−]. A weak negative correlation of intensity of +303 with −640 (r = −0.465; P < 0.0001) and +287 with −640 (r = −0.390; P = 0.0025) provided a semiempirical means of relating high HTG to low NO3−. There are a number of flavonoid glycosides in Arabidopsis. We also detected the unusual [M+ NO3]− for several of these in negative ion DI-ESI-MS. Thus, we observed [M+62]− at m/z 656.1489 associated with KRG, [M+62]− at m/z 672.1413 associated with quercetin-3-glucoside-7-rhamnoside, and [M+62]− at m/z 802.2092 associated with kaempferol-3-rhamnosylglucoside-7-rhamnoside.
Table 1.
Concentrations of NO3− and HTGs in leaf tissue under different stress conditions
| Concentration of nitrate, mg/g d.w. | Concentration of HTG [1]*, μg/g d.w. | Concentration of HTG [2]*, μg/g d.w. | ||||
| No days of treatment | 4 | 7 | 4 | 7 | 4 | 7 |
| Full nutrient supplied | 49.6 ± 4.8 | 36.1 ± 1.5 | n.d. | n.d. | n.d | n.d |
| Total nutrient deprived | 15.8 ± 1.0 | 3.6 ± 0.8 | 1155 ± 328 | 4729 ± 298 | 120 ± 61 | 1145 ± 298 |
| Nitrate deprived | 7.4 ± 2.4 | 0.2 ± 0.2 | 731 ± 240 | 2395 ± 366 | 383 ± 211 | 431 ± 152 |
| Ammonium substituted | 15.5 ± 4.4 | 2.7 ± 0.6 | 80 ± 41 | 2480 ± 218 | n.d. | 486 ± 56 |
| Potassium deprived | 32.6 ± 14.7 | 29.0 ± 1.9 | 44 ± 40 | 695 ± 170 | n.d. | 90 ± 57 |
| Salt stress | 36.3 ± 3.8 | 27.9 ± 2.4 | n.d. | n.d. | n.d. | n.d. |
| Oxidative stress | 41.2 ± 6.2 | 12.9 ± 1.7 | 170 ± 75 | 2583 ± 518 | 21 ± 20 | 571 ± 137 |
| Osmotic stress | 61.6 ± 4.0 | 72.3 ± 10.4 | n.d. | n.d. | n.d. | n.d. |
| Root damage | 23.0 ± 4.9 | 14.5 ± 2.7 | 119 ± 76 | 1920 ± 21 | 23 ± 21 | 529 ± 220 |
| Leaf damage | 39.1 ± 1.9 | 47.5 ± 11.5 | n.d. | n.d. | n.d. | n.d. |
| Cold treatment | 49.9 ± 11.6 | 21.8 ± 5.8 | 0.25 ± 0.25 | n.d. | n.d. | n.d. |
*Values represent average (± SD) of three biological and two technical replicates (n = 6); integration was achieved by summation of the NMR regions: δ5.725–5.685 [1] and δ5.635–5.585 [2]. Where HTG concentration was <100 μg/g, values stated are considered to be less accurate and are given as indications of the presence of a small peak. However, in all cases the presence of the HTG was confirmed by the presence of the relevant methyl group singlet in NMR and the 287/303 ions in positive ion ESI. n.d., not detected.
Fig. 4.
DI-ESI-MS analysis of KRR and NO3− ion adducts. (A) DI-ESI-MS (−) ion data showing effect of NO3− addition to pure KRR; (B) effect of increasing NO3− addition to an extract from an NO3−-depleted plant.
Analysis of the Biomarkers in Nitrate Reductase (NR) Mutants.
Mutants with deletions in NR genes can contain higher levels of free NO3− and thus served as a means to further validate the relationship between HTGs and NO3− starvation. We used the null mutant, nia2 (10% of wild-type NR) (18) and the double mutant nia1/nia2 (0.5% of wild-type NR) (19) and measured HTG [1] by NMR along with the KRR-NO3− adduct (m/z 640) in ESI-MS of leaves of plants grown on full media and then starved of NO3− for 4 d. The data (Table 2) showed that the single mutant nia2 produced less (69%) HTG than wild type when NO3− was withdrawn and retained more NO3− (higher m/z 640 intensity), whereas the double mutant nia1/nia2 did not produce HTGs at all and maintained high NO3− levels, despite the period of starvation.
Table 2.
Concentration of HTG [1] and intensity of nitrate status ion (m/z 640) in nitrate reductase mutants subjected to nitrate and starvation
| NO3− status | HTG [1], mg/g d.w. | m/z 640,intensity × 103 | |
| Col-0 | + | n.d. | 6.81 ± 0.7 |
| Col-0 | − | 1.54 ± 0.13 | 0.11 ± 0.16 |
| nia1/nia2 | − | n.d | 8.13 ± 0.67 |
| nia2 | − | 1.074 ± 0.055 | 1.57 ± 0.0103 |
n.d., not detected. Values represent mean (n = 3) ± SD.
Analysis of the HTG Biomarker Production in Other Stress Treatments.
The Araponics system and the ready detection of the HTG biomarkers allowed us to survey the effect of a range of stresses (SI Materials and Methods). The HTGs were quantified from the leaf NMR spectra. The data are shown in Table 1, along with measurements of [NO3−] in the same extracts. Wounding of roots, but not of leaves, induced the HTGs in leaves. Other root stresses such as NaCl treatment, osmotic stress (delivered through treatment with polyethylene glycol), and cold did not induce HTGs. K+ deprivation induced HTGs, but the magnitude was less than that of NO3− starvation or NH4+ substitution. However, oxidative stress induced by peroxide treatment of roots induced the HTGs in leaves, to levels similar to those from NO3− starvation. In general high HTG was always associated with low foliar NO3− (Table 1).
Feeding of MEP Pathway Intermediates.
Previously Page et al. (20) using gene silencing in Nicotiana benthamiana showed that deletion of HDR activity caused a diversion of HMBPP to the diol [3]. This was confirmed (20) by feeding labeled DXP to excised leaves. Using this experimental setup, we fed pathway intermediates (DXP, HMBPP, and diol [3]) to leaves of Arabidopsis, excised from plants that had been subjected to 4 d NO3− starvation (via both water only and NO3− deplete treatments). The metabolism of exogenous substrates was quantified by NMR after 8.5 and 24 h of incubation. DXP was metabolized efficiently (42% conversion after 24 h) to the diol and the HTGs, when plants had been starved of NO3−. (Fig. 5 and Table S2). No DXP or other products [e.g., deoxyxylulose (DX) or HMBPP] were detected. In NO3− sufficient leaves, DXP turnover to HTG and diol was observed at a much lower level (8% after 24 h). These results confirmed the biosynthesis of the diol and HTGs from HMBPP and a significant effect of NO3− starvation on flux into these compounds. Conversely, exogenous HMBPP was converted to diol [3] and HTGs in all plants regardless of nutrient status. Similarly, exogenous diol [3] was efficiently converted to the HTGs in all conditions (Fig. 5 and Table S2). These data indicate that the metabolism of exogenous HMBPP and diol is not subject to regulation, unlike HMBPP generated within the chloroplast from the DXP feed.
Fig. 5.
Metabolism of DXP, HMBPP, and diol [3] by leaves of nutrient-starved plants. Shown is turnover of substrates, quantified by NMR, after 8.5 and 24 h of feeding, to excised leaves, of control (N replete) and nutrient-deprived (4 d) (N deprived or water only) Arabidopsis. Stacked plots represent mean concentrations (n = 2). Concentrations of individual metabolites are depicted within the bars (solid, HTG major [1]; open, HTG minor [2]; shaded, diol [3]). Numerical data relating to the plots are given in Table S2. Error bars represent the SD of the combined HTGs.
Reprogramming of Root and Leaf Metabolism During a Variety of Root Stresses.
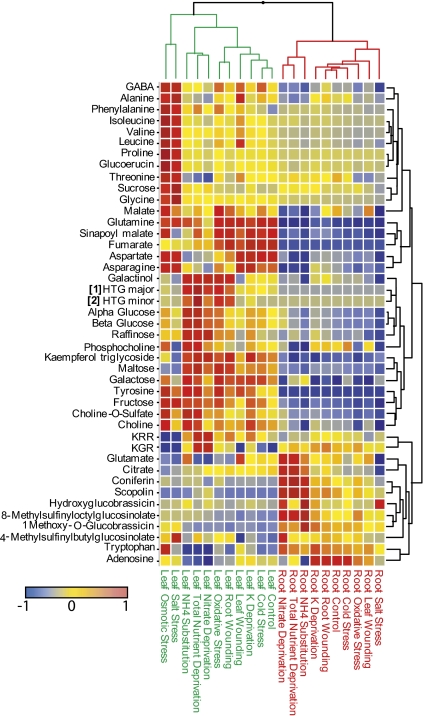
Data for key primary and secondary metabolites, including the HTGs were extracted from the metabolomic screen of roots and leaves under the various stress treatments and were compared by hierarchical cluster analysis as shown in Fig. 6. HTGs were not observed in roots. However, major changes in root metabolism as a consequence of NO3− deprivation were apparent, including an increase in coniferin and the induction of scopolin, a known coumarin in Arabidopsis roots, and also glucoraphanin. These compounds were identified by comparison with authentic samples isolated from Arabidopsis and characterized by NMR (Fig. S6). Levels of these metabolites in root tissue across all of the treatments, correlated with levels of HTG in leaf tissue (R2 > 0.6, P < 0.001). Associated metabolite changes in roots included small increases in sucrose and glucose and significant decreases in adenosine, malate, Ala, and Thr. Also evident from the metabolite map in Fig. 6 is the specificity of the changes induced by the different stresses. The accumulation of amino acids in leaves during root salt and osmotic stress is particularly striking as is the specificity of the accumulation of coniferin and scopolin in roots under NO3− deprivation.
Fig. 6.
Relative levels of key metabolites in root and leaf tissue of stressed Arabidopsis. Comparisons were generated via HCA using the complete linkage method with similarity based on Euclidian distance. Metabolite levels are based on characteristic chemical shift ranges from NMR or m/z intensities from DI-ESI-MS (−ve ion mode). For each metabolite, data have been normalized to unit variance and mean centered. 1H NMR shift ranges used are given in SI Materials and Methods.
Discussion
We have demonstrated how NMR-MS fingerprinting of hydroponically grown Arabidopsis plants can be used to discover and monitor metabolite biomarkers, as plants react to changing root environments. 1H NMR signals of the HTGs and their associated DI-ESI-MS ions were analyzed alongside a negative mode ion (m/z 640), reflecting NO3− status in the sample, to provide a “three-point check” allowing the tracking of HTG content in relation to NO3− status across roots and leaves. The production of HTGs in leaves was highly correlated with the production of scopolin and coniferin in roots of stressed plants, suggesting that a coordinated modulation of MEP and PP pathways in the two tissues was occurring. This phenomenon was observed for plants subjected to root oxidative stress, root wounding, or NO3− deprivation and to a lesser extent under K+ deficiency, but was not evident at all for other stresses, such as root saline or osmotic shock, which gave rise to a different palette of metabolite changes known to arise under these conditions (21). This result indicates that there are specific root–leaf signaling mechanisms that result in HTG and scopolin/coniferin coproduction. As all samples containing the HTG had lower foliar NO3−, the data suggests a role for NO3− sensing. Root wounding and oxidative stress both resulted in low leaf NO3− (Table 1), and it is assumed that root damage prevents the uptake of sufficient NO3− to leaves. The oxidative stress effect may be more complex than simple damage to the root NO3− assimilation pathway, but the observed outcome is low leaf NO3− (Table 1) and coproduction of the HTGs and PPs.
Studies using NH4+ as a replacement nitrogen source suggested that the induction of HTGs was related to NO3− rather than nitrogen per se and repeated deprivation and resupply indicated that Arabidopsis is able to switch back and forth from NO3− poor and NO3− rich states. Analysis of leaf tissue from the nia1,nia2 double mutant in which NR activity was reduced to 0.5% (19) also pointed to a direct NO3− relationship with HTG formation. When grown on a NO3− source, foliar NO3− accumulated to higher than wild-type levels and thus no HTG was produced under the 4-d starvation period. This result was in contrast to that for nia2 plants that retain 10–15% NR activity (18) and did not appear to retain enough NO3− before starvation to prevent HTG buildup on NO3− removal. These data also suggest that the observed NO3− effect is not attributable to an NR product such as nitrite.
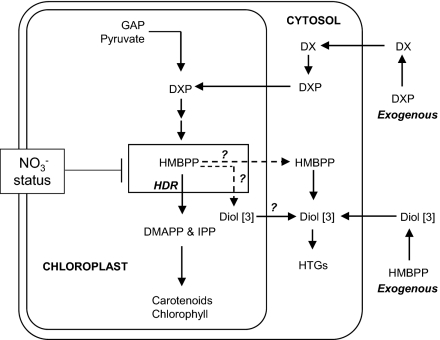
The discovery of the high levels of HTGs in Arabidopsis was unexpected, yet adds another dimension to our knowledge of isoprenoid metabolism in the model plant. The detection of the diol [3] and both isomers of the HTG suggested that the biosynthesis involves dephosphorylation of HMBPP followed by glycosylation at either end of the diol so formed. The likely biosynthetic route was investigated by feeding intermediates to detached leaves. Conversion of exogenous DXP to the HTGs was efficient in NO3− deplete plants (Fig. 5), inferring that the substrate entered the chloroplast where it was converted to HMBPP. Our DXP feeding results align with those of Page et al. (20), who observed conversion of labeled DXP to the diol [3] in intact leaves of N. benthamiana plants containing gene-silenced HDR. However, it is generally regarded that phosphorylated substrates such as DXP are not taken up by plant cells and that DX is a more suitable substrate for MEP pathway feeding studies (22). The incorporation of exogenous DXP into HMBPP in intact leaves (ref. 20 and this work) thus requires dephosphorylation of DXP to DX before uptake into the cell and then rephosphorylation to DXP as shown in our working model (Fig. 7). Such a phosphorylation of DX in the cytosol has been demonstrated in Arabidopsis (23), as has the existence of chloroplast import of DXP by a translocator (24).
Fig. 7.
Proposed routes to HTGs from endogenous and exogenous substrates.
Data from NO3− deprivation and NH4+ replacement studies, from repeated NO3− depletion and resupply experiments, and from the NO3−-accumulating nia1/nia2 double mutant all provide evidence for involvement of (low) NO3− sensing systems in the regulation of the MEP pathway in leaves and an associated diversion of lignin precursors to scopolin and coniferin in roots. It is known that plants need to balance C and N metabolism (6, 25) and transcriptomic studies of NO3−-limited Arabidopsis support this scenario (26). Involvement of light in the regulation of HDR gene expression is known (5) and the potential of a more complex regulation of this activity, which integrates photosynthesis with NO3− availability, can now be considered. This system would also be capable of regulating the production of coniferin and scopolin in Arabidopsis roots, which is NO3− sensitive (this work) and has previously been associated with light treatment (27). Indeed, scopolin production in tobacco roots was associated with low NO3− as early as 1970 (28).
The feeding study (Fig. 5 and Table S2) of DXP to intact leaves supports this model in that the pathway diverts HMBPP, generated in situ, to the HTGs via the diol, in NO3−-depleted leaves (42% conversion after 24 h), whereas in control leaves DXP turnover to HTGs was some five times lower (8% conversion after 24 h). This result is consistent with a repression of flux through HDR in NO3−-depleted plants. The observation of low (but not zero) conversion of DXP to HTGs in excised leaves of control plants can be attributed to an overflow due to an unnaturally large HMBPP pool, arising from the applied DXP.
In our working model (Fig. 7) HMBPP pool size determines the overflow and NO3− depletion restricts flow through HDR, causing HMBPP to accumulate and overflow to the shunt products. It is not clear whether dephosphorylation of excess HMBPP occurs in the chloroplasts and the resultant diol [3] moves to the cytosol where it is glycosylated or whether HMBPP can be exported from the chloroplasts and metabolized in the cytosol. Transporter function analysis (24) indicates that HMBPP, unlike IPP, is not transportable across the chloroplast membrane. Thus, dephosphorylation of HMBPP within the chloroplasts may be more likely. Further work, beyond the scope of this paper, is needed to identify a specific phosphatase activity in chloroplasts. Exogenous HMBPP, applied to leaves via cut petioles, produced similar levels of shunt products in both control and NO3−-starved plants. Because exogenous HMBPP is very unlikely to reach chloroplasts, an explanation (Fig. 7) of the feeding results of this compound (Fig. 5) involves nonspecific phosphatase action outside the cell and then glycosylation of the diol [3] in the cytosol. This “unnatural” metabolism of HMBPP was not influenced by NO3− status of the leaves (Fig. 5) and thus equally high levels of HTGs were produced in both control and NO3−-starved plants.
Although further evidence of involvement of NO3− in the regulation of HDR, at the gene or the enzyme level, will be needed, the data presented provide a compelling case for such sensing mechanisms playing a fundamental role in the reprogramming of metabolism that is necessary for plants to balance photosynthetic carbon flux with NO3− availability. Given the known role of light in regulation of the MEP pathway at HDR (5), an integrated NO3− and light signal transduction chain is a possibility that requires further investigation.
The biosynthetic shunt of HMBPP observed in Arabidopsis under stress has parallels with the formation of isoprene from dimethylallylalcohol diphosphate (DMAPP) under stress in trees (29). Although contrasting hypotheses concerning the role of isoprene as a carbon overflow or as a thermoprotectant have been the subject of much discussion (29, 30), some corollaries with HTG formation in Arabidopsis can be made. In Populus sp. a link between low NO3− and isoprene has been established (7) and other work indicates that DMAPP levels may govern the rate of emission (31, 32).
Conclusion
Our results suggest that an overflow mechanism is present in Arabidopsis that deals with undesired buildup of HMBPP via dephosphorylation and glycosylation. The overflow hypothesis put forward for isoprene emission from trees under stress (7, 30) can thus possibly be extended to other plants and other hemiterpenoids arising from HMBPP. This mechanism, in Arabidopsis, does not occur as a generalized stress response but instead appears to be induced by NO3− deprivation itself or scenarios that cause a lower NO3− status within the leaf, whether directly or indirectly. The high levels of HTGs and the coproduction of coniferin and scopolin under NO3− starvation provide ideal biomarkers to unravel the signaling pathways using the genetic resources available for Arabidopsis. Such efforts will benefit enormously from the methodology to rapidly quantify NO3−, HTGs, phenylpropanoids, and other key metabolites that we have described.
Materials and Methods
Plant Growth and Treatments.
Arabidopsis (Col-0) and mutant (nia2 and nia1/nia2) plants were grown using the Araponics system. For nutrient deprivation, resupply, and stress treatments, plants were moved to appropriate new media as detailed in SI Materials and Methods. Shoot and root tissues were harvested separately into liquid N2 and then lyophilized. Feeding studies were as described in Page et al. (20). Outer leaves excised from treated Arabidopsis were incubated with solutions of DXP, HMBPP and diol [3] introduced via the cut petioles.
Metabolomics.
Samples for NMR-MS screening were prepared and analyzed according to the protocols described in ref. 9. Full details are in SI Materials and Methods.
Synthesis of HTGs [1 and 2] and Diol [3].
Full details of the synthesis and associated structural characterization by NMR are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Ritsuo Nishida (Kyoto University) for NMR data of HTG [2]. We acknowledge one referee for helpful suggestions concerning the site of biosynthesis of the HTGs. Rothamsted receives grant-aided support from the Biotechnology and Biological Sciences Research Council, United Kingdom.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018875108/-/DCSupplemental.
References
- 1.D'Auria JC, Gershenzon J. The secondary metabolism of Arabidopsis thaliana: Growing like a weed. Curr Opin Plant Biol. 2005;8:308–316. doi: 10.1016/j.pbi.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Aubourg S, Lecharny A, Bohlmann J. Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Genet Genomics. 2002;267:730–745. doi: 10.1007/s00438-002-0709-y. [DOI] [PubMed] [Google Scholar]
- 3.Eisenreich W, Bacher A, Arigoni D, Rohdich F. Biosynthesis of isoprenoids via the non-mevalonate pathway. Cell Mol Life Sci. 2004;61:1401–1426. doi: 10.1007/s00018-004-3381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cordoba E, Salmi M, León P. Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J Exp Bot. 2009;60:2933–2943. doi: 10.1093/jxb/erp190. [DOI] [PubMed] [Google Scholar]
- 5.Botella-Pavía P, et al. Regulation of carotenoid biosynthesis in plants: Evidence for a key role of hydroxymethylbutenyl diphosphate reductase in controlling the supply of plastidial isoprenoid precursors. Plant J. 2004;40:188–199. doi: 10.1111/j.1365-313X.2004.02198.x. [DOI] [PubMed] [Google Scholar]
- 6.Tschoep H, et al. Adjustment of growth and central metabolism to a mild but sustained nitrogen-limitation in Arabidopsis. Plant Cell Environ. 2009;32:300–318. doi: 10.1111/j.1365-3040.2008.01921.x. [DOI] [PubMed] [Google Scholar]
- 7.Rosenstiel TN, Ebbets AL, Khatri WC, Fall R, Monson RK. Induction of poplar leaf nitrate reductase: A test of extrachloroplastic control of isoprene emission rate. Plant Biol (Stuttg) 2004;6:12–21. doi: 10.1055/s-2003-44722. [DOI] [PubMed] [Google Scholar]
- 8.Ward JL, Harris C, Lewis J, Beale MH. Assessment of 1H NMR spectroscopy and multivariate analysis as a technique for metabolite fingerprinting of Arabidopsis thaliana. Phytochemistry. 2003;62:949–957. doi: 10.1016/s0031-9422(02)00705-7. [DOI] [PubMed] [Google Scholar]
- 9.Ward JL, et al. The metabolic transition during disease following infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Plant J. 2010;63:443–457. doi: 10.1111/j.1365-313X.2010.04254.x. [DOI] [PubMed] [Google Scholar]
- 10.Boyes DC, et al. Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. Technical advance: Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 2000;23:131–142. doi: 10.1046/j.1365-313x.2000.00774.x. [DOI] [PubMed] [Google Scholar]
- 12.Halket JM. Derivatives for gas chromatography-mass spectrometry. In: Blau K, Halket J, editors. Handbook of Derivatives for Chromatography. 2nd Ed. Chichester, UK: Wiley; 1993. pp. 297–325. [Google Scholar]
- 13.Ward JL, Beale MH. Synthesis of (2E)-4-hydroxy-3-methylbut-2-enyl diphosphate, a key intermediate in the biosynthesis of isoprenoids. J Chem Soc Perkin Trans 1. 2002;6:710–712. [Google Scholar]
- 14.Nicoletti M, Tomassini L, Foddai S. A new hemiterpenoid glucoside from Ornithogalum montanum. Planta Med. 1992;58:472. doi: 10.1055/s-2006-961519. [DOI] [PubMed] [Google Scholar]
- 15.Kitajima J, Suzuki N, Ishikawa T, Tanaka Y. New hemiterpenoid pentol and monoterpenoid glycoside of Torillis japonica fruit, and consideration of the origin of apiose. Chem Pharm Bull (Tokyo) 1998;46:1583–1586. [Google Scholar]
- 16.Ohta N, Mori N, Kuwahara Y, Nishida R. A hemiterpene glucoside as a probing deterrent of the bean aphid, Megoura crassicauda, from a non-host vetch, Vicia hirsuta. Phytochemistry. 2006;67:584–588. doi: 10.1016/j.phytochem.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 17.Messerer M, Winterhalter P. (2Z)-4-hydroxy-2-methyl-2-buten-1-yl-β-D-glucoside and (2Z)-1-hydroxy-2-methyl-2-buten-4-yl-β-D-glucoside: Two new hemiterpene glucosides from Vitis vinifera leaves. Nat Prod Lett. 1995;5:241–244. [Google Scholar]
- 18.Wilkinson JQ, Crawford NM. Identification of the Arabidopsis CHL3 gene as the nitrate reductase structural gene NIA2. Plant Cell. 1991;3:461–471. doi: 10.1105/tpc.3.5.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkinson JQ, Crawford NM. Identification and characterization of a chlorate-resistant mutant of Arabidopsis thaliana with mutations in both nitrate reductase structural genes NIA1 and NIA2. Mol Gen Genet. 1993;239:289–297. doi: 10.1007/BF00281630. [DOI] [PubMed] [Google Scholar]
- 20.Page JE, et al. Functional analysis of the final steps of the 1-deoxy-D-xylulose 5-phosphate (DXP) pathway to isoprenoids in plants using virus-induced gene silencing. Plant Physiol. 2004;134:1401–1413. doi: 10.1104/pp.103.038133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez DH, Siahpoosh MR, Roessner U, Udvardi M, Kopka J. Plant metabolomics reveals conserved and divergent metabolic responses to salinity. Physiol Plant. 2008;132:209–219. doi: 10.1111/j.1399-3054.2007.00993.x. [DOI] [PubMed] [Google Scholar]
- 22.Wolfertz M, Sharkey TD, Boland W, Kühnemann F. Rapid regulation of the methylerythritol 4-phosphate pathway during isoprene synthesis. Plant Physiol. 2004;135:1939–1945. doi: 10.1104/pp.104.043737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hemmerlin A, et al. A cytosolic Arabidopsis D-xylulose kinase catalyzes the phosphorylation of 1-deoxy-D-xylulose into a precursor of the plastidial isoprenoid pathway. Plant Physiol. 2006;142:441–457. doi: 10.1104/pp.106.086652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flügge UI, Gao W. Transport of isoprenoid intermediates across chloroplast envelope membranes. Plant Biol (Stuttg) 2005;7:91–97. doi: 10.1055/s-2004-830446. [DOI] [PubMed] [Google Scholar]
- 25.Stitt M. Nitrate regulation of metabolism and growth. Curr Opin Plant Biol. 1999;2:178–186. doi: 10.1016/S1369-5266(99)80033-8. [DOI] [PubMed] [Google Scholar]
- 26.Scheible WR, et al. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004;136:2483–2499. doi: 10.1104/pp.104.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hemm MR, Rider SD, Ogas J, Murry DJ, Chapple C. Light induces phenylpropanoid metabolism in Arabidopsis roots. Plant J. 2004;38:765–778. doi: 10.1111/j.1365-313X.2004.02089.x. [DOI] [PubMed] [Google Scholar]
- 28.Armstrong GM, Rohrbaug LM, Rice EL, Wender SH. Effect of nitrogen deficiency on concentration of caffeoylquinic acids and scopolin in tobacco. Phytochemistry. 1970;9:945–948. [Google Scholar]
- 29.Sharkey TD, Wiberley AE, Donohue AR. Isoprene emission from plants: Why and how. Ann Bot (Lond) 2008;101:5–18. doi: 10.1093/aob/mcm240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanadze GA. Biogenic isoprene (a review) Russ J Plant Physiol. 2004;51:729–741. [Google Scholar]
- 31.Rosenstiel TN, Fisher AJ, Fall R, Monson RK. Differential accumulation of dimethylallyl diphosphate in leaves and needles of isoprene- and methylbutenol-emitting and nonemitting species. Plant Physiol. 2002;129:1276–1284. doi: 10.1104/pp.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vickers CE, Possell M, Nicholas Hewitt C, Mullineaux PM. Genetic structure and regulation of isoprene synthase in Poplar (Populus spp.) Plant Mol Biol. 2010;73:547–558. doi: 10.1007/s11103-010-9642-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.