Abstract
Characterized bacteria, unlike eukaryotes and some archaea, initiate replication bidirectionally from a single replication origin contained within a circular or linear chromosome. We constructed Escherichia coli cells with two WT origins separated by 1 Mb in their 4.64-Mb chromosome. Productive bidirectional replication initiated synchronously at both spatially separate origins. Newly replicated DNA from both origins was segregated sequentially as replication progressed, with two temporally and spatially separate replication termination events. Replication initiation occurred at a cell volume identical to that of cells with a single WT origin, showing that initiation control is independent of cellular and chromosomal oriC concentration. Cells containing just the ectopic origin initiated bidirectional replication at the expected cell mass and at the normal cellular location of that region. In all strains, spatial separation of sister loci adjacent to active origins occurred shortly after their replication, independently of whether replication initiated at the normal origin, the ectopic origin, or both origins.
Like most bacteria, Escherichia coli harbors a single circular chromosome within which replication is initiated at a single origin, oriC, and progresses bidirectionally toward the diametrically opposite replication terminus region (ter). The E. coli replication machinery assembles at the oriC, close to midcell at replication initiation, and in minimal medium, the two replisomes track independently around the chromosome (1). The spatial separation of many newly replicated sister genetic loci to opposite cell halves loci occurs sequentially 5 to 20 min after replication (1–6). Similarly, sequential replication-segregation has also been described in Caulobacter crescentus (7, 8) and Vibrio cholerae (9). Interlinking of newly replicated sisters (i.e., precatenation) may be responsible for the 5- to 20-min delay between replication and separation, because overexpression of topoisomerase IV, which plays a key role in decatenation, led to an approximate threefold decrease in the sister colocalization period of a locus 15 kb to the left of oriC (10). Consistent with this, inhibition of topoisomerase IV led to wholesale defects in sister chromosome segregation (10). Nevertheless, genetic loci in at least two discrete 150 kb regions, approximately 130 kb and 380 kb to the right of oriC, exhibited a further approximately 18 min delayed separation compared with neighboring loci (2–4). The mechanism that leads to this delayed separation of some sister loci on the right chromosome arm remains unclear and may not be linked to decatenation. Nevertheless, it was associated with an abrupt global transition in nucleoid morphology that may play a key role in sister nucleoid separation (2, 3).
In each of the three domains of life, replication is tightly regulated, so that no origin normally fires more than once per cell cycle. Because cell generation time can be modulated, overall rates of DNA synthesis within an organism or cell type must also be regulated. In Drosophila embryogenesis, as in other eukaryotes, a decreased S phase is usually accommodated by firing from an increased number of active origins, with the DNA synthesis rate of any pair of sister replication forks remaining constant (11). Similarly, in E. coli, overall DNA synthesis can be increased to reduce the generation time by increasing the number of active replication forks in the cell. For example, sister replisomes derived from a single initiation event take at least 40 min to replicate the whole chromosome (i.e., C-period), and cell division then follows approximately 20 min (i.e., D-period) after the completion of replication. When the generation time is shorter than the sum of C- and D-periods, initiation occurs synchronously at sister origins within a chromosome that is already undergoing replication (12–15).
In E. coli, the ATP-bound form of the abundant DnaA initiator protein controls replication initiation at oriC. A range of regulatory mechanisms ensures precise and controlled initiation timing by modulating DnaA binding and action on DNA (reviewed in refs. 16–18). Early data led to a model in which initiation occurs at a constant mass per chromosome origin (19), although the demonstration that cells containing oriC plasmids initiated DNA synthesis synchronously at the same mass as their plasmid-free parent, along with other data, showed that this model could not be strictly true (20, 21; reviewed in refs. 22, 23). Nevertheless, the mechanistic relationship, if any, between initiation, cell growth and mass remains unclear.
The processes that contribute to E. coli chromosome organization and segregation are poorly understood. A range of proteins and mechanisms that could facilitate chromosome segregation has been proposed, although no consensus mechanistic view has emerged. It has been proposed also that E. coli chromosomes could segregate spontaneously by using a self-avoidance mechanism driven by entropy (24, 25). In contrast, in Bacillus subtilis and C. crescentus, chromosome tethering mechanisms attach specific chromosome regions to cell poles (26–28), whereas dedicated partition systems facilitate chromosome segregation (reviewed in ref. 29). In E. coli, low copy number plasmids use similar partition systems (29), but none that contribute to E. coli chromosome segregation have been characterized. Independent tracking of sister replisomes around the chromosome could contribute directly to segregation (1).
To gain insights into the processes that govern replication and chromosome segregation, we analyzed E. coli cells with two identical functional replication origins separated by approximately 1 Mb (oriC-oriZ) and cells containing just the ectopic origin (oriZ), and compared them with WT cells (oriC). In oriC-oriZ cells, replication initiation occurred synchronously at both origins, which are located at the normal separate cellular locations of the loci associated with the origins. Genetic loci adjacent to oriC and to the insertion site of the ectopic oriC segregated sequentially as replication progressed, with two temporally and spatially separate replication termination events. Synchronous replication initiation at both origins occurred at a cell mass identical to that of WT cells with a single oriC, showing that the concentration of oriC within a chromosome does not influence the timing of replication initiation. The initial segregation pattern of the pairs of sister origins, in which sisters of a given locus lay side-by-side, is reminiscent of sister origin segregation in fast growing cells, when sisters only segregate to opposite cell halves and then to daughter cells, one to two generations after the initial segregation. Such behavior, which is incompatible with faithful chromosome segregation in the two-origin strain, was switched to a permissive pattern at the time of completion of replication of the smaller chromosome segment between oriC and oriZ. Cells with two origins or just the single ectopic oriC grew with relatively normal growth rates and cell cycle parameters, thereby demonstrating the robustness and adaptability of E. coli chromosome processing.
Results
Growth, Size, and Cell Cycle Parameters of E. coli with Two Distant Replication Origins and a Single Ectopic Origin.
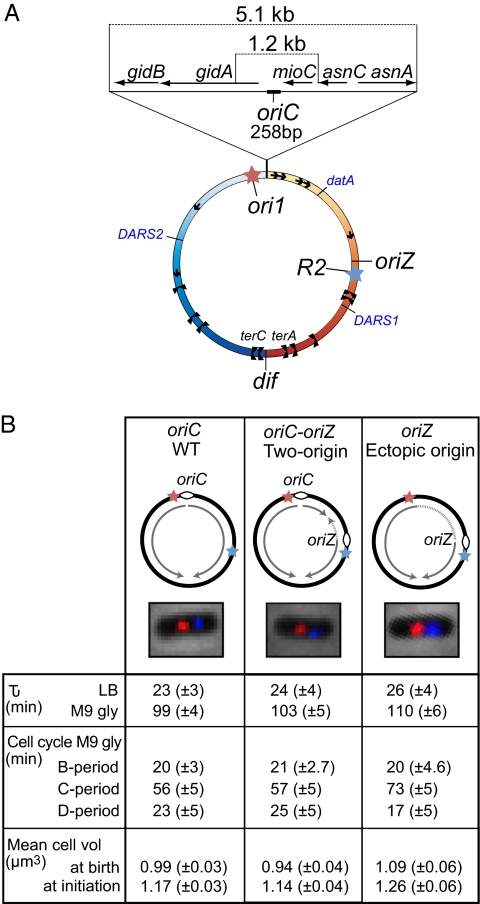
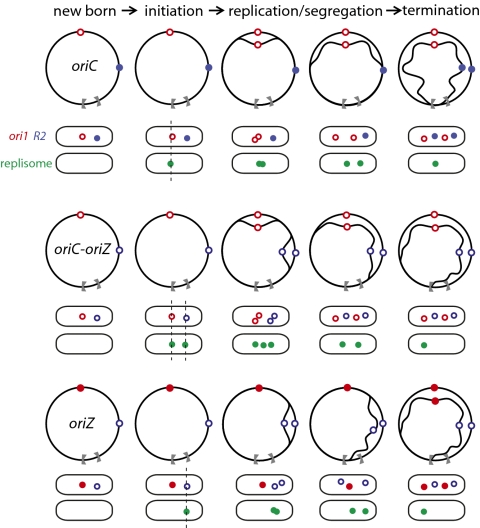
A copy of the 5.1-kb region containing oriC was inserted approximately 1 Mb away into the intergenic region at 344 kb on the E. coli genetic map, 21 kb upstream of lacZ (Fig. 1A). We name the ectopic origin locus oriZ. The oriC-oriZ strain, containing two copies of oriC, was further manipulated to have the 5.1-kb oriC region deleted from its original locus so that the only replication origin is at oriZ (Fig. 1B). We then characterized the cell cycle, replication, and segregation features of the oriC-oriZ strain with those of the oriZ strain and the oriC AB1157 parent. The cellular localization of oriC and oriZ was followed by using fluorescently labeled repressors bound to lacO and tetO operator arrays at ori1, 15 kb from oriC on the left replication arm, and at R2, 21 kb downstream of oriZ (6). Transposition of a smaller 1.2-kb oriC region (Fig. 1A) to the same ectopic position gave strains with indistinguishable properties to those described herein.
Fig. 1.
(A) Genetic map of the WT E. coli chromosome, with the left replichore in blue and the right replichore in orange. A zoom of the origin region shows the minimal initiation site oriC (258 bp) and its genetic context alongside the 5.1-kb and 1.2-kb DNA region, the segments that were relocated and/or deleted. oriZ (at 344 kb on the E. coli genetic map) is the site where the ectopic origin was inserted. The red and blue stars respectively indicate the insertion position of ori1 (lacO, at 3,908 kb) and R2 (tetO, at 366 kb) operator arrays used for chromosome localization. Replication termination sites (black dumbbell shapes), rRNA operons (black arrows), DnaA-binding DARS1 and DARS2 regions (812 kb and 2,967 kb, respectively), and the DnaA-binding cluster datA (3,494 kb) are shown, as is the dif site at which XerCD-FtsK recombination occurs. (B) Chromosome structure and representative newborn cells, with fluorescently marked ori1 (red) and R2 (blue) loci, of all three strains analyzed in detail. The replication arms inferred from the position of origins and terA and terC sites are represented by light gray arrows. Dashed lines indicate where replication and transcription are head-on. Lower: Doubling times (min; τ) in rich and minimal media, cell cycle parameters from flow cytometry (Fig. S1), and microscopy characterization (Figs. 3 and 4 and Fig. S2). B-period (min) is the time from birth to initiation of DNA replication, as measured by appearance of the replisome marker Ypet-DnaN. C-period (min) is the time of DNA synthesis assessed by replisome appearance to disappearance and by flow cytometry (C- plus D-period). D-period (min), the time from termination of DNA synthesis to division, was assessed by flow cytometry (C- plus D-period) and arithmetically as τ equal to the sum of B-, C-, and D-periods under our growth conditions. Cell volumes (in μm3) were measured by flow cytometry and direct microscopic measurement. During time-lapse analysis, cell doubling time on the agarose pads increased by 13% (oriC and oriC-oriZ strains) and 14% (oriZ); with a proportional increase in B-, C-, and D-periods, compared with growth in liquid medium. The values given have been compensated for this and reflect liquid growth values. The viabilities, determined as colony forming units per A600 were 1.95 × 109 (oriC) and 1.85 × 109 (oriC-oriZ and oriZ).
The doubling times and viabilities of all three strains were similar in minimal medium and in rich medium (Fig. 1B). Thus, the presence of two origins, or a single ectopic origin, in a single chromosome, had no substantial effect on generation time. We observed no loss of viability or abnormal cell morphology for any of the three strains, indicating that E. coli tolerates well the introduction of an additional ectopic origin and the subsequent deletion of the WT origin. We constructed oriC-oriZ and oriZ strains many times independently and found no evidence that any of the phenotypes we observed resulted from the accumulation of suppressor mutations.
We further characterized the three strains by examining cell size and cell cycle features. Flow cytometry profiles allowed estimation of the sum of C- plus D-period for each of the strains; and the time of initiation of DNA replication (Fig. 1B and Fig. S1). We used time-lapse microscopy to independently determine the time of replication initiation and the C-period, as assessed by the time of replisome (Ypet-DnaN) appearance and disappearance (Figs. 2 and 3 and Fig. S2 A and B) (1, 30). The values obtained by microscopy and flow cytometry were in broad agreement and additionally showed that the cell volume distributions for all three strains were very similar, with replication initiation occurring at similar cell volumes, and at the same time within the cell cycle. The flow cytometry analysis also showed that during growth in minimal glycerol medium, all three strains initiated and completed replication in the same generation.
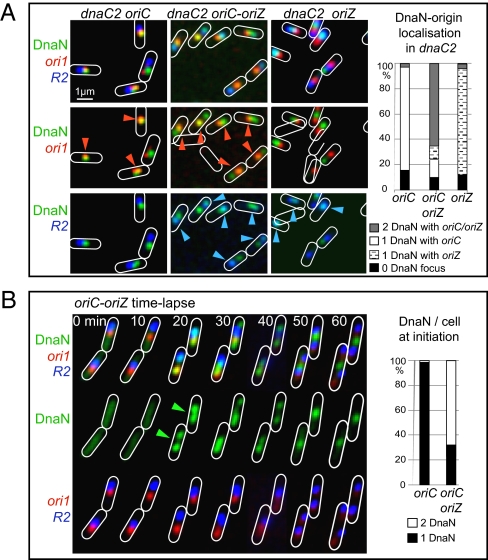
Fig. 2.
Synchronous initiation at both origins in oriC-oriZ cells. (A) Snapshot analysis of replisome (Ypet-DnaN) colocalization with genetic loci ori1 and R2, 10 min after initiation in temperature-sensitive dnaC2 derivatives of oriC, oriC-oriZ, and oriZ strains. Orange arrowheads indicate colocalization of replisome (green) and ori1 (red), and blue arrowheads indicate colocalization of replisome (green) and R2 (blue). Cell contours (white) have been added by using the phase-contrast images. The histograms show the proportion of different cell types in the three strains. Cells (n = 800–1,000) were analyzed for each strain. (B) Time-lapse analysis of replisome appearance in steady-state cells. A representative time-lapse image is shown for oriC-oriZ cells, in which replisome, ori1, and R2 are monitored simultaneously. Green arrowheads indicate replisome foci at initiation. The histograms show the number of replisome foci per cell within the time frame of Ypet-DnaN appearance, as observed by time-lapse microscopy. ori1 (lacO) was visualized by using LacI-mCherry (red); R2 (tetO) was visualized by using TetR-CFP (blue).
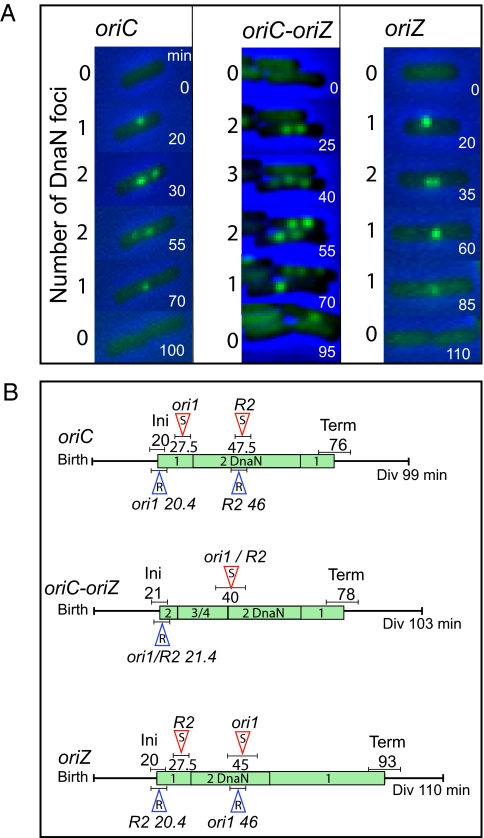
Fig. 3.
(A) Indicative time-lapse images of replisome (Ypet-DnaN) dynamics for oriC, oriC-oriZ, and oriZ cells growing on minimal medium with glycerol. Times (min), as well as the number of Ypet-DnaN foci in the corresponding frame, are indicated. (B) Cell cycle features of oriC, oriC-oriZ, and oriZ cells. Generation times in liquid were used to define the average time from birth to division. The mean replication C-period, as judged by the appearance and disappearance of Ypet-DnaN foci in time-lapse microscopy period in oriC (23 cells), oriC-oriZ (27 cells), and oriZ (24 cells), is labeled with green boxes, indicated with the number of resolvable foci (Fig. S2B). The times of replication initiation (Ini) and termination (Term) are indicated, with uncertainties indicated underneath by horizontal bars. The timings of ori1 and R2 separation, observed in time-lapse experiments (Fig. S2A), are labeled with empty red arrowheads (S, separation) and placed on top of the timeline with their uncertainties. The timings of ori1 and R2 replication were calculated on the basis of their genetic position and a mean replication rate for a 56-min C-period and is shown in empty blue arrowheads (R, replication), underneath the timeline with the uncertainties. In oriC-oriZ cells, 77% of cells with three replisomes had spatially separated ori1 and/or R2 sister foci (Fig. 4A). Times are in min.
Taken together, these observations show that the regulation of E. coli replication and of the cell cycle is maintained regardless of the presence of two identical functional origins or replication from an ectopic origin. This contrasts with the situation in B. subtilis, in which relocation of oriC to sites more than 250 kb distant from the normal position led to asynchronous initiation and perturbed initiation timing (31, 32). Importantly, we have demonstrated the cellular and chromosomal concentration of oriC is not a determinant in control of replication initiation, as originally proposed (19), as cell mass at initiation of the strain with two origins is identical to that of cells with one origin.
Synchronous Initiation of Replication from Both Origins.
To test whether the oriC-oriZ strain initiates replication from both origins and whether each of these function in every cell generation, we used fluorescence microscopy to analyze replisome assembly and localization at each of the origins. First, we tested whether oriZ fires synchronously with oriC at initiation in a dnaC2 temperature-sensitive strain, which allows synchronous replication initiation in a large population of cells (33). Exponentially growing cells at 30 °C were shifted to 37 °C at an optical density A600 of approximately 0.1 for 2 h to allow completion of ongoing rounds of replication, after which they were shifted back to 30 °C to allow replication initiation. Intracellular positions of ori1, R2, and replisomes were then analyzed 10 min after temperature downshift, when replication reinitiates (Fig. 2A). A single replisome focus colocalized with ori1 in oriC cells and with R2 in oriZ cells (82% and 86% of cells, respectively); the remaining cells were largely without a replisome focus (15% and 10% for the two strains, respectively), indicating that the replication has not yet initiated in those cells. The fewer than 3% of cells that had two replisome foci in these two strains had presumably undergone spatial separation of sister replisomes within the 10-min time frame (1).
In contrast, in the oriC-oriZ two-origin strain, two replisome foci appeared and colocalized with each origin in 67% of cells within 10 min of initiation (Fig. 2A). Furthermore, the replisomes appeared at the normal WT intracellular positions of ori1 and R2, confirming that independently acting replisomes assemble on origins irrespective of their cellular location, rather than being part of a replication factory that recruits origins to it (1, 30).
We were concerned that an accumulation of DnaA-ATP initiator protein in dnaC2 cells during incubation at the restrictive temperature could have led to initiation at both origins in the oriC-oriZ strain, whereas steady-state cells might not accumulate sufficient DnaA-ATP to allow productive initiation at both origins. Therefore, we analyzed replisome assembly at the two origins in exponentially growing cells by using time-lapse (Fig. 2B) and snapshot analysis (Fig. S2B). Time-lapse experiments demonstrated that replisome foci appeared synchronously at each of the two spatially separate origins within a 5-min time interval in 68% of initiation events in oriC-oriZ cells (Fig. 2B, green arrows); replisome appearance inevitably led to active replication and subsequent segregation of the newly replicated loci adjacent to each of the origins. In contrast, synchronous appearance of two replisome foci in oriC or oriZ cells was never observed.
Snapshot analysis of replisome foci confirmed these conclusions (Fig. S2B). Younger cells, between 2 and 2.5 μm in length, in which initiation happens, had an overrepresentation of cells with two or more replisome foci in the oriC-oriZ strain (51%), compared with WT cells (28%) or cells containing just oriZ (13%). A concomitant reduction in the proportion of single replisome focus cells in the oriC-oriZ strain was also evident. These data confirm that firing of both oriC and oriZ occurs within individual steady state oriC-oriZ cells.
To show that initiation at both origins in oriC-oriZ cells led to productive replication, newly replicated DNA was labeled with 5-ethynyl-2′-deoxyuridine (EdU) after synchronous initiation in dnaC2 cells (SI Materials and Methods and Fig. S2C). Visualization of the label showed that the majority of oriC-oriZ cells had two spatially separate fluorescent foci (54% after 8 min and 64% after 15 min of replication), whereas most oriC cells had a single focus (87% and 81%, respectively, at the two time points), confirming that replication in two-origin cells occurs at both spatially separate origins.
Replication Fork Progression in oriC-oriZ and oriZ Cells.
Because the presence of a second replication origin in oriC-oriZ cells, or changing the origin position in oriZ cells was likely to modify chromosome replication and subsequent segregation, we then studied the patterns of replication fork progression in these strains. Synchronous initiation at oriC and oriZ in oriC-oriZ cells would therefore be expected to lead to cells with four replisomes at four active forks until replication fork meeting and termination within the approximately 1 Mb segment, approximately 13 min after initiation, if replication of both small replichores occurs unimpeded (Fig. 1A). Consistent with this, time-lapse analysis showed that cells with at least three replisomes were only present for as long as 20 min after initiation (Fig. 3 A and B). Cells with four replisomes were rare (<2%) in both the time-lapse and snapshot analyses (described later), an observation that is not surprising, as it takes at least 5 min for independent replisomes to become spatially separate (1). We conclude that cells with at least three replisomes are undergoing replication of both chromosome segments that lie between oriC and oriZ and that replication of the smaller, approximately 1 Mb segment occurs without significant delay. This indicates that replication at both forks in the smaller segment contributes to duplication of the segment, and therefore that the counterclockwise (CCW) fork is not substantially impeded by “head-on” rRNA transcription (Fig. 1A). Indeed, if the CCW fork from oriZ was regularly stalling at the head-on rRNA cistron, we might have observed a significant number of four-replisome cells. The observation that the C-period of oriC-oriZ cells was almost identical to that of oriC cells is not surprising, given that a single fork replicates approximately 2.3 Mb in both strains. The time-lapse conclusions are supported by snapshot analysis, which revealed that 14% of 2,472 cells examined had at least three replisome foci (Fig. 4A).
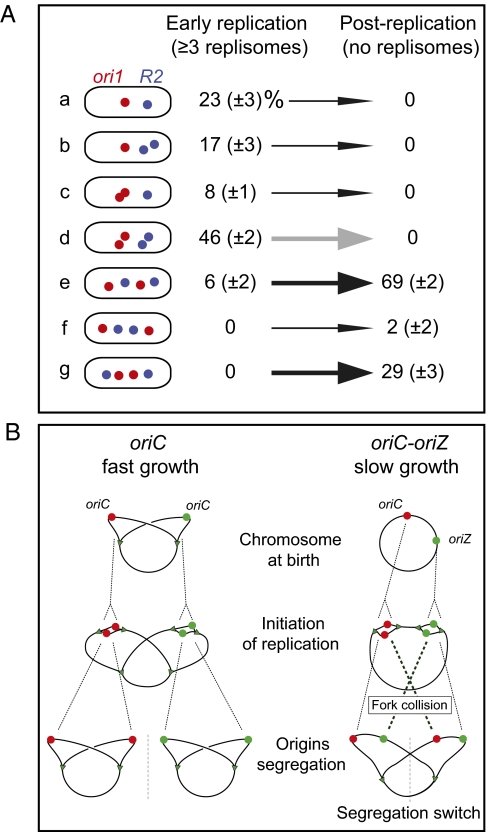
Fig. 4.
A switch in loci segregation pattern in oriC-oriZ cells after replication of the smaller 1-Mb segment between oriC and oriZ. (A) Patterns of ori1 and R2 segregation in snapshots of the 14% (n = 2,472) oriC-oriZ cells containing three or four replisomes, and therefore have the 1-Mb fragment between oriC and oriZ still replicating (Left). Only 6% of cells have <ori1 R2 ori1 R2> or <R2 ori1 ori1 R2> genetic locus patterns, whereas 71% have sister loci adjacent and 23% are noninformative because sisters of neither locus have spatially separated. Later in the cell cycle (Right), when chromosome replication is complete as judged by the disappearance of the replisome foci (13% of 1,894 cells), 98% of cells had the <ori1 R2 ori1 R2> (69%) or <R2 ori1 ori1 R2> (29%) pattern. Data were analyzed from three independent experiments; SD shown in brackets. The same trends were observed in time-lapse analysis (Fig. S2A). The thick arrows show the switch in patterns (black, increase; gray, decrease). (B) Comparison of segregation patterns of fast-growing WT oriC cells (born with two sister oriC copies) versus slow-growing oriC-oriZ cells. After initiation, sister origins in the fast-growing cells segregate to the same daughter cell after cell division, giving rise to the pattern. In oriC-oriZ cells, sister origins initially adopt the same initial pattern, but switch to after replication of the 1-Mb fragment between oriC and oriZ is complete. Sister origins are shown in the same color (green or red), replication forks are shown in green triangles, and gray dashed line indicates division sites.
In oriZ cells, we observed an almost twofold reduction in the proportion of two-replisome cells compared with oriC cells (snapshots, Fig. S2B). Furthermore, two-replisome cells were absent for the last 31 min of the C-period of the majority of oriZ cells, compared with the last approximately 10 min for oriC cells (time-lapse, Fig. 3). Analysis of replisome number and position with respect to R2 in oriZ cells showed no evidence for replication initiating at sites other than oriZ in oriZ cells. These observations indicate strongly that replication termination does not occur diametrically opposite to oriZ, with equal-sized CCW and clockwise (CW) replichores. Rather, it is consistent with the view that the CW fork reaches Tus-bound terC or a subsequent Tus-bound ter site (Fig. 1B), and eventually disassembles there. The CCW fork then would complete replication when it encounters the stalled fork at ter. We would therefore have anticipated that the C-period would be increased at least 50% from the approximately 56 min of oriC cells to more than 84 min, in the case of termination at terC, as a single fork would now need to replicate approximately 3.3 Mb of DNA. The more modest 17-min increase (30%) in C-period we observed could be explained by a compensating increased fork replication rate as a consequence of the CCW fork having available twice the amount of the normally limiting dNTPs for synthesis during much of the C-period (e.g., refs. 34, 35). Alternatively, CW forks could pass through one or two of the ter sites following terC, leading to marginally more equal replichores, as proposed for strains carrying inversions that unbalance replichore size (36). Other inversions encompassing oriC, which generated a replichore unbalance comparable to that proposed for oriZ cells, displayed viability defects (37), unlike the situation in oriZ cells. Nevertheless, these inversions additionally disrupted macrodomain organization, the integrity of which is important for normal cell viability (37, 38).
Patterns of Genetic Locus Segregation Are Determined by Replication.
We then examined how the positioning and segregation of ori1 and R2, adjacent to oriC and the oriZ insertion site, respectively, behaved with respect to replication fork progression in the three strains. Snapshot analysis was used to assess the behavior of large populations, and time-lapse analysis followed the behavior of the replisome and chromosome loci through time. The time-lapse analysis was facilitated by the use of an automated custom particle-tracking algorithm (Materials and Methods).
We observed that the time between the separation of sister ori1 loci and sister R2 loci was strictly dependent on the replication program of the chromosome. In oriC cells, replication of ori1 is expected to occur approximately 0.4 min after replication initiation, and we observed sister ori1 locus separation approximately 7.5 min after initiation, consistent with our earlier estimate of the sister origin colocalization period (1, 10). The R2 locus, 1,073 kb away from oriC, was expected to replicate approximately 26 min after replication initiation. Consistent with this, the time-lapse analysis showed that sister R2 loci separated, on average, approximately 27.5 min after initiation (Fig. 3B and Fig. S2A). In oriC-oriZ cells, synchronous replication initiation at both origins was followed by spatial separation of newly replicated sister ori1 and R2 loci within the same time-lapse frame (0–5-min difference), or with a difference of one time-lapse frame (5–10 min), showing that replisome appearance at each origin is followed by functional replication and subsequent sister separation a few minutes later (Figs. 3B and 4A and Fig. S2).
Similarly, we observed in oriZ cells that newly replicated sister R2 loci separate approximately 17.5 min earlier than ori1 sisters (Fig. 3B and Fig. S2 A and E); replication initiating at oriZ is expected to reach ori1 approximately 26 min later. Therefore, it appears that the progress of the CCW fork in oriZ cells is not substantially impeded by head-on transcription from the five ribosomal cistrons that this fork will inevitably encounter (Fig. 1A). We also note that in both oriC and oriZ cells, the sister loci proximal to the origin show a delay in their spatial separation compared with the distal locus.
Snapshot analysis supported the aforementioned observations: the average number of ori1 and R2 foci per cell were, respectively, 1.5 and 1.3 (oriC); 1.3 and 1.3 (oriC-oriZ); and 1.5 and 1.7 (oriZ; Fig. S2E). Therefore, the relative times of spatial separation of sister ori1 and R2 loci followed the order of their replication, with the first region to be replicated being the first to be segregated, as was shown previously for the same loci in WT cells (4, 6). Nevertheless, by displacing and/or duplicating origins, we are additionally able to conclude that the time of segregation of these sister chromosome loci is determined by the cell's replication program, rather than by intrinsic properties of chromosomal regions, or by a replication-independent segregation machinery.
Next, we examined ori1 and R2 locus positioning within cells throughout the cell cycle. Newborn cells of all three strains, containing nonreplicating chromosomes, had identical locus positioning, with ori1 close to midcell and R2 toward the quarter position (Fig. 1B and Fig. S2E). Therefore, changing origin position and number does not influence the cellular position of the genetic loci tested in nonreplicating chromosomes. In all three strains undergoing steady-state growth, ori1 was replicated close to midcell and ori1 sisters segregated toward the quarter positions, where they stayed until cell division (Fig. S2 A and E). Therefore, cellular ori1 positioning is not a consequence of ori1 being adjacent to an active origin.
Cellular positioning of single R2 loci in oriC-oriZ and oriZ cells was conserved when compared oriC cells, irrespective of whether the foci represented unreplicated or replicated loci. Nevertheless, the relative position of given R2 sisters in individual oriC-oriZ and oriZ cells, and their relationship to ori1 sisters, showed some differences, compared with oriC cells. In approximately 80% of sister oriC cells, one R2 sister positions closer to an old pole and one closer to a new pole, thereby generating the observed translational symmetry (<R2 ori1 R2 ori1>; Fig. 4) (6, 39). In both oriC-oriZ and oriZ cells, there was an increased tendency for the sister R2 loci to locate proximal to the old pole, thereby generating an increased proportion of <R2 ori1 ori1 R2> cells in snapshots (8% for oriC, 29% for oriC-oriZ, and 34% for oriZ; Fig. S2D). The increased migration apart of sister R2 loci when they are adjacent to an active origin, which was slightly more pronounced in oriZ cells compared with oriC-oriZ cells, was directly observed in time-lapse analysis (Fig. S2 A and D). The reason for this increased tendency of R2 sisters to move to the old poles, when an adjacent oriZ origin is active is unclear, although it is reminiscent of sister oriC bipolar migration to the outer poles when a functional Structural Maintenance of Chromosomes complex, MukBEF, is absent (40).
Switch in Locus Segregation in oriC-oriZ Cells.
When we examined ori1 and R2 positioning in oriC-oriZ cells harboring three or four replisomes, most of which are expected to have four replisomes active, we found evidence for a dramatic switch in segregation pattern as replication of the smaller, approximately 1 Mb segment is completed. In more than 90% of cells with three or more replisomes in which sisters of at least one genetic locus had separated (Fig. 4A, lines b–g), sister ori1 and/or R2 loci have not separated into opposite cell halves (Fig. 4A, lines b–d), and are therefore not positioned to allow their segregation into prospective daughter cells. In contrast, by the time replication is complete as judged by the disappearance of replisome foci (13% of 1,894 cells examined), all cells have adopted a configuration in which sisters now occupy separate cell halves and are thereby permissive for productive segregation (Fig. 4A, lines e–g). In cells with a single functional origin on one chromosome, the configuration in line d is hardly ever observed. Indeed, the initial configuration of sister loci, in which they lie adjacent to each other, is reminiscent of the situation in fast-growing cells with overlapping replication cycles, in which synchronous initiation at two or four origins, leads to sister pairs only being segregated to daughter cells one to two generations after their replication (Fig. 4B). The switch from an initial <ori1 ori1 R2 R2> pattern to permissive <ori1 R2 ori1 R2> or <R2 ori1 ori1 R2> patterns appears to wait for completion of replication of the chromosome segment lying between oriC and oriZ, as judged by the number of replisome foci. This switch in configuration was validated by time-lapse analysis (Fig. S2). We propose that the switch to a permissive pattern becomes possible only when a topological restraint to complete segregation, imposed by the incomplete replication of the small segment, is removed by the completion of replication. In fast-growing cells, in which replication initiates synchronously at two or more origins, such a constraint may play a crucial role in ensuring that newly replicated sisters are prevented from immediately segregating to opposite cells halves (Fig. 4B). Our own work has not revealed a similar “unlocking” of a topological restraint in replicating oriC cells, as judged by the delayed spatial separation of specific sister loci. Nor is it necessary, a priori, to invoke such an unlocking mechanism. Nevertheless, a delayed (∼18 min compared with most loci) spatial separation of sister loci in two 150 kb regions in the right chromosome arm, approximately 130 kb and 380 kb from oriC, has been observed by Kleckner, Austin, and their colleagues (2–4). The eventual separation of these sister loci was associated with an abrupt “splitting” of the nucleoid into a bilobed structure, a process proposed to play role in the segregation process (3).
Discussion
This work provides insight into control of replication initiation and the processes that influence the positioning and segregation of E. coli chromosome loci and the replication machinery. By characterizing replication initiation in cells containing two distantly separated yet functional replication origins, we have demonstrated that neither cellular nor chromosomal concentration of oriC is a determinant in control of replication initiation, as originally proposed (19), as cell volume at initiation of the oriC-oriZ strain with two origins is identical to that of oriC WT and oriZ cells. Although subsequent work, most of which used oriC-carrying plasmids, showed that initiation mass/oriC copy is not strictly constant (20–23), this is the first study of which we are aware in which cells with more than one WT origin within a bacterial chromosome has been extensively characterized.
We propose that, at all growth rates, total chromosomal DNA concentration in a cell at initiation, rather than oriC concentration, is a key factor in control of initiation by DnaA-ATP, because chromosomal DNA titrates most of the more than 1,000 DnaA-ADP and DnaA-ATP molecules, with only a small fraction being oriC-associated (reviewed in ref. 41). This hypothesis is not only consistent with the observation that the insertion of an additional origin sequence into the E. coli chromosome has no significant effect on the volume at which initiation occurs, but is broadly consistent with the calculated chromosomal DNA concentrations at which synchronous initiation at one, two, or four sister origins occurs [1:1:0.8, respectively (14); see http://simon.bio.uva.nl/cellcycle/index.html]. Furthermore, our own experimental data for slow- and fast-growing cells are also broadly consistent with initiation at a constant chromosomal DNA concentration (Fig. S1C). We note that the relative positioning of functional origins with datA and DARS chromosomal sequences, which bind DnaA and act in normal initiation control (Fig. 1A) (16, 41), is similar in each of the three strains. If regulatory inactivation of DnaA (16, 18) stimulates hydrolysis of DNA-bound DnaA-ATP at the fork as replication proceeds, the ratio of DnaA-ATP to DnaA-ADP should increase during D-periods. Therefore, irrespective of growth rate, the increase in concentration of DnaA-ATP associated with reduced DNA synthesis during a D-period may be a key event in controlling reinitiation, as well as being the time during which the final stages of chromosome segregation and cell division occur.
The demonstration that replication in oriC-oriZ cells is initiated synchronously at spatially separate origins, residing at the expected cellular positions of ori1 and R2, confirms that E. coli replication does not occur in replication factories containing sister replisomes in cells that initiate and complete replication in the same generation (1). Synchronous initiation at both origins is followed by productive replication and separation of sister ori1 and R2 loci, as judged by fluorescent labeling of newly replicated DNA at initiation and by sister foci separation. Indeed, separation of newly replicated sister ori1 and R2 loci occurred soon after locus replication, irrespective of whether replication initiated at oriC, oriZ, or both origins synchronously. As most other chromosomal loci appear to behave like ori1 and R2 (2–4), the data also extend support for the hypothesis that the timing of chromosome locus segregation is determined largely by the timing of replication, rather than by some intrinsic property of chromosome content or organization, perhaps associated with a dedicated segregation machinery. Nevertheless, delayed separation of a few specific regions may be instrumental in catalyzing nuceloid splitting (2).
The cellular location of the normal oriC region is independent of the presence of a functional origin, whereas inserting a functional origin into an ectopic locus does not perturb the cellular position of that locus in nonreplicating cells. Similarly, in B. subtilis, the positioning of the origin region remains the same after deletion of the initiation sequences (32), whereas insertion of oriC into a plasmid does not alter the plasmid's cellular location in E. coli (42).
Is chromosome organization in oriC, oriC-oriZ, and oriZ cells an intrinsic property of the chromosome and its replication pattern, or are there external factors involved in locus position and origin behavior, as in C. crescentus or during sporulation in B. subtilis? We have demonstrated functional expression of C. crescentus PopZ and ParB in E. coli and have shown that PopZ localizes to poles, where it can capture ParB in the absence of chromosomal parS sites (Fig. S3) (27, 28). Nevertheless, oriC-proximal parS sites bound by ParB, were not recruited to the poles by PopZ (Fig. S3). Therefore, it is not trivial to alter E. coli chromosome organization or behavior by introducing a polar tethering system. These observations indicate again that chromosome organization is not readily influenced by extrinsic factors.
In oriC cells, the location of oriC close to midcell could arise simply from the fact that replication “layers” equal amounts of DNA on either side of the newly replicated origins, because the replication arms (i.e., replichores) are of almost identical size. Similarly, in oriZ cells, if replication terminates within the normal ter, layering of two very different sized replichores, of approximately 3 Mb and 1 Mb (Fig. 1B), could have led to the R2 locus remaining at approximately the cell quarter position. Alternatively, a process other than replication may contribute to genetic locus positioning.
We observed no overriding problems in replicating through strong convergent transcription from the ribosomal RNA cistrons. In rich and minimal medium, generation times were similar to that of the WT strain, with no obvious filamentation or morphological abnormalities. The cell cycle parameters, measured in minimal medium, were also comparable despite the changed replication programs. In oriZ cells, it appears that the single CCW replicates through all five rRNA cistrons head-on, without compromising substantially the time of segregation of sister ori1 loci, or the overall C-period. Similarly, strains carrying inversions that include multiple rrn operons were shown to be fully viable in rich and minimal media, as long as a WT complement of helicases that facilitate replication fork progression through potential “roadblocks” was present (37, 43). We note that, in B. subtilis, head-on collision between rRNA transcription and replication have little effect on generation time in minimal medium (despite a ∼30% decrease in overall DNA elongation rate), but resulted in a greater than threefold increase in generation time in Luria Broth (LB) and as much as a sevenfold increase if combined with unbalancing of replichores (44). The apparent differences between E. coli and B. subtilis in dealing with head-on collisions between DNA polymerase and RNA polymerase may be because their DNA replication machineries have different compositions (45–47), and they use different nonreplicative helicases for removing roadblocks on DNA (43). Additionally, we showed that replication can effectively terminate in regions of the chromosome distant from ter sites and dif, showing that the converging replication fork machineries, along with other chromosome processing activities, can handle the final stages of replication and decatenation at sites distant from the normal ter and from the region of the cell where division will next occur. This is consistent with the demonstration that decatenation by topoisomerase IV occurs as replication proceeds around the chromosome (10).
We have demonstrated a dramatic robustness and flexibility of E. coli chromosome replication and segregation. Under laboratory conditions, cells can accommodate at least two functional origins, or an ectopic origin, without obvious detriment. This is despite oriC-oriZ and oriZ cells having altered replication-segregation patterns, with replication termination occurring distant from the normal termination sites and head-on collisions with the high levels of transcription from rRNA cistrons. Other work has also shown that E. coli can also accommodate a lifestyle with linear chromosomes (48), or with chromosomes carrying large inversions (36–38). Nevertheless, over evolutionary time, natural selection has favored gene and gene expression organizations that show common features when related to the normal single replication origin (e.g., refs. 44, 49). Key genes tend to be located close to replication origins so their cellular concentration remains constant irrespective of growth rate. Furthermore, strong and/or long transcriptional units are arranged to avoid head-on collisions with replication. In eukaryotes, too, replication is prevented from head-on collision with rRNA transcription by the presence of specific termination barriers (reviewed in ref. 50).
Materials and Methods
Bacterial Strains and Plasmids and Their Propagation.
Derivatives of E. coli K12 AB1157 growing in M9 glycerol (0.2%) supplemented with essential nutrients were used unless otherwise stated (6, 51). Ampicillin (100 μg/mL), kanamycin (25 μg/mL), gentamycin (15 μg/mL), hygromycin (50 μg/mL), and chloramphenicol (20 μg/mL) were added when required. Visualization of genetic loci using lacO and tetO arrays was as described by Wang et al. (6, 51). Fluorescent fusions to LacI and TetR were expressed at low levels to minimize perturbation of DNA replication and segregation (6, 51).
frt-CmR/KmR-frt-5.1kb oriC region (oriC integration fragment) or frt-CmR/KmR-5.1kb oriC region-frt (oriC deletion fragment) were cloned into pUC19 and further used as PCR template for λ-red recombination (52). The oriC integration fragment was integrated into position 344 kb on the genetic map, 21 kb upstream of lacZ, generating oriZ. The oriC deletion fragment was used to replace the WT 5.1-kb oriC region. The DNA region between the two frt sites (CmR/KmR genes with or without the 5.1-kb oriC region), were removed using Flp recombinase expressed from pCP20 (52). A smaller 1.2-kb oriC region (Fig. 1A) was manipulated the same way.
Flow Cytometry.
Flow cytometry was performed as described (36). Cells were treated with rifampicin (300 μg/mL) and cephalexin (100 μg/mL) for 4 h for runout experiments before fixation using ethanol (75%) and staining using Syto16 (0.2 μM). Analysis of the DNA content per cell was performed with a FACScan flow cytometer (Becton). Waseal software was used for plotting and analysis.
Microscopy.
Snapshot images were taken from exponentially growing cells (A600 ∼ 0.2). For time-lapse acquisition, cells were transferred from liquid culture to a slide mounted with 1% agarose in the same growth medium, and incubated at required temperature using an incubation chamber. Images were captured with a 5-min time interval. Microscopy was carried out on a Nikon Eclipse TE2000-U microscope equipped with a Photometrics Cool-Snap HQ CCD or a QuantEM camera. All images were taken and analyzed using MetaMorph 6.2 and ImageJ software.
Automatic tracking of the fluorescent signal was performed using a custom MATLAB routine adapted from the from the feature point tracker of Sbalzarini and Koumoutsakos (53). Identification of the cell poles was performed interactively by the users because automatic identification in bright-field images could not be reliably achieved. Three image sequences were then tracked: one consisting of artificial cell center “particles” and the other two the YFP and CFP channels containing the foci. As many as two foci per channel were assigned to each cell on the basis of their proximity to the cell center line running between the two poles. Output from the program consists of a table of coordinates for the cell poles, cell centers, and loci position, and, for each cell selected by the user, a kymograph summarizing the chromosome loci dynamics.
Supplementary Material
Acknowledgments
We thank L. Shapiro and C. Jacobs-Wagner for C. crescentus strains and plasmids. C.L. was supported by a European Molecular Biology Organization long-term fellowship. R.R-L. was supported by a fellowship of New College, Oxford, United Kingdom. This work was supported by the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 10383.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100874108/-/DCSupplemental.
References
- 1.Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ. Independent positioning and action of Escherichia coli replisomes in live cells. Cell. 2008;133:90–102. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates D, Kleckner N. Chromosome and replisome dynamics in E. coli: Loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joshi MC, et al. Escherichia coli sister chromosome separation includes an abrupt global transition with concomitant release of late-splitting intersister snaps. Proc Natl Acad Sci USA. 2011;108:2765–2770. doi: 10.1073/pnas.1019593108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen HJ, Li Y, Youngren B, Hansen FG, Austin S. Progressive segregation of the Escherichia coli chromosome. Mol Microbiol. 2006;61:383–393. doi: 10.1111/j.1365-2958.2006.05245.x. [DOI] [PubMed] [Google Scholar]
- 5.Niki H, Yamaichi Y, Hiraga S. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 2000;14:212–223. [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Liu X, Possoz C, Sherratt DJ. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 2006;20:1727–1731. doi: 10.1101/gad.388406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen RB, Shapiro L. Chromosome segregation during the prokaryotic cell division cycle. Curr Opin Cell Biol. 1999;11:726–731. doi: 10.1016/s0955-0674(99)00043-5. [DOI] [PubMed] [Google Scholar]
- 8.Viollier PH, et al. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fogel MA, Waldor MK. Distinct segregation dynamics of the two Vibrio cholerae chromosomes. Mol Microbiol. 2005;55:125–136. doi: 10.1111/j.1365-2958.2004.04379.x. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Reyes-Lamothe R, Sherratt DJ. Modulation of Escherichia coli sister chromosome cohesion by topoisomerase IV. Genes Dev. 2008;22:2426–2433. doi: 10.1101/gad.487508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shinomiya T, Ina S. Analysis of chromosomal replicons in early embryos of Drosophila melanogaster by two-dimensional gel electrophoresis. Nucleic Acids Res. 1991;19:3935–3941. doi: 10.1093/nar/19.14.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper S, Helmstetter CE. Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol. 1968;31:519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- 13.Skarstad K, Steen HB, Boye E. Escherichia coli DNA distributions measured by flow cytometry and compared with theoretical computer simulations. J Bacteriol. 1985;163:661–668. doi: 10.1128/jb.163.2.661-668.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helmstetter C, Cooper S, Pierucci O, Revelas E. On the bacterial life sequence. Cold Spring Harb Symp Quant Biol. 1968;33:809–822. doi: 10.1101/sqb.1968.033.01.093. [DOI] [PubMed] [Google Scholar]
- 15.Boye E, Løbner-Olesen A, Skarstad K. Limiting DNA replication to once and only once. EMBO Rep. 2000;1:479–483. doi: 10.1093/embo-reports/kvd116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katayama T, Ozaki S, Keyamura K, Fujimitsu K. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol. 2010;8:163–170. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- 17.Waldminghaus T, Skarstad K. The Escherichia coli SeqA protein. Plasmid. 2009;61:141–150. doi: 10.1016/j.plasmid.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Kaguni JM. DnaA: controlling the initiation of bacterial DNA replication and more. Annu Rev Microbiol. 2006;60:351–375. doi: 10.1146/annurev.micro.60.080805.142111. [DOI] [PubMed] [Google Scholar]
- 19.Donachie WD. Relationship between cell size and time of initiation of DNA replication. Nature. 1968;219:1077–1079. doi: 10.1038/2191077a0. [DOI] [PubMed] [Google Scholar]
- 20.Helmstetter CE, Leonard AC. Coordinate initiation of chromosome and minichromosome replication in Escherichia coli. J Bacteriol. 1987;169:3489–3494. doi: 10.1128/jb.169.8.3489-3494.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wold S, Skarstad K, Steen HB, Stokke T, Boye E. The initiation mass for DNA replication in Escherichia coli K-12 is dependent on growth rate. EMBO J. 1994;13:2097–2102. doi: 10.1002/j.1460-2075.1994.tb06485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donachie WD, Blakely GW. Coupling the initiation of chromosome replication to cell size in Escherichia coli. Curr Opin Microbiol. 2003;6:146–150. doi: 10.1016/s1369-5274(03)00026-2. [DOI] [PubMed] [Google Scholar]
- 23.Dasgupta S, Løbner-Olesen A. Host controlled plasmid replication: Escherichia coli minichromosomes. Plasmid. 2004;52:151–168. doi: 10.1016/j.plasmid.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Jun S, Mulder B. Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proc Natl Acad Sci USA. 2006;103:12388–12393. doi: 10.1073/pnas.0605305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jun S, Wright A. Entropy as the driver of chromosome segregation. Nat Rev Microbiol. 2010;8:600–607. doi: 10.1038/nrmicro2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ben-Yehuda S, Rudner DZ, Losick R. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science. 2003;299:532–536. doi: 10.1126/science.1079914. [DOI] [PubMed] [Google Scholar]
- 27.Bowman GR, et al. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell. 2008;134:945–955. doi: 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell. 2008;134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerdes K, Howard M, Szardenings F. Pushing and pulling in prokaryotic DNA segregation. Cell. 2010;141:927–942. doi: 10.1016/j.cell.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 30.Reyes-Lamothe R, Sherratt DJ, Leake MC. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moriya S, et al. Effects of oriC relocation on control of replication initiation in Bacillus subtilis. Microbiology. 2009;155:3070–3082. doi: 10.1099/mic.0.030080-0. [DOI] [PubMed] [Google Scholar]
- 32.Berkmen MB, Grossman AD. Subcellular positioning of the origin region of the Bacillus subtilis chromosome is independent of sequences within oriC, the site of replication initiation, and the replication initiator DnaA. Mol Microbiol. 2007;63:150–165. doi: 10.1111/j.1365-2958.2006.05505.x. [DOI] [PubMed] [Google Scholar]
- 33.Maisnier-Patin S, Nordström K, Dasgupta S. Replication arrests during a single round of replication of the Escherichia coli chromosome in the absence of DnaC activity. Mol Microbiol. 2001;42:1371–1382. doi: 10.1046/j.1365-2958.2001.02718.x. [DOI] [PubMed] [Google Scholar]
- 34.Morigen OI, Odsbu I, Skarstad K. Growth rate dependent numbers of SeqA structures organize the multiple replication forks in rapidly growing Escherichia coli. Genes Cells. 2009;14:643–657. doi: 10.1111/j.1365-2443.2009.01298.x. [DOI] [PubMed] [Google Scholar]
- 35.Atlung T, Hansen FG. Effect of different concentrations of H-NS protein on chromosome replication and the cell cycle in Escherichia coli. J Bacteriol. 2002;184:1843–1850. doi: 10.1128/JB.184.7.1843-1850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lesterlin C, Pages C, Dubarry N, Dasgupta S, Cornet F. Asymmetry of chromosome Replichores renders the DNA translocase activity of FtsK essential for cell division and cell shape maintenance in Escherichia coli. PLoS Genet. 2008;4:e1000288. doi: 10.1371/journal.pgen.1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esnault E, Valens M, Espéli O, Boccard F. Chromosome structuring limits genome plasticity in Escherichia coli. PLoS Genet. 2007;3:e226. doi: 10.1371/journal.pgen.0030226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lesterlin C, Mercier R, Boccard F, Barre FX, Cornet F. Roles for replichores and macrodomains in segregation of the Escherichia coli chromosome. EMBO Rep. 2005;6:557–562. doi: 10.1038/sj.embor.7400428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, Wang X, Reyes-Lamothe R, Sherratt D. Replication-directed sister chromosome alignment in Escherichia coli. Mol Microbiol. 2010;75:1090–1097. doi: 10.1111/j.1365-2958.2009.06791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Danilova O, Reyes-Lamothe R, Pinskaya M, Sherratt D, Possoz C. MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol Microbiol. 2007;65:1485–1492. doi: 10.1111/j.1365-2958.2007.05881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leonard AC, Grimwade JE. Initiating chromosome replication in E. coli: It makes sense to recycle. Genes Dev. 2009;23:1145–1150. doi: 10.1101/gad.1809909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niki H, Hiraga S. Subcellular localization of plasmids containing the oriC region of the Escherichia coli chromosome, with or without the sopABC partitioning system. Mol Microbiol. 1999;34:498–503. doi: 10.1046/j.1365-2958.1999.01611.x. [DOI] [PubMed] [Google Scholar]
- 43.Boubakri H, de Septenville AL, Viguera E, Michel B. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 2010;29:145–157. doi: 10.1038/emboj.2009.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srivatsan A, Tehranchi A, MacAlpine DM, Wang JD. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet. 2010;6:e1000810. doi: 10.1371/journal.pgen.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Donnell M. Replisome architecture and dynamics in Escherichia coli. J Biol Chem. 2006;281:10653–10656. doi: 10.1074/jbc.R500028200. [DOI] [PubMed] [Google Scholar]
- 46.Pomerantz RT, O'Donnell M. What happens when replication and transcription complexes collide? Cell Cycle. 2010;9:2537–2543. doi: 10.4161/cc.9.13.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanders GM, Dallmann HG, McHenry CS. Reconstitution of the B. subtilis replisome with 13 proteins including two distinct replicases. Mol Cell. 2010;37:273–281. doi: 10.1016/j.molcel.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 48.Cui T, et al. Escherichia coli with a linear genome. EMBO Rep. 2007;8:181–187. doi: 10.1038/sj.embor.7400880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rocha EP. The replication-related organization of bacterial genomes. Microbiology. 2004;150:1609–1627. doi: 10.1099/mic.0.26974-0. [DOI] [PubMed] [Google Scholar]
- 50.Labib K, Hodgson B. Replication fork barriers: Pausing for a break or stalling for time? EMBO Rep. 2007;8:346–353. doi: 10.1038/sj.embor.7400940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, Possoz C, Sherratt DJ. Dancing around the divisome: Asymmetric chromosome segregation in Escherichia coli. Genes Dev. 2005;19:2367–2377. doi: 10.1101/gad.345305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sbalzarini IF, Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J Struct Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]