Abstract
Activation of genes by heavy metals, notably zinc, cadmium and copper, depends on MTF-1, a unique zinc finger transcription factor conserved from insects to human. Knockout of MTF-1 in the mouse results in embryonic lethality due to liver decay, while knockout of its best characterized target genes, the stress-inducible metallothionein genes I and II, is viable, suggesting additional target genes of MTF-1. Here we report on a multi-pronged search for potential target genes of MTF-1, including microarray screening, SABRE selective amplification, a computer search for MREs (DNA-binding sites of MTF-1) and transfection of reporter genes driven by candidate gene promoters. Some new candidate target genes emerged, including those encoding α-fetoprotein, the liver-enriched transcription factor C/EBPα and tear lipocalin/von Ebner’s gland protein, all of which have a role in toxicity/the cell stress response. In contrast, expression of other cell stress-associated genes, such as those for superoxide dismutases, thioredoxin and heat shock proteins, do not appear to be affected by loss of MTF-1. Our experiments have also exposed some problems with target gene searches. First, finding the optimal time window for detecting MTF-1 target genes in a lethal phenotype of rapid liver decay proved problematical: 12.5-day-old mouse embryos (stage E12.5) yielded hardly any differentially expressed genes, whereas at stage 13.0 reduced expression of secretory liver proteins probably reflected the onset of liver decay, i.e. a secondary effect. Likewise, up-regulation of some proliferation-associated genes may also just reflect responses to the concomitant loss of hepatocytes. Another sobering finding concerns γ-glutamylcysteine synthetasehc (γ-GCShc), which controls synthesis of the antioxidant glutathione and which was previously suggested to be a target gene contributing to the lethal phenotype in MTF-1 knockout mice. γ-GCShc mRNA is reduced at the onset of liver decay but MTF-1 null mutant embryos manage to maintain a very high glutathione level until shortly before that stage, perhaps in an attempt to compensate for low expression of metallothioneins, which also have a role as antioxidants.
INTRODUCTION
The transcriptional activator MTF-1 (metal-responsive transcription factor 1) is a highly conserved protein involved in the transcriptional response to heavy metal exposure (1–6). We have cloned and characterized MTF-1 from several species, including human, mouse, pufferfish (Fugu rubripes) and Drosophila melanogaster, and thereby shown evolutionary conservation of the MTF-1 gene (2,7,8; B.Zhang and W.Schaffner, unpublished results). MTF-1 contains six zinc fingers of the C2H2 type as a DNA-binding domain and at least three distinct transcriptional activation domains. It binds to a number of so-called metal-responsive elements (MREs) present in the promoters of metallothionein I and II (MT-I and MT-II) and other heavy metal-inducible genes (9). These MRE motifs contain a well-defined 7 bp core sequence (TGCRCNC) (4,6) and to date there is no experimental evidence that MTF-1 can utilize other DNA motifs to promote transcription in vivo. Metallothioneins are small, cysteine-rich proteins involved in heavy metal detoxification and other cell stress responses (10,11). There are four types of metallothioneins in mammals, designated MT-I–MT-IV (10,12–14; reviewed in 15). Whereas expression of MT-I and MT-II is stress-inducible and ubiquitous, MT-III and MT-IV expression is tissue-specific and is induced by stress only weakly. We have previously shown that MTF-1 directs basal and heavy metal-induced expression of the MT-I and MT-II genes (4). In addition, MTF-1 plays a role in the cellular responses to oxidative stress and hypoxia (5,16,17), implying a more general role for this transcription factor in cellular stress responses. We have generated mice lacking functional MTF-1 as a result of targeted gene disruption. These mice die in utero around day 14 of gestation due to an acute decay of hepatocytes (16). To date, the MT-I and MT-II genes are the best characterized target genes of MTF-1. Mice with disruption of both the MT-I and MT-II genes are more sensitive to metal stress, but do not show any obvious phenotype under normal laboratory conditions (18–20). This finding strongly suggests that MTF-1 has additional target genes besides MT-I and MT-II and that a failure to properly regulate these genes may be responsible for the lethal phenotype of MTF-1 null mice. One apparent candidate was γ-glutamylcysteine synthetasehc (γ-GCShc), a subunit of a key enzyme for glutathione synthesis whose gene promoter contains MREs (16). However, in the light of the results presented here, the relevance of this gene for development of the observed phenotype has to be questioned (see Discussion).
Even though further candidate genes were proposed (16), none of them could convincingly explain the knockout phenotype. This led us to perform a specific screen for MTF-1 target genes by comparing gene expression profiles between wild-type and MTF-1 knockout embryos or embryonic livers. Firstly, we chose selective amplification via biotin- and restriction-mediated enrichment (SABRE), a novel selective amplification procedure for the detection of differentially expressed mRNAs (21). This protocol combines the differential display technique (22) with selection procedures based on subtractive hybridization library techniques (23). In a different approach to find MTF-1 target genes we used Affymetrix gene chip microarrays to identify differentially expressed genes in E13.0 embryos or in E12.5 embryonic livers. Finally, we searched databases for the presence of MRE-containing promoters and further characterized putative target genes by band shift and transient transfection assays. Our studies reveal α-fetoprotein (AFP), C/EBPα and tear lipocalin as especially likely target genes of MTF-1 and suggest possible mechanisms leading to embryonic lethality in MTF-1 null mutant mice.
MATERIALS AND METHODS
Microarray assay
Whole mouse embryos or embryonic livers were prepared at stage E13.0 or E12.5 and immediately frozen in liquid nitrogen until isolation of RNA. Preparation of labeled targets for hybridization and array hybridization of Affymetrix Mu11 oligonucleotide microarrays were performed essentially as described (24), with the following modifications. cDNA was synthesized directly from total RNA using the Superscript Choice Gene Chip Kit (Gibco) and in vitro transcription reactions were performed using the Megascript T7 System (Ambion). For testing whole embryos, two independent E13.0 MTF-1 null mutants and two MTF-1 wild-type embryos were used in one experiment. For experiments using E12.5 embryonic livers, 16 embryonic livers from MTF-1 null mutant and wild-type embryos were prepared. The RNA was then isolated from independent pools of eight livers from each genotype. Array hybridization and scanning procedures were performed at Hoffmann-La Roche (Basel, Switzerland) according to the supplier’s protocols. Genes were considered to be differentially expressed if there was at least a 2-fold difference in expression levels between wild-type and knockout samples.
Animal breeding and embryo isolation
Mice heterozygous for MTF-1 were mated following superovulation and the morning at which a vaginal plug was detected was defined as embryonic day E0.5. Following isolation of embryos, yolk sac DNA was used for genotyping as previously described (16,25). Isolation of ‘late’ E12.5 embryos was performed at 5 p.m., isolation of E13.0 embryos was performed at 12 p.m. (7 h later than late E12.5).
Selective amplification via biotin- and restriction-mediated enrichment (SABRE)
SABRE was performed essentially as described (21), with the following modifications. Briefly, for our experiments mRNA was prepared from E13.0 MTF-1 wild-type and knockout embryos with a Qiagen RNA/DNA Maxi Kit and Qiagen Oligotex mRNA Mini Kit, according to the supplier’s information. Three micrograms of cDNA was then digested with 28 U Sau3AI. An aliquot of 1 µg of the digested cDNA pool was ligated to a double-stranded linker oligo in a 20:1 molar ratio of linker to digested cDNA using 10 Weiss units of T4 DNA ligase. Linker oligo:
5′-GGTCCATCCAACC-3′
3′- TGCCAGGTAGGTTGGCTAG-5′
The ligation reactions were loaded onto a 1% agarose minigel and cDNA pools ranging from 150 bp to 2 kb were isolated. Wild-type cDNA was PCR amplified with a 5′-biotinylated primer containing a BamHI restriction site. cDNA from knockout embryos was PCR amplified with a non-biotinylated primer containing a mutated BamHI restriction site. Wild-type primer, 5′-C(9)-biotin-CCAGGATCCAACCGATC-3′; knockout primer, 5′-GGTCCATCCAACCGATC-3′.
For PCR Pharmacia Ready-to-Go PCR beads were used with the following mix: 1 µg cDNA, 5 µl DMSO, 20 pmol wild-type primer (or 60 pmol knockout primer), 1 µCi [α-32P]dATP and water to 50 µl final volume. PCR conditions were 4 min initial melting at 94°C, followed by 30 cycles of 1 min at 94°C, 2 min at 52°C and 2 min at 72°C, followed by a 10 min final elongation step at 72°C. PCR products were then purified by phenol extraction and 10 µg knockout (driver) product was co-precipitated with 300 ng wild-type (tester) product. PERT hybridization of the mixture was performed according to Miller and Riblet (26). Following hybridization, single-stranded cDNA was digested with S1 nuclease and the whole reaction was mixed with 50 µl of Dynal M-280 streptavidin magnetic beads. After 2 h incubation at room temperature on a wheel, the beads were extracted using a magnetic concentrator and tester homohybrids were removed by BamHI digestion. After 3 h digestion, a new PCR cycle was started as described above. After five consecutive cycles, the products were loaded on a sequencing gel and selectively amplified cDNA bands were cut out of the gel and cloned into pBluescript. Following transformation of bacteria, plasmid inserts were sequenced from plasmid minipreps using T7 or T3 primers.
Transfections and S1 nuclease mapping
Transfections and S1 nuclease mapping of transcripts were performed as described previously (2,27). For cloning of C/EBPα sequences a 0.6 kb fragment from the rat C/EBPα upstream promoter sequence (–1916 to –1302) was amplified from rat genomic DNA using primers 5′-atcgagctcgtagaaagactgaagatccttctc-3′ and 5′-tgagtcgacagttcattctccacctggatctcgg-3′. For cloning of tear lipocalin sequences, a 0.3 kb fragment from the human tear lipocalin promoter sequence (–523 to –213) was amplified from human genomic DNA using primers 5′-gacgagctcgaggagggtcgttactgcacac-3′ and 5′-tcggtcgacatttcacctgaagggaaactg-3′. Both fragments were inserted into the SalI and SacI sites of the OVEC vector (28).
Electrophoretic mobility shift assay (EMSA)
EMSAs were performed as described (2,16). The oligos used for the experiments were as follows.
For AFP: MRE2, 5′-cgagacacacatgcacacgcacacagg-3′ and
3′-tcgagctctgtgtgtacgtgtgcgtgtgtccagct-5′;
MRE4, 5′-cgagggttctgcacacggtttctcag-3′ and
3′-tcgagctcccaagacgtgtgccaaagagtcagct-5′;
MRE5, 5′-cgagagtttagatgcgcactgggaag-3′ and
3′-tcgagctctcaaatctacgcgtgacccttcagct-5′.
For C/EBPα:
MRE2, 5′-cgagggtcttcggggtgcaaaaacgagg-3′ and
3′-tcgagctcccagaagccccacgtttttgctccagct-5′;
MRE4, 5′-cgagcttgtttatgcgctctggaccgaag-3′ and
3′-tcgagctcgaacaaatacgcgagacctggcttcagct-5′;
MRE5, 5′-cgagattaatgggtcgcgtgcatctggcg-3′ and
5′-tcgagctctaattacccagcgcacgtagaccgcagct-5′.
For lipocalin:
MRE1, 5′-cgaggtcgttactgcacaccttgctcag-3′ and
3′-tcgagctccagcaatgacgtgtggaacgagtcagct-5′;
MRE2, 5′-cgagtctttaaatgcacacgggcatcgg-3′ and
3′-tcgagctcagaaatttacgtgtgcccgtagccagct-5′;
MRE4, 5′-cgaggttacggctgcacacgtccccagg-3′ and
3′-tcgagctccaatgccgacgtgtgcaggggtccagct-5′;
MRE5, 5′-cgagaggaggcctgcacacccttcactg-3′ and
3′-tcgagctctcctccggacgtgtgggaagtgacagct-5′.
Measurement of embryonic glutathione levels
Embryonic glutathione amounts were determined in late E12.5 embryonic livers according to Sack et al. (29). The following modifications were applied. Tissue samples were homogenized using a rotor-stator homogenizer. The samples were then ultracentrifuged at 100 000 g at 4°C for 1 h in the absence of sulfosalicylic acid. For standardization, sample protein amounts were determined with a Bradford Protein Assay Kit (Bio-Rad).
RESULTS
The first round of target gene searches was done with the SABRE protocol, which is essentially a combination of the subtraction library and differential display techniques (21). Basically, two populations of mRNAs were prepared from either three MTF-1 wild-type or three MTF-1 knockout embryos at stage E13.0. This stage was chosen because histological analyses of embryos suggested that E12.5 might be too early to see the effects of a lack of MTF-1 at the transcriptional level, while E13.5 might be too late due to the onset of liver decay (16; see Discussion). After five complete cycles of SABRE, the reactions were loaded on a sequencing gel and 20 differentially amplified bands were cut out, cloned into the pBluescript vector and sequenced for identification of the insert. Between four and 15 clones were analyzed from each eluted band. Three of the 20 candidate bands consisted of a homogenous population of cDNA fragments, of which two represented mouse AFP and one represented the mouse X chromosome inactivation transcript (XIST). The other bands represented mixed populations of at least two different cDNA fragments. The XIST result may merely represent an unequal distribution of sexes among the embryos used for generation of the cDNAs. Alternatively, MTF-1 might have sex-specific functions in heavy metal homeostasis, which would be compatible with the finding of an increased resistance of female versus male MT-I/MT-II knockout mice to cadmium intoxication (19). The finding of down-regulation of AFP in the MTF-1 knockout embryos is interesting because of the various biological functions of AFP and its preferential expression in the liver (30). Other genes that were found using SABRE include BiP, which encodes an endoplasmic reticulum lumenal chaperone (31) and a gene coding for a homolog of the rat synaptic vesicle protein SV2 (32), as well as several ESTs. All of them may represent genuine target genes of MTF-1, since they were found repeatedly or at least did not show up in the controls. Whereas the promoter of SV2 has not so far been characterized, the human BiP promoter does not contain MREs and thus is probably not a direct target of MTF-1. The AFP result, which in the light of the MTF-1 knockout phenotype was the most interesting (see Discussion), was additionally checked by semi-quantitative RT–PCR (data not shown). This experiment confirmed a reduced abundance of AFP mRNA in E13.0 MTF-1 knockout embryos in comparison to wild-type littermates. Consistent with a role for MTF-1 in expression of AFP is the presence of several MRE consensus motifs in the rat AFP promoter and upstream enhancers (Fig. 1). Taken together, these findings make AFP a good candidate target gene of MTF-1.
Figure 1.
Rat AFP as a putative target gene of MTF-1. (A) Five MREs with a perfect 7 bp core consensus sequence are located in the upstream regulatory sequence of the AFP gene. Six additional putative MREs with 1 nt mismatch are not indicated. (B) MREs used for EMSA are underlined. (C) EMSA with indicated MREs from AFP regulatory sequences. MRE 2 and 4 bind MTF-1 with high affinity, whereas MRE 5 does not bind under the conditions tested. The specificity of the interaction was verified by addition of excess unlabeled oligonucleotide with a very strong consensus site (MRE-s).
As a complementary system to find further MTF-1 target genes another systematic screening protocol, the Affymetrix Gene Chip system, was also utilized. The results with this oligonucleotide-based microarray system have proven reliable, i.e. they did not show any deviation from northern blots whenever RNA from identical sources was used (U.Certa, unpublished results). Analysis of our acquired data showed that ∼30% of the 13 000 genes on the array yielded a specific signal. More than 99% of the genes did not show significant differences in expression levels between genotypes. Within the group of genes with equal expression levels were classical reference genes such as actin and tubulin, as well as genes whose independence from MTF-1 expression was not necessarily expected, such as heat shock proteins, members of the AP-1 family of transcription factors and enzymes involved in free radical scavenging processes (Table 1). As a first differentially expressed gene we found MT-II, a well-established target gene of MTF-1. Other genes identified represent secretory liver proteins, including AFP and albumin. The finding that mRNAs for secretory liver proteins are reduced in E13.0 embryos does not necessarily mean that the corresponding genes are genuine target genes of MTF-1, since a reduced mRNA level might be the result of damage or loss of hepatocytes in the knockout embryos. In an attempt to clarify this point we screened the acquired data for the presence of any other liver-specific genes with equal expression levels in wild-type and knockout embryos. Indeed, some genes, like ferritin, Cyp4A14 (a cytochrome P450 family member) and others, were equally expressed, but their suitability to serve as liver-specific control genes is questionable, since their expression pattern is either not precisely known or not strictly hepatocyte-specific (33,34). Unfortunately, other genes with purely hepatocyte-restricted expression did not yield a specific signal in the data set. However, this is not unexpected since these genes are usually expressed at a low level. The question whether differential gene expression had been influenced by the onset of liver decay was therefore addressed by another microarray screening using samples from an earlier stage, namely ‘late’ E12.5 embryonic livers. In this screening several ubiquitously expressed genes, as well as genes with liver-restricted expression, did not show significantly different expression levels in MTF-1 knockout versus wild-type samples (data not shown). One of these genes encoded albumin, suggesting that it is not a target gene of MTF-1, at least at E12.5. In contrast, AFP transcripts were found to be down-regulated to ∼60% of the expression levels observed in wild-type livers. The other genes encoding secretory liver proteins, which in the first screening had emerged as differentially expressed, unfortunately belonged to the group of genes for which no specific signal was obtained in the second screening. In the case of AFP a dependence on MTF-1 is likely, but its physiological relevance for the early MTF-1 knockout phenotype remains to be clarified (see Discussion). In this second screening the gene for vacuolar adenosine triphosphatase subunit b showed a modest reduction in expression in MTF-1 knockout livers. This enzyme is involved in the regulation of cytosolic pH in hepatocytes (Table 2; 35).
Table 1. Stress defense genes with unaltered expression level in E13.0 MTF-1–/– embryos (microarray).
| Heat shock proteins |
| Small heat shock protein hsp 25 |
| hsp 47 |
| hsp 60 |
| hsp 90α (hsp86) |
| hsp 70 |
| heat shock cognate 71 kDa protein |
| Transcription factors |
| HIF-1α |
| NF-κB, p50 subunit |
| Jun D |
| c-jun |
| Detoxification enzymes |
| Copper-zinc superoxide dismutase |
| Manganese superoxide dismutase |
| Glutathione peroxidase |
| Free radical scavengers |
| Thioredoxin |
Table 2. MTF-1 target genes obtained in the screenings (established and putative).
| Gene |
Source of evidence |
MRE in regulatory regiona |
Binding of MTF-1 to oligob |
| Stress-responsive genes | |||
| MT-II | Detailed in vivo and in vitro evidence (4,16; E13.0 microarray) | 6 MREs in promoter (M) | ++++ |
| AFP | SABRE, E13.0/E12.5 microarray | 5 MREs in promoter/enhancers (R) | +++ |
| Glucose 6-phosphate dehydrogenase | Database search | 5 MREs in promoter (H) | |
| Transforming growth factor β-1 | Database search | 3 MREs in promoter (M) | |
| Cytosolic zinc-binding protein | Database search | 1 MRE in promoter (R) | |
| Tear lipocalin | Cell culture data and in vitro evidence | 5 MREs in promoter (H) | +++ |
| AMD-1 S-adenosylmethionine decarboxylase | Database search | 6 MREs in promoter (M) | |
| BiP (hsp 70 family) | SABRE | Noc | |
| Vacuolar adenosine triphosphatase subunit b | E12.5 microarray | Missing sequence information | |
| Secretory liver proteins | |||
| Fetuin | E13.0 microarray | 3 MREs in promoter (M) | |
| Corticoid-binding globulin | E13.0 microarray | 1 MRE in promoter (M) | |
| Plasma retinol-binding protein | E13.0 microarray | Noc | |
| Liver-enriched transcription factors | |||
| HNF-3α | Database search | 3 MREs in promoter (M) | |
| HNF-3γ | Database search | 5 MREs in promoter (R) | |
| Hfh-1L | Database search | 8 MREs in promoter (M) | |
| C/EBPα | Database search | 5 MREs in 5′-upstream region (R) | +++ |
| Signal pathway factors | |||
| ALK-6 (TGFβ receptor type I) | E13 microarray | Missing sequence information | |
| Inhibin/activin bc subunit | Database search | 1 MRE in promoter (M) | |
| FGF receptor | E13 microarray | Missing sequence information | |
| Gas3/PMP22 | E13 microarray | 2 MREs in promoter (M) | |
| Notch-1 | Database search | 3 MREs in promoter (M) | |
| Tissue-specific proteins | |||
| SV2 | SABRE | Missing sequence information | |
| Neuronal nicotinic acetylcholine receptor | Database search | 4 MREs in promoter (M) | |
| Dopamine receptor D1 | Database search | 1 MRE (R) | |
| Testes-specific kinase 1 | Database search | 6 MREs in promoter (M) | |
| Synaptonemal complex protein I | Database search | 3 MREs in promoter (M) | |
| Other transcripts | |||
| XIST | SABRE | 3 MREs in promoter (M) | |
| ESTs | |||
| ms20h07 | E13.0 microarray | Missing sequence information | |
| mg75d07 | E13.0 microarray | Missing sequence information | |
| mq10d02 | E13.0 microarray | Missing sequence information | |
| va1e02 | E13.0 microarray | Missing sequence information | |
| vk53c08 | E13.0 microarray | Missing sequence information | |
| vn65b01 | E13.0 microarray | Missing sequence information | |
| vl04h06 | E13.0 microarray | Missing sequence information | |
| C76222 | E13.0 microarray | Missing sequence information | |
| Genes transcriptionally up-regulated in MTF-1 knockout embryos/livers | |||
| Cell division control protein 3 | E13.0 microarray | Missing sequence information | |
| NF-YB transcription factor | E13.0 microarray | Noc | |
| Activin receptor II(B) | E13.0 microarray | 7 MREs in upstream region (–1.8 kb) (H) | |
| Hepatopoietin receptor (ERV1) | E12.5 microarray | Missing sequence information | |
a(M), mouse; (H), human; (R), rat. Human or rat were considered when mouse sequence was unavailable.
bMREs were tested only from the indicated subset of genes.
cA remote cis-regulatory region with MREs cannot be excluded. Alternatively, indirect transcriptional regulation via MTF-1 is possible.
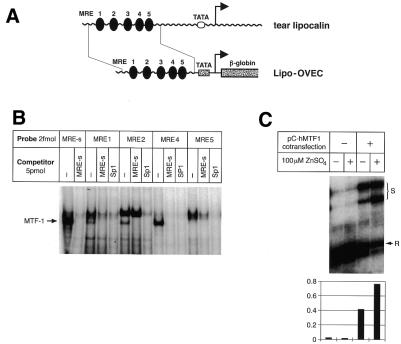
Another candidate target gene, encoding tear albumin/lipocalin, was analyzed by different means. This protein is mainly expressed in tear glands and lingual glands. It belongs to the lipocalin superfamily of lipophilic ligand-carrier proteins and may have a general role in binding lipophilic substances, including toxic ones. Both the human and porcine tear albumin genes contain two conserved MRE sequence motifs in their upstream promoter regions (36). We have analyzed the binding properties of these MREs in EMSAs and found them to bind to MTF-1 (Fig. 2). Furthermore, we tested fusion constructs in which the human tear lipocalin upstream region was fused to the OVEC reporter gene. In cell transfection experiments the tear lipocalin promoter could indeed confer zinc/MTF-1-responsive expression on the reporter gene (Fig. 2).
Figure 2.
Human tear lipocalin as a putative target gene of MTF-1. (A) The human tear lipocalin locus. Its proximal promoter contains five MRE sequence motifs. A 0.3 kb fragment from this region was cloned into the OVEC reporter gene. (B) The EMSA shows that four MREs from the tear lipocalin promoter region bind to MTF-1 with high affinity. (C) S1 nuclease mapping of the lipocalin–OVEC reporter transcript. The signal (S) was normalized to the reference (R) signal (co-transfection of CMV-REF).
C/EBPα and C/EBPβ belong to the family of b-Zip transcription factors. Both are also enriched in the liver (37,38), where they play different and, at least in part, antagonistic roles in cell differentiation and proliferation. We found a cluster of MREs at –1.9 kb from the main transcription start of the C/EBPα gene. These upstream regulatory sequences were also tested in transfection experiments and could confer zinc/MTF-1-dependent expression on a reporter gene (Fig. 3).
Figure 3.
The rat C/EBPα promoter contains six MREs. (A) The rat C/EBPα promoter. A 0.6 kb fragment containing five MREs was cloned into the OVEC reporter gene. (B) Different MREs from the C/EBPα promoter region bind to MTF-1 with high affinity. (C) S1 nuclease mapping of transcripts from the C/EBPα reporter construct. S, signal; R, reference.
Besides the genes down-regulated in the absence of MTF-1, three genes emerged which are expressed at higher levels in the MTF-1 knockout. The first gene is a homolog of the yeast cell division control protein 3 (cdc3), which is a member of the conserved eukaryotic family of septin proteins, involved in cytokinesis (39). The second gene, NF-YB, codes for a subunit of a CCAAT box-binding protein (40). NF-Y/CBF1 is a basal transcription factor, but nevertheless may well be relevant for the liver phenotype of MTF-1 knockout embryos. Over the years evidence has accumulated that expression of hepatocyte-specific genes is mainly controlled by a few families of liver-specific transcriptional activators, in combination with some ubiquitous and basal transcription factors (41,42). In addition, functional binding sites for general transcription factors, such as NF-Y/CBF1, and stress-responsive ubiquitous factors, like AP-1, are present in typical promoters of liver-specific genes and their cognate transcription factors are required for high transcription rates of liver-specific genes (41). The third gene is a receptor for hepatopoietin, a novel polypeptide mitogen specific for hepatocytes (43), which was up-regulated in E12.5 MTF-1 knockout livers ∼5-fold. All three genes may be up-regulated as part of an effort by the MTF-1-deficient embryo to compensate for the loss of hepatocytes at the onset of liver decay.
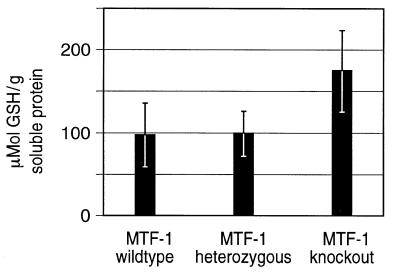
γ-GCS is the key enzyme in the biosynthesis of glutathione (GSH). Previously we found by northern blot analysis that γ-GCShc mRNA is down-regulated in E13.5 MTF-1 knockout embryos (16) and we thus speculated that an insufficient GSH level could contribute to the MTF-1 knockout phenotype. To clarify the relevance of this finding we measured GSH levels in embryonic livers using a recently developed, HPLC-based method (29) adapted to our purpose. GSH was determined from several liver samples from late E12.5 MTF-1 knockout, heterozygous and wild-type embryos as well as from an adult wild-type liver. The measured GSH levels were in the same range as those in other studies using rat hepatocytes (44): wild-type and heterozygous livers contained ∼100 µmol GSH/g soluble protein. However, in contrast to our expectation, the GSH level in knockout E12.5 embryonic livers turned out to be at least as high, if not higher, than in the wild-type (Fig. 4).
Figure 4.
GSH levels in embryonic livers at late E12.5 stage. GSH levels were determined in MTF-1 wild-type (n = 3), heterozygous (n = 9) and knockout (n = 3) liver samples via a HPLC-based method (29). No significant reduction in GSH in MTF-1 knockout livers was observed. Note that the GSH level in knockout embryos at E12.5 is at least as high, if not higher, than in wild-type and heterozygous embryos (even though mRNA levels for γ-GCShc are reduced in MTF-1 knockout embryos at later stages; 16).
DISCUSSION
We used several methods to find genes differentially expressed between MTF-1 wild-type and knockout embryos at day 12.5/13.0 of gestation. Our studies identified several new putative target genes of MTF-1 which are compatible with either of two distinct models that could explain the liver decay phenotype of MTF-1 null mutant embryos. As we speculated earlier (16), insufficient expression of genes involved in the stress response may lead to an accumulation of endogenously generated toxic metabolites, leading to metabolic collapse of the liver. Alternatively, MTF-1 may be essential for control of a specific stage of liver development and thereafter would be required only to handle particular stress situations.
Each of the methods used in our study has its advantages and drawbacks, which might explain why hardly any of the candidate target genes of MTF-1 scored positive in all screenings (45). Furthermore, it appears that the critical time window for an executional role of MTF-1 is quite narrow, constituting a major challenge for the elucidation of the exact reason for embryonic liver decay in MTF-1 knockout embryos. While E12.5 embryos may well be too young to observe clear cut effects of MTF-1 deficiency, E13.5 embryos already suffer from the onset of liver decay, which confounds primary and secondary effects. Therefore, we reasoned that an intermediate stage should be most suitable; SABRE and one of the microarray screenings were done with E13.0 RNA. Such embryos are still viable and tissues other than the liver are not detectably affected by the lack of MTF-1 (nevertheless, even the E13.0 results may be influenced by the onset of liver decay; see below). The finding of MT-II in the microarray screening with E13.0 embryos was reassuring evidence that MTF-1 directly controls expression of this gene (Table 2; 4,16). On the other hand, MT-I did not emerge as a differentially expressed gene in our screenings (Table 3). Since MT-I is a well-characterized target gene of MTF-1 (4,16), its expression scored absent most likely due to a low expression level of MT-I in the samples or for technical reasons (45). Several additional differentially expressed genes were identified. One of them, AFP, was found in all screenings to be less abundantly expressed in MTF-1 knockout versus wild-type embryos. Nevertheless, in a parallel collaborative study, where expression of AFP in the endoderm cells of the visceral yolk sac at E12 was measured, no difference between MTF-1 null mutant embryos and the wild-type was found (46). This suggests that the contribution of MTF-1 to the complex transcriptional regulation of AFP (see also below) is not equally important in all tissues, at least not at this early stage of development. AFP is an interesting protein for several reasons. First of all, it is mainly expressed in embryonic hepatocytes and its expression is controlled at the transcriptional level (30). As a member of the albuminoid gene superfamily it is the major embryonic protein responsible for maintenance of the colloid osmotic pressure. Its down-regulation may therefore explain the late stages of the MTF-1 knockout phenotype, which are characterized by generalized edema and disseminated bleeding (16). AFP plays various additional roles, including binding to, and scavenging of, heavy metals and reactive oxygen species. In addition, AFP influences developmental processes and interacts with growth factors and the apoptotic signaling cascade (30). Hence, AFP might very well contribute to a diminished ability of MTF-1 knockout embryos to cope with accumulation of harmful agents during liver development. Within the time window that MTF-1 knockout embryos die the size of the liver increases several-fold. Most importantly, the hepatocytes begin to express many new enzymes and secretory proteins required for liver function (47). The embryonic liver gains metabolic functions at this stage, which implies the need to detoxify endogenously generated metabolites. Support for a role of MTF-1 in transcriptional regulation of AFP is the lower expression of AFP in E12.5 livers of MTF-1 knockout embryos and the finding of putative MREs in its upstream regulatory sequences (see below). At this early stage a general disturbance of hepatocyte structure cannot be observed either histologically (16) or at the molecular level. Several well-characterized cis-acting control regions govern AFP expression (Fig. 1). Three remote enhancers, located between –7.6 and –1 kb in the rat, have unique but overlapping patterns of activity. Post-natal repression of AFP transcription is caused by a silencer region located between the promoter and the three remote enhancers (48). Three perfect MREs and several additional potential MREs are located within the enhancer regions (see Fig. 1). MTF-1 could influence AFP expression directly or indirectly, by activating other transcription factors, such as C/EBPα, known to bind to the AFP promoter and enhancers (49). Indeed, C/EBPα emerged as another candidate target gene of MTF-1 (Fig. 3).
Table 3. MTF-1 target genes described in the literature.
| Gene |
Source of evidence |
MRE in regulatory regiona |
Binding of MTF-1 to oligob |
| Stress-responsive genes | |||
| MT-I | Detailed in vivo and in vitro evidence (4,16) | 6 MREs in promoter (M) | ++++ |
| ZnT-1 | Detailed in vivo and in vitro evidence (59) | 3 MREs in promoter (M) | |
| γ-GCS | Conflicting in vivo data, but promoter responsive to MTF-1 in transient transfections (16) | 1 MRE in promoter, 1 in leader (H) | +/+++ |
| γ-Glutamyltranspeptidase | Database search (16) | 3 MREs in promoter (M) | |
| PlGF | In vivo and in vitro evidence (60) | 2 MREs in promoter (M) |
a(M), mouse; (H), human. Human was considered when mouse sequence was unavailable.
bMREs were tested only from the indicated subset of genes.
C/EBPα, which contains several MREs in its upstream regulatory sequence, is the founding member of a family of b-Zip transcription factors. Both C/EBPα and C/EBPβ are liver-enriched factors, where they exert some antagonistic effects. While C/EBPβ is associated with hepatic cell proliferation in embryonic cells and regenerating liver, C/EBPα is involved in maintenance of the differentiated, non-proliferating state of liver and other cells such as adipocytes. Ectopic expression of C/EBPα in various transfected cell types, including fibroblasts, blocks their proliferation, apparently by inducing a high level of the anti-proliferative protein p21. In support of this notion, C/EBPα knockout mice display a low level of p21 and an extended proliferation period of their liver cells (50). Likewise, liver regeneration after partial hepatectomy is accompanied by sustained induction of C/EBPβ with concomitant down-regulation of C/EBPα (51). Furthermore, C/EBPα knockout mice lack hepatic glycogen at birth, confirming a role of C/EBPα in cellular energy metabolism. Finally, and perhaps most relevant to our findings, C/EBPα is also involved in the cellular stress response, as indicated by its induction by infectious agents in the so-called acute phase inflammatory response.
Tear lipocalin, another candidate target gene of MTF-1, was found because of a clustering of MREs in its upstream regulatory region. Also referred to as tear albumin or von Ebner’s gland protein, it is mainly expressed in lachrymal (tear) and lingual glands, including parotid salivary glands and sublingual von Ebner’s glands. More recently, expression was also found in the nasal mucosa and the prostate gland. Lipocalin is able to bind a number of lipophilic compounds such as retinol, cholesterol, fatty acids, phospholipids and fatty alcohols (52,53) and potentially toxic molecules as well. In addition, it inhibits cysteine proteases, plays a role in inflammatory processes and may be part of a general defense against infectious agents (54,55). Tear lipocalin, by binding and neutralizing bitter tasting organic compounds, can also modulate the taste of food. Thus lipocalin has a role in coping with several forms of cell stress and a link to MTF-1 via its MREs in the promoter region (36; Fig. 2) makes sense. Furthermore, the fact that the prostate gland is the organ with the highest zinc content (56) may imply a connection of tear lipocalin to prostate function via MTF-1/zinc metabolism. Nevertheless, tear lipocalin is not apparently expressed in the liver and therefore is probably not involved in development of the MTF-1 knockout phenotype.
We have previously noted a similarity of the MTF-1 null phenotype to the phenotypes of mice deficient for c-jun, p65/RelA or HGF/c-met (16). This similarity, though not identity, does not necessarily mean that MTF-1 acts upstream or downstream of one or several of these genes. Although binding sites for NF-κB are present in the MTF-1 promoter, MTF-1 is expressed ubiquitously and its promoter reveals the characteristics of a classical housekeeping gene (57). Therefore, most probably none of these factors is epistatic to MTF-1. Indeed, MTF-1 expression is not changed in c-jun knockout fibroblasts and vice versa (16). More likely is the possibility that certain target genes are regulated by a combination of these factors and that insufficient expression of one or several of these target genes is responsible for the initiation of hepatocyte decay. AP-1, NF-κB and MTF-1 all have in common that they are involved in different aspects of cellular anti-stress defense mechanisms. Nevertheless, MTF-1 is the only one of these genes that seems to be essential for hepatocyte development only and the knockout of which is not accompanied by other phenotypic manifestations (16). Therefore, at least some gene(s) involved in the hepatocyte decay in MTF-1 knockout embryos is likely to be a MTF-1-specific target. However, to date it is still unclear whether the MTF-1 knockout phenotype is caused by an inability of hepatocytes to cope with toxic metabolites or whether MTF-1 plays a decisive role in liver development around day E12.5–E13.5. Conditional knockout mice may provide answers to these open questions.
We have reported before that γ-glutamyltranspeptidase and γ-GCShc both contain MRE motifs in their promoters and were potential MTF-1 target genes. Indeed, transcript levels of γ-GCShc (a GSH synthesizing enzyme) were found to be reduced in E13.5 MTF-1 knockout embryos (16). Although a reduced level of GSH would provide an explanation for the increased susceptibility of MTF-1 knockout primary cells to H2O2 (16), the role of γ-GCS as a target gene of MTF-1 has to be critically reviewed in the light of our new data. First, it became obvious that stage E13.5 observations are not very informative, as the liver is already clearly affected by the lack of MTF-1. Second, our finding that GSH levels in E12.5 embryonic livers is at least as high, if not higher, in MTF-1 knockouts than in the wild-type (Fig. 4) is a compelling argument against a crucial role of γ-GCShc in development of the MTF-1 knockout phenotype. From these results we conclude that the MTF-1 phenotype cannot be explained by a decrease in γ-GCShc expression, although we cannot exclude additional unknown functions of this enzyme besides synthesis of GSH. In contrast, there might even be overproduction of GSH in MTF-1 knockout livers to compensate for the loss of metallothionein, which has functions as an antioxidant molecule overlapping with those of GSH.
Previous studies suggested that MTF-1 may regulate expression of zinc transporter-1 (ZnT-1), a ubiquitously expressed zinc export pump (16,58). Indeed, a recent study convincingly shows that ZnT-1 is a target gene of MTF-1 in vivo, at least in the mid-gestation visceral yolk sac and placenta (59). Nevertheless, there is no direct evidence for involvement of ZnT-1 in development of the MTF-1 knockout phenotype. In spite of this missing link to the onset of liver decay in MTF-1 null mutant mice, one can conclude that MTF-1 coordinates the regulation of genes involved in zinc homeostasis and protection against metal toxicity, such as the metallothioneins and ZnT-1.
Recently it was found by Brian Murphy and colleagues that MTF-1 is also activated by hypoxia (17). The same group identified a particularly interesting target gene of MTF-1, placental growth factor (PlGF), though with unclear relevance for the lethal knockout phenotype. In collaboration with our group they have shown that PlGF is activated in a cell type-specific manner and that its basal and hypoxia-inducible expression is dependent on MTF-1 (60). PlGF encodes an angiogenic factor (61) with complementary activities to vascular endothelial growth factor (VEGF). Hypoxia is a well-established pathophysiological condition, found for example in the majority of malignant solid tumors. Such conditions act as potent inducers of neovascularization by initiating expression of the VEGF genes (62). These new findings also imply a role of MTF-1 in tumor growth and therapy resistance in vivo. In agreement with this, high expression of metallothioneins correlates with a poor prognosis and progressive disease in a number of human tumors (63,64). In addition, metallothionein expression is cell cycle dependent (65). Hence, MTF-1 may be critical for modulating gene expression associated with some malignant phenotypes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Brian J.Murphy (Menlo Park, CA), Oliver Clay (Naples) and Bo Zhang for communication of unpublished results, Till Strassen and Dr Antonia Manova for excellent technical support, and Dr Ueli Schibler (Geneva) for advice on the SABRE technique. We also thank Drs Hansruedi Büeler and Lee Martin for critical reading of the manuscript. This work was supported by the Schweizerische Nationalfonds and the Kanton Zurich.
DDBJ/EMBL/GenBank accession no. NM008636
References
- 1.Westin G. and Schaffner,W. (1988) A zinc-responsive factor interacts with a metal-regulated enhancer element (MRE) of the mouse metallothionein-I gene. EMBO J., 7, 3763–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radtke F., Heuchel,R., Georgiev,O., Hergersberg,M., Gariglio,M., Dembic,Z. and Schaffner,W. (1993) Cloned transcription factor MTF-1 activates the mouse metallothionein I promoter. EMBO J., 12, 1355–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radtke F., Georgiev,O., Müller,H.-P., Brugnera,E. and Schaffner,W. (1995) Functional domains of the heavy metal-responsive transcription factor MTF-1. Nucleic Acids Res., 23, 2277–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heuchel R., Radtke,F., Georgiev,O., Stark,G., Aguet,M. and Schaffner,W. (1994) The transcription factor MTF-1 is essential for basal and heavy metal-induced metallothionein gene expression. EMBO J., 13, 2870–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalton T.P., Li,Q., Liang,L. and Andrews,G.K. (1996) Oxidative stress activates metal-responsive transcription factor-1 binding activity. Occupancy in vivo of metal response elements in the metallothionein-I gene promoter. J. Biol. Chem., 271, 26233–26241. [DOI] [PubMed] [Google Scholar]
- 6.Chen X., Chu,M. and Giedroc,D.P. (1999) MRE-binding transcription factor-1: weak zinc-binding finger domains 5 and 6 modulate the structure, affinity and specificity of the metal-response element complex. Biochemistry, 38, 12915–12925. [DOI] [PubMed] [Google Scholar]
- 7.Brugnera E., Georgiev,O., Radtke,F., Heuchel,R., Baker,E., Sutherland,G.R. and Schaffner,W. (1994) Cloning, chromosomal mapping and characterisation of the human metal-regulatory transcription factor MTF-1. Nucleic Acids Res., 22, 3167–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Auf der Maur A., Belser,T., Elgar,G., Georgiev,O. and Schaffner,W. (1999) Characterization of the transcription factor MTF-1 from the Japanese pufferfish (Fugu rubripes) reveals evolutionary conservation of the heavy metal stress response. Biol. Chem., 380, 175–180. [DOI] [PubMed] [Google Scholar]
- 9.Stuart G.W., Searle,P.F., Chen,H.Y., Brinster,R. and Palmiter,R.D. (1984) A 12-base-pair DNA motif that is repeated several times in metallothionein gene promoters confers metal regulation to a heterologous gene. Proc. Natl Acad. Sci. USA, 81, 7318–7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kägi J.H.R. (1991) Overview of metallothionein. Methods Enzymol., 205, 613–626. [DOI] [PubMed] [Google Scholar]
- 11.Andrews G.K. (2000) Regulation of metallothionein gene expression by oxidative stress and metal ions. Biochem. Pharmacol., 59, 95–104. [DOI] [PubMed] [Google Scholar]
- 12.Uchida Y., Takio,K., Titani,K., Ihara,Y. and Tomonaga,M. (1991) The growth inhibitory factor that is deficient in the Alzheimer’s disease brain is a 68 amino acid metallothionein-like protein. Neuron, 7, 337–347. [DOI] [PubMed] [Google Scholar]
- 13.Palmiter R.D., Findley,S.D., Whitmore,T.E. and Durnam,D.M. (1992) MT-III, a brain-specific member of the metallothionein gene family. Proc. Natl Acad. Sci. USA, 89, 6333–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quaife C.J., Findley,S.D., Erickson,J.C., Froelick,G.J., Kelly,E.J., Zambrowicz,B.P. and Palmiter,R.D. (1994) Induction of a new metallothionein isoform (MT-IV) occurs during differentiation of stratified squamous epithelia. Biochemistry, 33, 7250–7259. [DOI] [PubMed] [Google Scholar]
- 15.Vasak M. and Hasler,D.W. (2000) Metallothioneins: new functional and structural insights. Curr. Opin. Chem. Biol., 4, 177–183. [DOI] [PubMed] [Google Scholar]
- 16.Günes C., Heuchel,R., Georgiev,O., Müller,K.-H., Lichtlen,P., Blüthmann,H., Marino,S., Aguzzi,A. and Schaffner,W. (1998) Embryonic lethality and liver degeneration in mice lacking the metal-responsive transcriptional activator MTF-1. EMBO J., 17, 2846–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy B.J., Andrews,G.K., Bittel,D., Discher,D.J., McCue,J., Green,C.J., Yanovsky,M., Giacca,A., Sutherland,R.M., Laderoute,K.R. and Webster,K.A. (1999) Activation of metallothionein gene expression by hypoxia involves metal response elements and metal transcription factor-1. Cancer Res., 59, 1315–1322. [PubMed] [Google Scholar]
- 18.Michalska A.E. and Choo,K.H.A. (1993) Targeting and germ-line transmission of a null mutation at the metallothionein I and II loci in mouse. Proc. Natl Acad. Sci. USA, 90, 8088–8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masters B.A., Kelly,E.J., Quaife,C.F., Brinster,R.L. and Palmiter,R.D. (1994) Targeted disruption of metallothionein I and II genes increases sensitivity to cadmium. Proc. Natl Acad. Sci. USA, 91, 584–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly E.J., Quaife,C.J., Froelick,G.J. and Palmiter,R.D. (1996) Metallothionein I and II protect against zinc deficiency and zinc toxicity in mice. J. Nutr., 126, 1782–1790. [DOI] [PubMed] [Google Scholar]
- 21.Lavery D.J., Lopez Molina,L., Fleury,O.F. and Schibler,U. (1997) Selective amplification via biotin- and restriction-mediated enrichment (SABRE), a novel selective amplification procedure for detection of differentially expressed mRNAs. Proc. Natl Acad. Sci. USA, 94, 6831–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang P. and Pardee,A.B. (1992) Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science, 257, 967–971. [DOI] [PubMed] [Google Scholar]
- 23.Zeng J., Gorski,R.A. and Hamer,D. (1994) Differential cDNA cloning by enzymatic degrading subtraction (EDS). Nucleic Acids Res., 22, 4381–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webb G.C., Akbar,M.S., Zhao,C. and Steiner,D.F. (2000) Expression profiling of pancreatic β cells: glucose regulation of secretory and metabolic pathway genes. Proc. Natl Acad. Sci. USA, 97, 5773–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lichtlen P., Georgiev,O., Schaffner,W., Aguzzi,A. and Brandner,S. (1999) The heavy metal-responsive transcription factor-1 (MTF-1) is not required for neural differentiation. Biol. Chem., 380, 711–715. [DOI] [PubMed] [Google Scholar]
- 26.Miller R.D. and Riblet,R. (1995) Improved phenol emulsion DNA reassociation technique (PERT) using thermal cycling. Nucleic Acids Res., 23, 2339–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kemler I., Bucher,E., Seipel,K., Muller-Immergluck,M.M. and Schaffner,W. (1991) Promoters with the octamer DNA motif (ATGCAAAT) can be ubiquitous or cell type-specific depending on binding affinity of the octamer site and Oct-factor concentration. Nucleic Acids Res., 19, 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westin G., Gerster,T., Müller,M.M., Schaffner,G. and Schaffner,W. (1987) OVEC, a versatile system to study transcription in mammalian cells and cell free extracts. Nucleic Acids Res., 15, 6787–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sack R., Willi,A. and Hunziker,P.E. (2000) Determination of total glutathione in cell lysates by high performance liquid chromatography with o-phthalaldehyde pre-column derivatization in the presence of tris(2-carboxyethyl)-phosphine. J. Liq. Chromatogr. Relat. Tech., 23, 2947–2962. [Google Scholar]
- 30.Mizejewski G.J. (1997) α-Fetoprotein as a biologic response modifier: relevance to domain and subdomain structure. Proc. Soc. Exp. Biol. Med., 215, 333–362. [DOI] [PubMed] [Google Scholar]
- 31.Gething M.J. (1999) Role and regulation of the ER chaperone BiP. Semin. Cell Dev. Biol., 10, 465–472. [DOI] [PubMed] [Google Scholar]
- 32.Son Y.J., Scranton,T.W., Sunderland,W.J., Baek,S.J., Miner,J.H., Sanes,J.R. and Carlson,S.S. (2000) The synaptic vesicle protein SV2 is complexed with an α5-containing laminin on the nerve terminal surface. J. Biol. Chem., 275, 451–460. [DOI] [PubMed] [Google Scholar]
- 33.Richardson D.R. and Ponka,P. (1997) The molecular mechanisms of the metabolism and transport of iron in normal and neoplastic cells. Biochim. Biophys. Acta, 1331, 1–40. [DOI] [PubMed] [Google Scholar]
- 34.Heng Y.M., Kuo,C.S., Jones,P.S., Savory,R., Schulz,R.M., Tomlinson,S.R., Gray,T.J. and Bell,D.R. (1997) A novel murine P-450 gene, Cyp4a14 is part of a cluster of Cyp4a and Cyp4b, but not of CYP4F, genes in mouse and humans. Biochem. J., 325, 741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wadsworth S.J. and van Rossum,G.D. (1994) Role of vacuolar adenosinetriphosphatase in the regulation of cytosolic pH in hepatocytes. J. Membr. Biol., 142, 21–34. [DOI] [PubMed] [Google Scholar]
- 36.Holzfeind P., Merschak,P., Wojnar,P. and Redl,B. (1997) Structure and organization of the porcine LCN1 gene encoding tear lipocalin/von Ebner’s gland protein. Gene, 202, 61–67. [DOI] [PubMed] [Google Scholar]
- 37.Darlington G.J., Wang,N. and Hanson,R.W. (1995) C/EBPα: a critical regulator of genes governing integrative metabolic processes. Curr. Opin. Genet. Dev., 5, 565–570. [DOI] [PubMed] [Google Scholar]
- 38.Calkhoven C.F., Müller,C. and Leutz,A. (2000) Translational control of C/EBPα and C/EBPβ isoform expression. Genes Dev., 14, 1920–1932. [PMC free article] [PubMed] [Google Scholar]
- 39.Kim H.B., Haarer,B.K. and Pringle,J.R. (1991) Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J. Cell Biol., 112, 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maity S.N. and de Crombrugghe,B. (1998) Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem. Sci., 23, 174–178. [DOI] [PubMed] [Google Scholar]
- 41.Xanthopoulos K.G. and Mirkovitch,J. (1993) Gene regulation in rodent hepatocytes during development, differentiation and disease. Eur. J. Biochem., 216, 353–360. [DOI] [PubMed] [Google Scholar]
- 42.Jump D.B., Badin,M.V. and Thelen,A. (1997) The CCAAT box binding factor, NF-Y, is required for thyroid hormone regulation of rat liver S14 gene transcription. J. Biol. Chem., 272, 27778–27786. [DOI] [PubMed] [Google Scholar]
- 43.Wang G., Yang,X.M., Zhang,Y., Wang,Q.M., Chen,H., Wei,H., Xing,G.C., Xie,L., Hu,Z.Y., Zhang,C., Fang,D.C., Wu,C.T. and He,F.C. (1999) Identification and characterization of receptor for mammalian hepatopoietin that is homologous to yeast ERV1. J. Biol. Chem., 274, 11469–11472. [DOI] [PubMed] [Google Scholar]
- 44.Tsuboi S. (1999) Elevation of glutathione level in rat hepatocytes by hepatocyte growth factor via induction of γ-glutamylcysteine synthetase. J. Biochem., 126, 815–820. [DOI] [PubMed] [Google Scholar]
- 45.Lipshutz R.J., Fodor,S.P.A., Gingeras,T.R. and Lockhart,D.J. (1999) High density synthetic oligonucleotide arrays. Nat. Genet., 21, 20–24. [DOI] [PubMed] [Google Scholar]
- 46.Andrews G.K., Lee,D.K., Ravindra,R., Lichtlen,P., Sirito,M., Sawadogo,M. and Schaffner,W. (2001) The transcription factors MTF-1 and USF1 cooperate to regulate mouse metallothionein-I expression in response to the essential metal zinc in visceral endoderm cells during early development. EMBO J., 20, 1114–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zaret K.S. (1996) Molecular genetics of early liver development. Annu. Rev. Physiol., 58, 231–251. [DOI] [PubMed] [Google Scholar]
- 48.Ramesh T.M., Ellis,A.W. and Spear,B.T. (1995) Individual mouse α-fetoprotein enhancer elements exhibit different patterns of tissue-specific and hepatic position-dependent activities. Mol. Cell. Biol., 15, 4947–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bois-Joyeux B. and Danan,J.L. (1994) Members of the CAAT/enhancer-binding protein, hepatocyte nuclear factor-1 and nuclear factor-1 families can differentially modulate the activities of the rat α-fetoprotein promoter and enhancer. Biochem. J., 301, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Timchenko N.A., Harris,T.E., Wilde,M., Bilyeu,T.A., Burgess-Beusse,B.L., Finegold,M.J. and Darlington,G.J. (1997) CCAAT/enhancer binding protein α regulates p21 protein and hepatocyte proliferation in newborn mice. Mol. Cell. Biol., 17, 7353–7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skrtic S., Ekberg,S., Wallenius,V., Enerbäck,S., Hedin,L. and Jansson,J.O. (1997) Changes in expression of CCAAT/enhancer binding protein α (C/EBPα) and C/EBPβ in rat liver after partial hepatectomy but not after treatment with cyproterone acetate. J. Hepatol., 27, 903–911. [DOI] [PubMed] [Google Scholar]
- 52.Redl B., Holzfeind,P. and Lottspeich,F. (1992) cDNA cloning and sequencing reveals human tear prealbumin to be a member of the lipophilic-ligand carrier protein superfamily. J. Biol. Chem., 267, 20282–20287. [PubMed] [Google Scholar]
- 53.Glasgow B.J., Abduragimov,A.R., Farabakhsh,Z.T., Faull,K.F. and Hubbell,W.L. (1995) Tear lipocalins bind a broad array of lipid ligands. Curr. Eye Res., 14, 363–372. [DOI] [PubMed] [Google Scholar]
- 54.Holzfeind P., Merschak,P., Rogatsch,H., Culig,Z., Feichtinger,H., Klocker,H. and Redl,B. (1996) Expression of the gene for tear lipocalin/von Ebner’s gland protein in human prostate. FEBS Lett., 395, 95–98. [DOI] [PubMed] [Google Scholar]
- 55.van’t Hoff W., Blankenvoorde,M.F.J., Veerman,E.C.I. and Nieuw Amerongen,A.V. (1997) The salivary lipocalin von Ebner’s gland protein is a cysteine proteinase inhibitor. J. Biol. Chem., 272, 1837–1841. [DOI] [PubMed] [Google Scholar]
- 56.Costello L.C., Liu,Y., Zou,J. and Franklin,R.B. (1999) Evidence for a zinc uptake transporter in human prostate cancer cells which is regulated by prolactin and testosterone. J. Biol. Chem., 274, 17499–17504. [DOI] [PubMed] [Google Scholar]
- 57.Auf der Maur A., Belser,T., Wang,Y., Günes,C., Lichtlen,P., Georgiev,O. and Schaffner,W. (2000) Characterization of the mouse gene for the heavy metal-responsive transcription factor MTF-1. Cell Stress Chaperones, 5, 196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palmiter R.D. and Findley,S.D. (1995) Cloning and characterization of a mammalian zinc transporter that confers resistance to zinc. EMBO J., 14, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langmade S.J., Ravindra,R., Daniels,P.J. and Andrews,G.K. (2000) The transcription factor MTF-1 mediates metal regulation of the mouse ZnT1 gene. J. Biol. Chem., 275, 34803–34809. [DOI] [PubMed] [Google Scholar]
- 60.Green C.J., Lichtlen,P., Huynh,N.T., Yanovsky,M., Laderoute,K.R., Schaffner,W. and Murphy,B.J. (2001) Placenta growth factor gene expression is induced by hypoxia in fibroblasts: a central role for metal transcription factor MTF-1. Cancer Res., 61, in press. [PubMed] [Google Scholar]
- 61.Maglione D., Guerriero,V., Viglietto,G., Delli-Bovi,P. and Persico,M.G. (1991) Isolation of a human placenta cDNA coding for a protein related to the vascular permeability factor. Proc. Natl Acad. Sci. USA, 88, 9267–9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sutherland R.M., Ausserer,W.A. Murphy,B.J. and Laderoute,K.R. (1996) Tumor hypoxia and heterogenity: challenges and opportunities for the future. Semin. Radiat. Oncol., 6, 59–70. [DOI] [PubMed] [Google Scholar]
- 63.Jasani B. and Schmid,K.W. (1997) Significance of metallothionein overexpression in human tumors. Histopathology, 31, 211–214. [DOI] [PubMed] [Google Scholar]
- 64.Moussa M., Kloth,D., Peers,G., Cherian,M.G., Frei,J.V. and Chin,J.L. (1997) Metallothionein expression in prostatic carcinoma: correlation with Gleason grade, pathologic stage, DNA content and serum level of prostate-specific antigen. Clin. Invest. Med., 20, 371–380. [PubMed] [Google Scholar]
- 65.Nagel W.W. and Vallee,B.L. (1995) Cell cycle regulation of metallothionein in human colonic cancer cells. Proc. Natl Acad. Sci. USA, 92, 579–583. [DOI] [PMC free article] [PubMed] [Google Scholar]