Abstract
Polycistronic pre-mRNAs from Caenorhabditis elegans are processed by 3′ end formation of the upstream mRNA and SL2-specific trans-splicing of the downstream mRNA. These processes usually occur within an ∼100-nucleotide region and are mechanistically coupled. In this paper, we report a complex in C. elegans extracts containing the 3′ end formation protein CstF-64 and the SL2 snRNP. This complex, immunoprecipitated with αCstF-64 antibody, contains SL2 RNA, but not SL1 RNA or other U snRNAs. Using mutational analysis we have been able to uncouple SL2 snRNP function and identity. SL2 RNA with a mutation in stem/loop III is functional in vivo as a trans-splice donor, but fails to splice to SL2-accepting trans-splice sites, suggesting that it has lost its identity as an SL2 snRNP. Importantly, stem/loop III mutations prevent association of SL2 RNA with CstF-64. In contrast, a mutation in stem II that inactivates the SL2 snRNP still permits complex formation with CstF-64. Therefore, SL2 RNA stem/loop III is required for both SL2 identity and formation of a complex containing CstF-64, but not for trans-splicing. These results provide a molecular framework for the coupling of 3′ end formation and trans-splicing in the processing of polycistronic pre-mRNAs from C. elegans operons.
Keywords: Spliced leader RNA, polycistronic transcription, cleavage, polyadenylation
Between 15% and 25% of Caenorhabditis elegans genes are organized in polycistronic transcription units, or operons (Zorio et al. 1994; T. Blumenthal, D. Evans, C. Link, A. Guffanti, D. Lawson, J. Thierry-Mieg, D. Thierry-Mieg, K. Duke, and S. Kim, in prep.). Polycistronic pre-mRNAs from these operons are processed into monocistronic mRNAs by cleavage and polyadenylation at the 3′ ends of upstream gene mRNAs accompanied by trans-splicing at the 5′ ends of downstream gene mRNAs. In general, these two processes occur within a 100-nucleotide region (Blumenthal and Steward 1997) and are mechanistically coupled (Kuersten et al. 1997).
In C. elegans, 3′ end formation is dependent on an AAUAAA signal (Kuersten et al. 1997; Liu et al. 2001). It is expected that this sequence is bound by cleavage and polyadenylation specificity factor (CPSF), as it is in mammalian cells (for reviews, see Colgan and Manley 1997; Keller and Minvielle-Sebastia 1997; Zhao et al. 1999). Presumably, C. elegans 3′ end formation also requires cleavage stimulation factor (CstF), which binds a U-rich or GU-rich sequence downstream of the cleavage site. Homologs of each of the subunits of both mammalian CstF and CPSF are present in the C. elegans genome (C.J. Wilusz and T. Blumenthal, unpubl.).
Trans-splicing generates 5′ ends of mRNAs in trypanosomes and many animals (Murphy et al. 1986; Sutton and Boothroyd 1986; Krause and Hirsh 1987; Rajkovic et al. 1990; Tessier et al. 1991; Stover and Steele 2001; Vandenberghe et al. 2001). A spliced leader (SL) exon is donated to the 5′ ends of mRNAs by a short RNA donor called SL RNA. The SL RNA exists as a ribonucleoprotein (RNP) particle (Thomas et al. 1988; Van Doren and Hirsh 1988; Maroney et al. 1990; Goncharov et al. 1999) that includes the Sm core proteins (Lerner and Steitz 1979). Unlike the other U snRNPs, which are capable of catalyzing repeated splicing reactions, the SL snRNP is consumed during the trans-splicing reaction.
C. elegans possesses two distinct SL RNAs, SL1 RNA (Krause and Hirsh 1987) and SL2 RNA (Huang and Hirsh 1989). SL1 RNA is present at ∼7–10 times the level of SL2 RNA (S. Kuersten, R. Conrad, and T. Blumenthal, unpubl.). Nevertheless, mRNAs from most genes located in downstream positions in operons receive almost exclusively SL2 (Spieth et al. 1993), suggesting that a mechanism exists to regulate which spliced leader a particular pre-mRNA receives (Huang and Hirsh 1989). SL1 and SL2 have different sequences, but sequence complementarity between the SL RNAs and the pre-mRNA is not a factor in trans-splice site choice (Blumenthal and Steward 1997). SL1 and SL2 RNAs have many similar features—both are small (∼100 and 110 nucleotides, respectively), possess a trimethylguanosine (TMG) cap, associate with Sm proteins (Krause and Hirsh 1987; Thomas et al. 1988; Van Doren and Hirsh 1988; Huang and Hirsh 1989), and can be folded into a characteristic secondary structure (Bruzik et al. 1988). This structure consists of three stem/loops, with the trans-splice site located on the 3′ side of stem I, and the Sm-binding site located between stems II and III (see Fig. 2A, below). These similarities between the SL1 and SL2 RNAs make the question of substrate specificity even more intriguing.
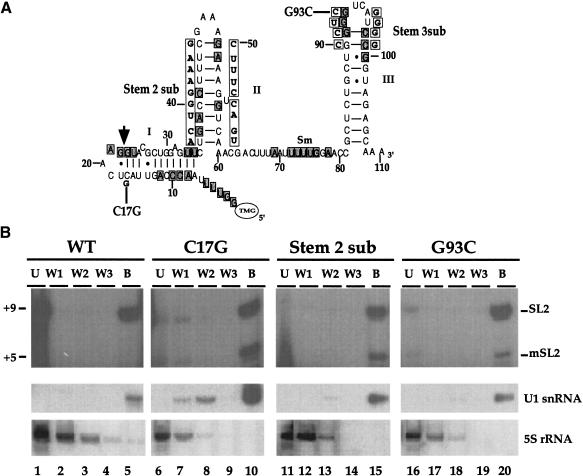
Figure 2.
SL2 RNA mutations still allow core snRNP formation. (A) The mutations studied are shown (boxed and in boldface type) adjacent to the predicted SL2α RNA structure. Nucleotide positions that are conserved in 100% of known SL2 RNA genes (boxed and shaded; Evans et al. 1997) are shown. The trans-splice site is marked by an arrow. The stems are numbered as they are referred to in the text and the Sm-binding site is indicated. (B) snRNPs were immunoprecipitated from wild-type or transgenic C. elegans crude embryonic extracts with a human anti-Sm serum. The RNAs contained in the unbound (U), washes (W1-W3), and bound (B) fractions were analyzed by primer extension in the presence of ddCTP to analyze SL2 RNA and by Northern blotting for the controls. The Northern blot was hybridized to a U1 snRNA probe as a positive control, stripped, and hybridized to a 5S rRNA probe as a negative control.
Sequence analysis of SL2 RNA genes from the nematodes Dolichorhabditis and C. elegans identified several conserved features, including the 5′ end of the spliced leader, the trans-splice site, part of stem II, the Sm-binding site, and the top of stem loop III (Evans et al. 1997). Whereas the sequence of the spliced leader is dispensable, the primary sequence of stem II and loop III are required for SL2 trans-splicing in vivo (Evans and Blumenthal 2000).
Several observations have suggested that SL2 trans-splicing specificity is coupled to 3′ end processing upstream. Mutation of the AAUAAA polyadenylation signal reduced the SL2 specificity of trans-splicing to the downstream gene mRNA (Kuersten et al. 1997). Conversely, strengthening the poly(A) signal, by changing AGUAAA to AAUAAA, resulted in increased SL2 trans-splicing (Liu et al. 2001). Furthermore, a mutational analysis of an operon intercistronic region revealed a second sequence required for SL2 trans-splicing: a U-rich region, located downstream of the poly(A) site (Huang et al. 2001). The location and sequence of this site suggest it may serve as a CstF-binding site. Although mutation of this sequence had only a small effect on 3′ end formation, it did prevent SL2 trans-splicing to the downstream gene mRNA (Huang et al. 2001).
Therefore, we hypothesized that a physical interaction exists between the 3′ end formation machinery and the SL2 trans-splicing machinery. In an attempt to detect such an interaction, we performed immunoprecipitations using an antibody to C. elegans CstF-64, the homolog to the RNA-binding subunit of mammalian CstF (MacDonald et al. 1994). We report here that SL2 RNA, but not other snRNAs, is precipitated with αCstF-64. A mutational analysis indicates that association of CstF-64 correlates with SL2 snRNP specificity. Changes in the third stem/loop of SL2 RNA both prevent SL2 trans-splicing in vivo and immunoprecipitation with αCstF-64. Interestingly, a complete replacement of the third stem/loop interferes with SL2 identity, but it does not prevent snRNP function. Therefore, a molecular framework for SL2 trans-splicing is suggested in which association with CstF allows the less abundant SL2 snRNP to be preferentially used at trans-splice sites located downstream of a poly(A) site.
Results
Precipitation of SL2 RNA by CstF-64 antibody
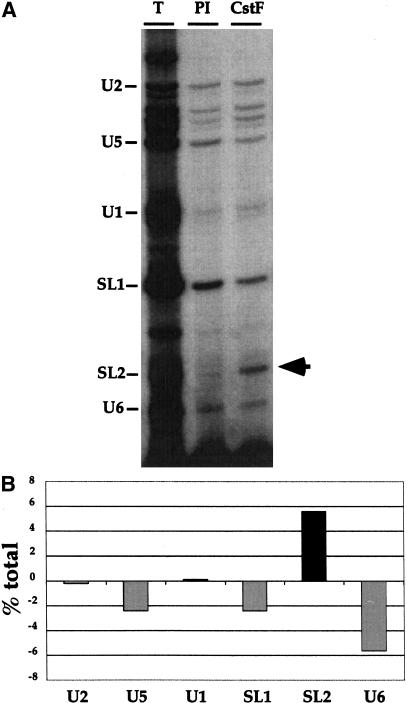
To investigate the relationship between the 3′ end formation machinery and SL2 trans-splicing, we focused our attention on CstF because the U-rich region required for SL2 trans-splicing was consistent in location and sequence with a mammalian CstF-binding site (Huang et al. 2001). Therefore, the C. elegans homolog of CstF-64 was cloned, expressed in Escherichia coli, and the recombinant protein used to generate polyclonal antibodies. These antibodies recognized a single band of the expected size in C. elegans extracts on Western blots and were able to immunoprecipitate CstF-64 (data not shown). C. elegans crude embryonic extracts were immunoprecipitated with this antibody and snRNAs found in a complex with CstF-64 were identified by primer extension (Fig. 1). Only SL2 RNA was precipitated at a level significantly above that obtained using the pre-immune control serum. SL1 RNA and the other U snRNAs (U1, U2, U5, and U6) were present at similar levels in the pre-immune control and αCstF-64 immunoprecipitates. This result suggests the existence of a specific complex containing both CstF-64 and SL2 RNA.
Figure 1.
SL2 RNA is specifically immunoprecipitated by αCstF64. (A) U snRNPs were immunoprecipitated from a wild-type C. elegans crude embryonic extract with an antibody raised against the C. elegans homolog of the mammalian 64-kD subunit of the 3′ end formation factor CstF, as well as a pre-immune control antibody. RNA was extracted and analyzed by primer extension with a pool of oligonucleotides, each specific for one of the snRNAs. The oligonucleotides were designed to result in termination at different locations on the gel as indicated. The unlabeled bands below U2 represent U2 breakdown products (data not shown). (T) Total before immunoprecipitation (IP); (PI) preimmune control IP; (CstF) αCstF-64 IP. (B) The results from A were quantified using a Molecular Dynamics PhosphorImager. The values shown are the percent of the total of each RNA immunoprecipitated after subtracting the signal from the pre-immune control.
SL2 RNA mutations that inhibit operon processing allow core snRNP formation
To determine whether the SL2 RNA sequences required for trans-splicing in vivo (Evans and Blumenthal 2000) were also required for the CstF-64 interaction, we performed immunoprecipitations from crude embryonic extracts made from transgenic strains expressing SL2 RNAs with mutations in either stem II or the loop closing stem III. The SL2α RNA gene, marked with a single C substituted with G at position 17 for identification (Fig. 2A), was expressed in vivo from an extrachromosomal tandem array (Evans and Blumenthal 2000). Primer extension in the presence of ddCTP results in termination products from the marked SL2 RNA (+5) and the endogenous SL2 RNA (+9) that can be resolved, allowing the fate of the marked SL2 RNA to be followed.
We first tested whether the mutant SL2 RNAs could be assembled into snRNPs. Extracts made from strains carrying the mutant SL2 RNA genes were immunoprecipitated with a human serum that recognizes the core (or Sm) proteins required for U snRNA maturation (Lerner et al. 1981; De Robertis et al. 1982; Matter et al. 1982; Fritz et al. 1984; Mattaj and De Robertis 1985). As expected, this serum precipitates the U1 snRNA, but not the 5S RNA (Fig. 2B). The data in Figure 2B show that the antiserum effectively precipitated both endogenous wild-type and marked (C17G) SL2 snRNP (Fig. 2B, lane 10). Furthermore, mutation of stem II or loop III (Stem 2 sub and G93C, Fig. 2A) did not prevent core snRNP formation, as these mutant SL2 RNAs were also bound by this serum (Fig. 2B, +5 product in lanes 15 and 20). Therefore, the defects of Stem 2 sub and G93C RNAs in SL2 trans-splicing are not the result of a defect in core snRNP formation. The apparently lower level of G93C in the immunoprecipitate is attributable to a lower level of expression of this construct (see Fig. 3). Quantitation of the data in Figure 2 shows that 69% of the C17G and 58% of the G93C SL2 RNA are precipitated by the anti-Sm serum.
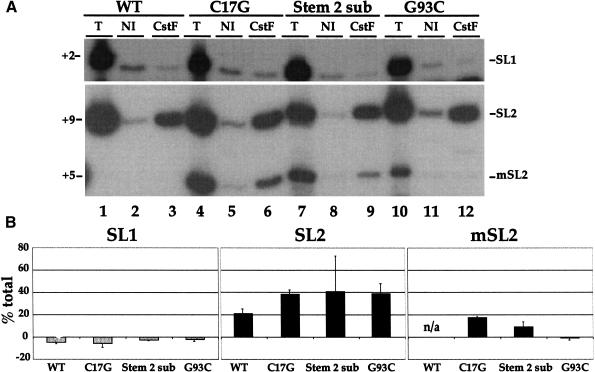
Figure 3.
Mutation of the third stem/loop prevents interaction of the SL2 snRNP with CstF64. (A) Immunoprecipitations of C. elegans crude embryonic extracts from untransformed or transgenic strains were performed with αCstF-64 and non-immune control serum. The presence of the SL1 and SL2 RNAs was determined by primer extension in the presence of ddCTP. (B) Quantification of the results shown in A. The values are the percent of the total signal bound to the CstF beads after subtraction of the non-immune control.
Interaction of SL2 RNA with CstF-64 requires an intact third loop
SL2 RNA interacts with CstF-64 (Fig. 1), and mutations of stem II and loop III prevent trans-splicing (Evans and Blumenthal 2000) but not core snRNP assembly (Fig. 2B). To determine whether these mutations affect formation of the complex containing CstF, immunoprecipitations with the CstF-64 antibody were performed with the C. elegans crude embryonic extracts from strains expressing these mutated SL2 RNAs (Fig. 3). Samples were analyzed by primer extension in the presence of ddCTP, as in Figure 2B. The marked SL2 RNA (C17G) was able to form a complex containing CstF-64 (Fig. 3A, lane 6). Furthermore, when the second stem was mutated, SL2 RNA was still able to form a complex with CstF-64 (Fig. 3A, lane 9). Interestingly, however, complex formation was undetectable when the marked SL2 RNA also contained a single nucleotide substitution mutation in loop III (G93C; Fig. 3A, lane 12). Similar results were observed with a larger mutation to stem/loop III in which the entire loop was mutated (Stem 3 sub; Evans and Blumenthal 2000; data not shown). Therefore, the sequence of loop III, but not stem II, is required for complex formation with CstF-64.
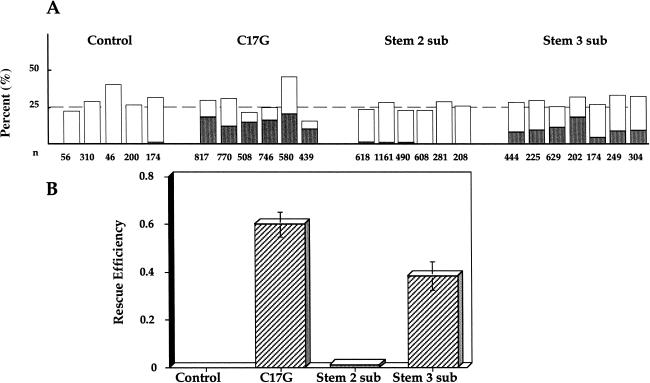
Stem 3 sub mutant SL2 RNA is able to partially rescue rrs-1
We showed previously that the Stem 3 sub mutation prevents trans-splicing to gpd-3, a natural SL2 acceptor (Evans and Blumenthal 2000). But does the mutation completely prevent any trans-splicing of the mutant SL2 RNA or has the mutant SL2 RNA simply lost its identity, resulting in the loss of trans-splicing specificity? To test these possibilities, we exploited the C. elegans rrs-1 mutant, in which all 110 copies of the 5S rRNA/SL1 RNA tandem gene cluster are deleted (Ferguson et al. 1996). Animals homozygous for this deletion die as embryos, but transgenic expression of SL2 RNA allows partial rescue, resulting in embryos that hatch but often die as larvae (Ferguson et al. 1996). We investigated whether the SL2 RNA genes carrying either the Stem 2 sub or the Stem 3 sub mutations resulted in functional SL RNAs by looking for rescue of the rrs-1 phenotype.
Without rescue, the rrs-1 heterozygotes produce ∼25% homozygous rrs-1 progeny that die as embryos (Fig. 4). Rescue of all the rrs-1/rrs-1 embryos to the larval stage should produce 25% larvae that are Unc (Uncoordinated). In Figure 4A, the percent stunted Unc larvae and the percent dead embryos are plotted for the transgenic progeny from each strain with a potentially rescuing SL2 construct. Figure 4B shows the rescue efficiency calculated from the data in Figure 4A. As expected, injection mixes lacking an SL2 plasmid failed to rescue (Fig. 4, control), and ∼25% of the transgenic progeny died as embryos. Inclusion of the C17G plasmid resulted in efficient partial rescue, producing GFP-positive stunted larvae in each strain, with 60% efficiency. Inclusion of plasmids encoding Stem 2 sub or Stem 3 sub had radically different effects on rescue efficiency. Stem 2 sub was completely unable to rescue, demonstrating that this mutation blocks trans-splicing. In contrast, Stem 3 sub was able to rescue almost as well (∼40% efficiency) as C17G. This result suggests that Stem 3 sub has lost its ability to interact productively with SL2-specific trans-splice acceptors at downstream positions in operons, but not its ability to trans-splice in general. We expect that if the entire loop of stem III can be replaced without loss of snRNP function, the single base change of G93C would function as well. We conclude that loop III is required for both interaction of SL2 snRNP with CstF and for SL2 specificity of trans-splicing, but is not required for SL snRNP function in general. In contrast, mutation of stem II results in a nonfunctional SL snRNP that still forms a complex with CstF.
Figure 4.
rrs-1 rescue by transgenic SL2. (A) Percentage of transgenic (GFP+) progeny that are dead embryos (open bars) or stunted larvae (shaded bars) are shown for each stable transgenic line. The total number of GFP+ progeny scored (n) is indicated for each line below the bar. (B) Rescue efficiency for each set of transgenic lines in A is presented as a fraction, where the rescue efficiency is calculated as an average (with standard deviation shown) from the number of stunted larvae divided by the 25% expected for 100% rescue of the rrs-1unc-76/rrs-1 unc-76 embryonic lethal class.
Discussion
Trypanosomes, flatworms, and perhaps even most nematodes apparently use only a single type of SL RNA to process pre-mRNAs. In contrast, pre-mRNA processing in C. elegans and its relatives in the Rhabditida family of nematodes use two different SL RNAs, SL1 and SL2. SL1 trans-splicing usually occurs quite close to the 5′ ends of pre-mRNAs and is specified by an outron, an intron-like sequence that ends in a functional 3′ splice site, but lacks an upstream 5′ splice site (Conrad et al. 1991, 1993, 1995). Although SL2 RNA is seven- to 10-fold less abundant than SL1 RNA, it is used preferentially to process the products of downstream genes in operon polycistronic pre-mRNAs. Therefore, a mechanism must exist for specifically using SL2 RNA at trans-splice sites between genes in multicistronic pre-mRNAs. In this report, we present evidence for a complex that contains both the SL2 RNA and the 3′ end formation factor CstF-64 that is likely to be responsible for this trans-splicing specificity.
A physical interaction between 3′ end formation and SL2 trans-splicing machineries
Elements of C. elegans polycistronic pre-mRNAs required for SL2 trans-splicing were investigated previously by expressing the mai-1/gpd-2/gpd-3 operon in transgenic worms under the control of a heat shock promoter. These experiments provided several lines of evidence suggesting an interaction between SL2 trans-splicing and 3′ end formation. First, mutation of the CPSF binding site, AAUAAA, of gpd-2 resulted in loss of splicing specificity to the gpd-3 trans-splice site located 110 nucleotides downstream (Kuersten et al. 1997). Second, replacing the AGUAAA variant sequence of the mai-1 gene with the canonical AAUAAA increased SL2 trans-splicing to the gpd-2 trans-splice site 127 nucleotides downstream almost 10-fold (Liu et al. 2001). These results provided support for some sort of role for CPSF in SL2 trans-splicing. Third, linker scan analysis of the intercistronic region between gpd-2 and gpd-3 identified a U-rich region beginning 29 nucleotides downstream of the site of polyadenylation of gpd-2 mRNA, which when mutated, resulted in a complete loss of SL2 trans-splicing to gpd-3 mRNA (Huang et al. 2001). The location of this element, as well as its base composition, is consistent with it being a binding site for the 3′ end formation factor CstF. Therefore, it was surprising to discover that mutation of the U-rich region had no effect on 3′ end formation of gpd-2 mRNA (Huang et al. 2001), but did eliminate SL2 trans-splicing to the downstream gene. An analysis of several other C. elegans intercistronic regions revealed in each case a conserved U-rich sequence in a location consistent with those of known CstF-binding sites. Whether these sequences actually serve as CstF-binding sites awaits further experimentation.
To test the hypothesis that a direct interaction occurs between 3′ end formation machinery and downstream trans-splicing machinery, we performed immunoprecipitations to look for snRNPs associated with CstF-64. We found that SL2 RNA is in a complex that includes CstF-64 (Fig. 1) consistent with the idea that CstF, bound to the U-rich site shown previously to be required for SL2 trans-splicing, is responsible for the preferential use of SL2 at these sites. We believe that the requirement for the CPSF-binding site, albeit a less stringent one, may be attributable to its function in increasing the affinity of CstF for the U-rich sequence, as mammalian CstF and CPSF have been shown to bind cooperatively to pre-mRNA (Murthy and Manley 1995). Therefore, CPSF interaction with AAUAAA may be the primary interaction required for 3′ end formation, whereas its role in SL2 trans-splicing may be ancillary. In contrast, the CstF interaction with the U-rich sequence is of primary importance for SL2 trans-splicing, but not for 3′ end formation.
Requirement for the loop III sequence in SL2-specific trans-splicing and for CstF complex formation
We showed previously that mutation of a single nucleotide in the highly conserved loop III of SL2 RNA completely prevented trans-splicing to the SL2 accepting gpd-3 mRNA (Evans and Blumenthal 2000). Here we show that this mutation, G93C, also prevents the SL2 snRNP from forming a complex that includes CstF-64 (Fig. 3). Although, trans-splicing of G93C to several SL1-accepting pre-mRNAs (e.g., act-1, rpl-37) was not detected as assayed by ddC primer extension (data not shown), it is possible that this was caused by the inability of this RNA to compete with endogenous SL1 snRNP. To examine whether stem III mutants could trans-splice to SL1-accepting pre-mRNAs in the absence of competing SL1 RNA, we tested our most severe stem III mutation, Stem 3 sub, for ability to rescue rrs-1. The fact that this mutant RNA was able to partially rescue rrs-1 demonstrated that mutation of loop III results in the loss of SL2 identity, as opposed to the loss of trans-splicing. Therefore, correct identity of the SL2 RNA correlates with its ability to form a complex with CstF-64. The importance of loop III in establishing SL2 identity and formation of a complex with CstF-64 is consistent with the high degree of conservation of the nucleotides in loop III of all SL2 RNAs sequenced (Evans et al. 1997). This is in contrast to the lack of conservation of SL1 RNA loop III and its apparent lack of importance in splicing to SL1 trans-splice sites (Evans and Blumenthal 2000).
The role of stem II in trans-splicing
The Stem 2 sub mutation prevented trans-splicing to an SL2-accepting mRNA, gpd-3, in vivo (Evans and Blumenthal 2000). Furthermore, it was unable to rescue rrs-1, suggesting that this region of the SL2 RNA is required for trans-splicing (Fig. 4). Previous studies have demonstrated a similar requirement for stem II in SL RNAs. Several nucleotides in the second stem of the SL RNA from the nematode Ascaris lumbricoides were shown to be required for in vitro trans-splicing (Denker et al. 1996). Stem II has also been shown to be important in trypanosome SL RNA function (Lucke et al. 1996; Strum and Campbell 1999) and protein binding (Xu et al. 2001). Therefore, the finding that the Stem 2 sub mutation eliminates snRNP function is not surprising. Interestingly, however, this mutated SL2 RNA has an intact stem/loop III and was able to form a complex containing CstF-64 (Fig. 3). Our results with the two mutant SL2 RNAs separate SL2 snRNP function from SL2 snRNP identity. The stem II mutation results in a snRNP that does not function, but nevertheless interacts with CstF. In contrast, the loop III mutation makes a functional snRNP that neither interacts with CstF nor works at SL2-accepting trans-splice sites. This suggests that stem/loop III contains a site that allows the SL2 snRNP to form a complex with CstF-64.
Mechanism of SL2 trans-splicing specificity
How does the much less abundant SL2 snRNP compete so effectively with the SL1 snRNP for trans-splicing to pre-mRNAs encoded by downstream genes in operons? Interaction with the 3′ end formation machinery is likely to be the answer. A schematic diagram of one possible model of SL2 trans-splicing is presented in Figure 5. In this model, the cooperative binding of CPSF to the AAUAAA 3′ end formation signal and CstF to the U-rich sequence between the genes determines the site of cleavage. SL2 RNA is specifically recruited to the trans-splice site via its interaction with the 3′ end formation machinery. We believe that the interaction is likely to be with CstF, rather than CPSF, because the proposed binding site for CstF, the U-rich sequence, is absolutely required for SL2 trans-splicing, whereas mutation of the CPSF-binding site merely reduces its efficiency. Furthermore, CPSF is presumably separated by 3′ end formation from the downstream RNA that is subjected to trans-splicing, whereas CstF is not. We present evidence that CstF-64 and SL2 RNA participate in the same complex, but not that they directly contact one another. We also do not know whether other CstF subunits or CPSF are part of this complex.
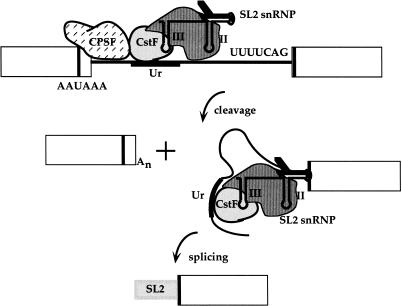
Figure 5.
A two-step model for polycistronic pre-mRNA processing. In the first step, CPSF and CstF collaborate to cleave the pre-mRNA between the two binding sites, as in vertebrate 3′ end formation. CPSF is bound to the AAUAAA and CstF is bound to the U-rich region just downstream. This region, called Ur, has been shown to be required for SL2-specific trans-splicing (Huang et al. 2001). In the second step, the mature 5′ end of the downstream mRNA is generated by trans-splicing with the SL2 snRNP, delivered to the nearby trans-splice site via its interaction with CstF. For simplicity, competing SL1 RNPs are not shown, however, SL1 RNA is seven- to 10-fold more abundant than SL2 RNA. The model illustrates a direct interaction between CstF and the SL2 snRNP. However, it remains possible that the interaction is indirect and is bridged by an unidentified factor.
We suggest that CstF may have a dual role in facilitating SL2 trans-splicing. First, following cleavage at the 3′ end formation site, the CstF bound to the U-rich sequence protects the downstream RNA from degradation by 5′ to 3′ exonucleases, and therefore allows time for trans-splicing to take place. Second, by the interaction with the SL2 snRNP shown here, CstF provides a binding site for the SL2 snRNP to bring it preferentially to sites following 3′ end formation sites. Trans-splicing adds a cap and so prevents degradation of the downstream mRNA. We have previously observed that some trans-splice sites between genes in operons receive a mixture of SL2 and SL1. In general, those intercistronic regions with a greater distance between genes tend to result in a decreased SL2/SL1 ratio (Zorio et al. 1994). We suggest that close proximity between the CstF-binding site, which must be near the 3′ end of the upstream gene, and the trans-splice site of the downstream gene is required for efficient use of the SL2 snRNP. This may be one explanation for the strong tendency of genes in operons to be ∼100 bp apart.
Interactions between splicing and 3′ end formation
Several instances of connections between splicing and 3′ end formation machinery have been reported previously. An interaction between polyadenylation and trans-splicing has been observed in the trypanosomes (LeBowitz et al. 1993; Matthews et al. 1994; Schurch et al. 1994; Lopez-Estrano et al. 1998), which also use 3′ end formation and trans-splicing to process polycistronic pre-mRNAs (Johnson et al. 1987; Muhich and Boothroyd 1988; Tschudi and Ullu 1988). Interestingly, a pyrimidine-rich element was identified in the trypanosome intercistronic region that was required for both trans-splicing and 3′ end formation (Matthews et al. 1994). However, in these organisms 3′ end formation is abolished by blocking trans-splicing (Ullu et al. 1993). In this paper we report a physical interaction between the 3′ end formation and trans-splicing machineries in C. elegans. It will be interesting to determine whether an interaction similar to that described for C. elegans exists in trypanosomes. Exon definition interactions have also been observed between 3′ end formation and cis-splicing of the last intron in pre-mRNA (Niwa et al. 1990; Niwa and Berget 1991). This connection has been shown to occur through a direct interaction between the 3′ splice site recognition factor, U2AF65, and poly(A) polymerase (Vagner et al. 2000). In this report, we present evidence that the connection between 3′ end formation and trans-splicing in polycistronic pre-mRNA processing may occur through formation of a complex containing the SL2 snRNP and CstF. In this case, 3′ end formation upstream is connected to splicing downstream, in contrast to exon definition, where splicing upstream is connected to 3′ end formation downstream.
Materials and methods
CstF-64 antibody production
F56A8.6, the C. elegans homolog of the mammalian 3′ end formation factor CstF-64, was identified by BLASTP. A cDNA encoding this protein (yk75h12) was used as template for PCR with oligonucleotides to the 5′ and 3′ ends of the gene as primers (5′, CCCCAAGCTTATGTCCGGGGGATAC; 3′, AAGCA CAAATACTCGAGAGACGAG). The resulting PCR product was digested with HindIII and XhoI and cloned into pFlag-MAC (Eastman Kodak) also digested with HindIII and XhoI. This results in an amino-terminal fusion of the protein with the Flag epitope. This recombinant protein was expressed in E. coli and affinity purified using M2 affinity gel (Eastman Kodak) following the manufacturer's protocol. The purified protein was used as an antigen to produce polyclonal antibodies in rabbits.
Strains and extracts
The SL2 mutant strains used in these experiments (C17G, BL1301; Stem 2 sub, BL1318; Stem 3 sub, BL1304; G93C, BL1329) and the protocol for preparing C. elegans crude embryonic extracts were described previously (Evans and Blumenthal 2000).
Immunoprecipitations
Aliquots of GammaBind Plus Sepharose (750 μL packed volume, Amersham Pharmacia) were washed five times with an equal volume of phosphate buffered saline (PBS). An equal volume of αCstF-64 antiserum or control serum (pre-immune or non-immune rabbit serum, Sigma) was added and the mixture was incubated at 4°C for 3 h. The antibody/bead mixture was washed four times with an equal volume of PBS, followed by three washes with an equal volume Buffer E (20 mM Tris at pH 8.0, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM DTT, 20% glycerol). Beads were blocked with Buffer E plus 0.1% nonfat dry milk for 1 h at 4°C, washed three times with Buffer E, and resuspended in an equal volume of Buffer E. To a 1.5 mL Eppendorf tube, the following were added in order: 500 μL Buffer E, 1 μL RNasin (Promega, 40 U/μL), 150 μL blocked beads, and 20 μL of crude C. elegans embryonic extract. These tubes were incubated for 5 h at 4°C with mixing. Beads were pelleted for 30 sec in a tabletop nanofuge. The unbound fraction was removed and the beads were washed three times with Buffer E plus 0.5% NP-40. The washed beads were resuspended in 200 μL 2× proteinase K buffer (20 mM Tris at pH 7.4, 25 mM EDTA, 300 mM NaCl, 2% SDS), plus 180 μL TE, plus 20 μL proteinase K (10 mg/mL). This mixture was incubated at 65°C for 20 min with frequent mixing. The beads were pelleted by centrifugation at 13,000 rpm for 1 min. The supernatant was extracted with phenol/chloroform, followed by chloroform, and ethanol precipitated with glycogen as a carrier molecule. Precipitated nucleic acids were pelleted, washed with 70% ethanol, and resuspended in 1× RQ1 DNase buffer and digested with RQ1 DNase (Promega) for 20 min at 37°C. The RNA was phenol/chloroform and chloroform extracted, ethanol precipitated with glycogen as a carrier molecule, pelleted, washed with 70% ethanol, and dried in a speed-vac. For the experiment depicted in Figure 1, RNA was selectively precipitated with LiCl instead of the treatment with RQ1 DNase described above.
Immunoprecipitations of snRNPs containing core or Sm proteins were performed using the Sm antibody as described previously (Evans and Blumenthal 2000).
Primer extensions
Primer extensions were carried out as described previously (Evans and Blumenthal 2000). The mixed oligonucleotide probe used in Figure 1 consisted of an equal mixture of primers complementary to U1 (AGCCCGCACCAAAAGTGC), U2 (GGGCCGAGCCCGGCAGCAGTGC), U5 (GACCCGCTTTC TCAAGG), U6 (TCCAATTTTAGTATATGTTC), SL1 (CGC CAAATTTCTTTGGGTCAG), and SL2 (GAACTCCAGCG TACC). Primer extension in the absence of a ddNTP results in products of 86, 180, 121, 30, 56, and 37 nucleotides, respectively. SL2 primer extensions shown in Figures 2 and 3 were performed using the oligonucleotide SL2 short PE in the presence of ddCTP (Evans and Blumenthal 2000).
rrs-1 rescue
JR41 heterozygous worms (rrs-1 unc-76/unc-61) were injected with plasmids containing 5S RNA, sur-5::gfp and either C17G, Stem 2 sub, or Stem 3 sub all at 100 ng/μL. Control injections contained 5S RNA and sur-5::gfp plasmids, but no marked SL2 RNA plasmid. Stable transgenic lines were selected by expression of GFP, checked for segregation of JR41 parental markers, that is, embryonic lethality and Unc-61 adults. From each stable line, four to eight heterozygous adults were cloned, transferred after 24 h and the transgenic GFP-expressing progeny were scored as growing larvae, unhatched embryos, or stunted larvae using a Leica M26 steomicroscope with epifluorescence.
Acknowledgments
We thank Y. Kohara for cDNA clone yk75h12; S. Kuersten and I. Mattaj for the anti-Sm serum; K. Ferguson and J. Rothman for the rrs-1 strain; M. Han for the sur-5:/:gfp fusion construct; and A.M. Deshpande for the 5S rRNA plasmid and suggesting the rrs-1 experiment. The C. elegans crude embryonic extract protocol was initially developed by S. Kuersten. In addition, we thank D. Zorio, A.M. Deshpande, and R. Singh for critical reading of the manuscript. This work was supported by research grant GM42432 from the National Institute of General Medical Sciences.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL blumenthal@uchsc.edu; FAX (303) 315-8215.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.920501.
References
- Blumenthal T, Steward K. RNA processing and gene structure. In: Riddle D, Blumenthal T, Meyer B, Priess J, editors. C. elegans II. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 117–145. [PubMed] [Google Scholar]
- Bruzik JP, Van Doren K, Hirsh D, Steitz J A. Trans-splicing involves a novel form of small nuclear ribonucleoprotein particles. Nature. 1988;335:559–562. doi: 10.1038/335559a0. [DOI] [PubMed] [Google Scholar]
- Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes & Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- Conrad R, Thomas J, Spieth J, Blumenthal T. Insertion of part of an intron into the 5′ untranslated region of Caenorhabditis elegans gene converts it into a trans-spliced gene. Mol Cell Biol. 1991;11:1921–1926. doi: 10.1128/mcb.11.4.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R, Liou RF, Blumenthal T. Conversion of a trans-spliced C. elegans gene into a conventional gene by introduction of a splice donor site. EMBO J. 1993;12:1249–1255. doi: 10.1002/j.1460-2075.1993.tb05766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R, Lea K, Blumenthal T. SL1 trans-splicing specified by AU-rich synthetic RNA inserted at the 5′ end of Caenorhabditis elegans pre-mRNA. RNA. 1995;1:164–170. [PMC free article] [PubMed] [Google Scholar]
- De Robertis EM, Lienhard S, Parisot R. Intracellular transport of microinjected 5S and small nuclear RNAs. Nature. 1982;295:572–577. doi: 10.1038/295572a0. [DOI] [PubMed] [Google Scholar]
- Denker JA, Maroney PA, Yu Y-T, Kanost RA, Nilsen TW. Multiple requirements for nematode spliced leader RNP function in trans-splicing. RNA. 1996;2:746–755. [PMC free article] [PubMed] [Google Scholar]
- Evans D, Blumenthal T. Trans-splicing of polycistronic Caenorhabditis elegans pre-mRNAs: Analysis of the SL2 RNA. Mol Cell Biol. 2000;20:6659–6667. doi: 10.1128/mcb.20.18.6659-6667.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D, Zorio D, MacMorris M, Winter CE, Lea K, Blumenthal T. Operons and SL2 trans-splicing exist in nematodes outside the genus Caenorhabditis. Proc Natl Acad Sci. 1997;94:9751–9756. doi: 10.1073/pnas.94.18.9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KC, Heid PJ, Rothman JH. The SL1 trans-spliced leader RNA performs an essential embryonic function in Caenorhabditis elegans that can also be supplied by SL2 RNA. Genes & Dev. 1996;10:1543–1556. doi: 10.1101/gad.10.12.1543. [DOI] [PubMed] [Google Scholar]
- Fritz A, Parisot RD, Newmeyer D, De Robertis EM. Small nuclear U-ribonucleoproteins in Xenopus laevis development. J Mol Biol. 1984;178:273–285. doi: 10.1016/0022-2836(84)90144-x. [DOI] [PubMed] [Google Scholar]
- Goncharov I, Palfi Z, Bindereif A, Michaeli S. Purification of the spliced leader ribonucleoprotein particle from Leptomonas collosoma revealed the existence of an Sm protein in trypanosomes. J Biol Chem. 1999;274:12217–12221. doi: 10.1074/jbc.274.18.12217. [DOI] [PubMed] [Google Scholar]
- Huang T, Kuersten S, Deshpande A, Spieth J, MacMorris M, Blumenthal T. Intercistronic region required for polycistronic pre-mRNA processing in Caenorhabditis elegans. Mol Cell Biol. 2001;21:1111–1120. doi: 10.1128/MCB.21.4.1111-1120.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X-Y, Hirsh D. A second trans-spliced RNA leader sequence in the nematode Caenorhabditis elegans. Proc Natl Acad Sci. 1989;86:8640–8644. doi: 10.1073/pnas.86.22.8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PJ, Looter JM, Borst P. Inactivation of transcription by UV irradiation of T. brucei provides evidence for a multicistronic transcription unit including a VSG gene. Cell. 1987;51:273–281. doi: 10.1016/0092-8674(87)90154-1. [DOI] [PubMed] [Google Scholar]
- Keller W, Minvielle-Sebastia L. A comparison of mammalian and yeast pre-mRNA 3′-end processing. Curr Opin Cell Biol. 1997;9:329–336. doi: 10.1016/s0955-0674(97)80004-x. [DOI] [PubMed] [Google Scholar]
- Krause M, Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuersten S, Lea K, MacMorris M, Spieth J, Blumenthal T. Relationship between 3′ end formation and SL2-specific trans-splicing in polycistronic Caenorhabditis elegans pre-mRNA processing. RNA. 1997;3:269–278. [PMC free article] [PubMed] [Google Scholar]
- LeBowitz JH, Smith HQ, Rusche L, Beverly SM. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes & Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA, Jr, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: Probes for molecular biology and autoimmune disease. Proc Natl Acad Sci. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR, Steitz JA. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci. 1979;76:5495–5499. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Huang T, MacMorris M, Blumenthal T. Interplay between AAUAAA and the trans-splice site in processing of a Caenorhabditis elegans operon pre-mRNA. RNA. 2001;7:176–181. doi: 10.1017/s1355838201002333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Estrano C, Tschudi C, Ullu E. Exonic sequences in the 5′ untranslated region of α-tubulin mRNA modulate trans splicing in Trypanosoma brucei. Mol Cell Biol. 1998;18:4620–4628. doi: 10.1128/mcb.18.8.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucke S, Xu G-L, Palfi Z, Cross M, Bellofatto V, Bindereif A. Spliced leader RNA of trypanosomes: In vivo mutational analysis reveals extensive and distinct requirements for trans-splicing and cap4 formation. EMBO J. 1996;15:4380–4391. [PMC free article] [PubMed] [Google Scholar]
- MacDonald CC, Wilusz J, Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol. 1994;14:6647–6654. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Hannon GJ, Denker JA, Nilsen TW. The nematode spliced leader RNA participates in trans-splicing as an Sm snRNP. EMBO J. 1990;9:3667–3673. doi: 10.1002/j.1460-2075.1990.tb07578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj IW, De Robertis EM. Nuclear segregation of U2 snRNA requires binding of specific snRNP proteins. Cell. 1985;40:111–118. doi: 10.1016/0092-8674(85)90314-9. [DOI] [PubMed] [Google Scholar]
- Matter L, Schopfer K, Wilhelm JA, Nyffenegger T, Parisot RF, De Robertis EM. Molecular characterization of ribonucleoprotein antigens bound by antinuclear antibodies: A diagnostic evaluation. Arth Rheum. 1982;25:1278–1283. doi: 10.1002/art.1780251102. [DOI] [PubMed] [Google Scholar]
- Matthews KR, Tschudi C, Ullu E. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNAs in trypanosomes. Genes & Dev. 1994;8:491–501. doi: 10.1101/gad.8.4.491. [DOI] [PubMed] [Google Scholar]
- Muhich ML, Boothroyd JC. Polycistronic transcripts in trypanosomes and their accumulation during heat shock: Evidence for a precursor role in mRNA synthesis. Mol Cell Biol. 1988;8:3837–3846. doi: 10.1128/mcb.8.9.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Watkins KP, Agabian N. Identification of a novel Y branch structure as an intermediate in trypanosome mRNA processing: Evidence for trans-splicing. Cell. 1986;47:517–525. doi: 10.1016/0092-8674(86)90616-1. [DOI] [PubMed] [Google Scholar]
- Murthy KG, Manley JL. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes & Dev. 1995;9:2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- Niwa M, Berget SM. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes & Dev. 1991;5:2086–2095. doi: 10.1101/gad.5.11.2086. [DOI] [PubMed] [Google Scholar]
- Niwa M, Rose SD, Berget SM. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes & Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- Rajkovic A, Davis RE, Simonsen JN, Rottman RM. A spliced leader is present on a subset of mRNAs from the human parasite Schistosoma mansoni. Proc Natl Acad Sci. 1990;87:8879–8883. doi: 10.1073/pnas.87.22.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurch N, Hehl A, Vassella E, Braun R, Roditi I. Accurate polyadenylation of procyclin mRNAs in Trypanosoma brucei is determined by pyrimidine-rich elements in the intergenic regions. Mol Cell Biol. 1994;14:3668–3675. doi: 10.1128/mcb.14.6.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieth J, Brooke G, Kuersten S, Lea K, Blumenthal T. Operons in C. elegans: Polycistronic mRNA precursors are processed by trans-splicing of SL2 to downstream coding regions. Cell. 1993;73:521–532. doi: 10.1016/0092-8674(93)90139-h. [DOI] [PubMed] [Google Scholar]
- Stover NA, Steele RE. Trans-spliced leader addition to mRNAs in a cnidarian. Proc Natl Acad Sci. 2001;98:5693–5698. doi: 10.1073/pnas.101049998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm NR, Campbell DA. The role of intron structures in trans-splicing and cap 4 formation for the Leishmania spliced leader RNA. J Biol Chem. 1999;274:19361–19367. doi: 10.1074/jbc.274.27.19361. [DOI] [PubMed] [Google Scholar]
- Sutton RE, Boothroyd JC. Evidence for trans-splicing in trypanosomes. Cell. 1986;47:527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier L-H, Keller M, Chan R, Fournier R, Weil JH, Imbault P. Short leader sequences may be transferred from small RNAs to pre-mature mRNAs by trans-splicing in Euglena. EMBO J. 1991;10:2621–2625. doi: 10.1002/j.1460-2075.1991.tb07804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JD, Conrad RC, Blumenthal T. The C. elegans trans-spliced leader RNA is bound to Sm and has a trimethylguanosine cap. Cell. 1988;54:533–539. doi: 10.1016/0092-8674(88)90075-x. [DOI] [PubMed] [Google Scholar]
- Tschudi C, Ullu E. Polygene transcripts are precursors to calmodulin mRNAs in trypanosomes. EMBO J. 1988;7:455–463. doi: 10.1002/j.1460-2075.1988.tb02833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E, Matthews KR, Tschudi C. Temporal order of RNA-processing reactions in trypanosomes: rapid trans splicing precedes polyadenylation of newly synthesized tubulin transcripts. Mol Cell Biol. 1993;13:720–725. doi: 10.1128/mcb.13.1.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S, Vagner C, Mattaj IW. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes & Dev. 2000;14:403–413. [PMC free article] [PubMed] [Google Scholar]
- Vandenberghe AE, Meedel TH, Hastings KEM. mRNA 5′-leader trans-splicing in the chordates. Genes & Dev. 2001;15:294–303. doi: 10.1101/gad.865401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren K, Hirsh D. Trans-spliced leader RNA exists as small nuclear ribonucleoprotein particles in Caenorhabditis elegans. Nature. 1988;335:556–559. doi: 10.1038/335556a0. [DOI] [PubMed] [Google Scholar]
- Xu P, Wen L, Benegal G, Wang X, Buck GA. Identification of a spliced leader RNA binding protein from Trypanosoma cruzi. Mol Biochem Parasitol. 2001;112:39–49. doi: 10.1016/s0166-6851(00)00341-8. [DOI] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DAR, Cheng NN, Blumenthal T, Spieth J. Operons as a common form of chromosomal organization in C. elegans. Nature. 1994;372:270–272. doi: 10.1038/372270a0. [DOI] [PubMed] [Google Scholar]