Abstract
Erythroid Krüppel-like factor (EKLF) plays an important role in erythroid development by stimulating β-globin gene expression. We have examined the details by which the minimal transactivation domain (TAD) of EKLF (EKLFTAD) interacts with several transcriptional regulatory factors. We report that EKLFTAD displays homology to the p53TAD and, like the p53TAD, can be divided into two functional subdomains (EKLFTAD1 and EKLFTAD2). Based on sequence analysis, we found that EKLFTAD2 is conserved in KLF2, KLF4, KLF5, and KLF15. In addition, we demonstrate that EKLFTAD2 binds the amino-terminal PH domain of the Tfb1/p62 subunit of TFIIH (Tfb1PH/p62PH) and four domains of CREB-binding protein/p300. The solution structure of the EKLFTAD2/Tfb1PH complex indicates that EKLFTAD2 binds Tfb1PH in an extended conformation, which is in contrast to the α-helical conformation seen for p53TAD2 in complex with Tfb1PH. These studies provide detailed mechanistic information into EKLFTAD functions as well as insights into potential interactions of the TADs of other KLF proteins. In addition, they suggest that not only have acidic TADs evolved so that they bind using different conformations on a common target, but that transitioning from a disordered to a more ordered state is not a requirement for their ability to bind multiple partners.
Keywords: hematopoiesis, NMR spectroscopy, transcriptional activators, intrinsically unstructured domain, transcription factor IIE
The Krüppel-like factors (KLF) are regulatory proteins that function in a number of tissue-specific processes (1, 2). All KLF proteins contain a DNA-binding domain (DBD) consisting of three zinc fingers positioned at their carboxyl-terminal end that enables them to bind to “GT-box” or “CACCC” sites (1, 2). KLF proteins vary considerably at their amino-terminal region, and they can be divided into subgroups based on this region (1, 2). The amino-terminal regions of several KLF proteins contain transcriptional regulatory domains including acidic transactivation domains (TADs), Sin-3 interacting repressor domains, and CtBP2 interacting repressor domains (PVLS/T motifs) (2).
Erythroid Krüppel-like factor (EKLF/KLF1) is an activator that plays a key role in erythroid development (3, 4). EKLF stimulates β-globin gene expression and helps enable the switch from the fetal γ-globin to the adult β-globin (5–9). EKLF specifically binds to CAACC elements (1, 3) and either the absence of EKLF or mutations in the β-globin CAACC element leads to severe β-thalassemia (5, 6, 10). In addition, EKLF plays a role in generating an open chromatin structure on the adult β-globin gene through interactions with transcriptional regulatory factors (11–16).
The human EKLF protein has 362 amino acids and contains two functional domains: the carboxyl-terminal DBD (residues 292–362) and an amino-terminal TAD (residues 1–271) (3, 17). Like the DBD, the TAD plays an important role in EKLF’s ability to activate β-globin expression (16–18). Initial experiments demonstrated that the first 100 residues constituted the minimal TAD of EKLF (EKLFTAD) (17). Subsequent experiments demonstrated that the first 40 residues within EKLFTAD play a role in recruiting cofactors required for β-globin activation and that residues 50 to 90 are essential for β-globin gene activation (17). EKLFTAD is highly acidic, and the combination of acidic and hydrophobic amino acids within this domain is similar to that found in the TAD of other activators including the tumor suppressor protein p53 and the herpes simplex viral protein 16 (VP16) (19, 20). The presence of two critical regions within EKLFTAD is also consistent with the fact that the TAD of both p53 and VP16 contain two subdomains each capable of activating transcription (21, 22). In p53TAD, the two subdomains (p53TAD1 and p53TAD2) participate in distinct interactions with general transcription factors, the mediator complex, MDM2, and the histone-acetyl transferase (HAT) CREB-binding protein (CBP)/p300 (23–25).
Despite the importance of the EKLFTAD, mechanistic details by which it interacts with other proteins to regulate EKLF function are limited. However, like other acidic TADs, it is thought that EKLFTAD participates in interactions with transcriptional regulatory factors such as general transcription factors, chromatin remodeling complexes, and HATs (12–14, 16). In this article, we demonstrate that EKLFTAD displays sequence homology to the p53TAD and contains two functional subdomains (EKLFTAD1 and EKLFTAD2). Sequence comparisons indicate that EKLFTAD2 is conserved in other KLF proteins and directly binds the amino-terminal PH domain of the Tfb1/p62 subunit of TFIIH (Tfb1PH/p62PH) and four domains of CBP/p300. NMR structural analysis of an EKLFTAD2/Tfb1PH complex demonstrates that EKLFTAD2 binds Tfb1PH in a unique manner that is distinct from the TAD of either p53 or VP16 and that Trp73 is a key residue at the interface (26, 27). Our studies suggest that acidic TADs have evolved so that they are able to bind a common target using different mechanisms and that coupled folding and binding is not a strict requirement for acidic TADs to bind to multiple partners.
Results
EKLFTAD Resembles the TAD of p53.
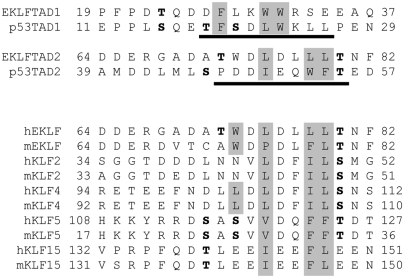
Based on mutational analysis and sequence comparison with p53 (Fig. 1), human EKLFTAD can be divided in two subdomains EKLFTAD1 (residues 1–40) and EKLFTAD2 (residues 51–90) (17). To test if these subdomains can function independently, we determined if they are capable of activating transcription when tethered to a heterologous DBD as previously observed for p53TAD1 (residues 1–40) and p53TAD2 (residues 40–73). Similar to other studies in yeast (22), p53TAD1 and p53TAD2 are both able to stimulate transcription of a lacZ reporter gene when fused to the LexA-DBD, and the two subdomains of p53TAD activate transcription at levels just slightly lower (81% and 71%, respectively) than the positive control (Fig. S1). Likewise, EKLFTAD1 and EKLFTAD2 activate transcription when fused to the LexA-DBD at levels 74% and 60% relative to the positive control (Fig. S1). These experiments demonstrate that EKLFTAD1 and EKLFTAD2 are both able to independently activate transcription in the yeast system when tethered to the LexA-DBD.
Fig. 1.
EKLFTAD1 and EKLFTAD2 are homologous to p53TAD1 and p53TAD2. (Top) Sequence alignments of EKLFTAD1 and EKLFTAD2 from human EKLF with p53TAD1 and p53TAD2 from human p53. The residues of p53 that form helices in the p53TAD1/MDM2 complex (23) and the p53TAD2/Tfb1PH complex (27) are underlined in black. The hydrophobic residues at the binding interfaces of the p53TAD1/MDM2 complex and the p53TAD2/Tfb1PH are in gray. Several known or potential phosphorylation sites are in bold. (Lower) Sequence alignment of regions of other KLF proteins (mouse and human) that share homology with EKLFTAD2. The KLF proteins are members of either KLF group 3 (EKLF, KLF2, KLF4) or KLF group 4 (KLF5, KLF15) based on phylogenetic analysis (1). Key hydrophobic residues are shaded gray and potential phosphorylation sites are in bold.
EKLFTAD2 Is Conserved in Other KLF Proteins.
The KLF family can be divided into subgroups based on either phylogenetic analysis or on the presence of specific domains at the amino-terminal end of the protein (1, 2). In an attempt to identify other KLF members with acidic TADs related to either EKLFTAD1 or EKLFTAD2, we analyzed the amino acid sequences of all known human and mouse KLF proteins. Our searches failed to identify KLF proteins containing a region with significant homology to the region of EKLFTAD1 that aligns with the region of p53TAD1 that forms a helix when bound to MDM2 (23). However, we did identify four KLF proteins (KLF2, KLF4, KLF5, and KLF15) containing a region with significant homology to the region of EKLFTAD2 that resembles the region of p53TAD2 that forms a helix when bound to Tfb1PH (Fig. 1) (27). This result is consistent with the fact that EKLF, KLF2, KLF4, KLF5, and KLF15 are members of group 3 and group 4 of the human KLF proteins based on the phylogenetic analysis (1).
EKLFTAD2 Binds Tfb1PH/p62PH.
Recent studies have demonstrated that TFIIH is a rate-limiting factor during the elongation phase of β-globin gene expression (28, 29), and this is consistent with the fact that patients with mutations in TFIIH suffer from β-thalassemia (30). Together, these studies indicate that TFIIH plays a crucial role in β-globin gene expression. Recently, we have shown that the TADs of p53 and VP16 interact with the amino-terminal PH domain of the Tfb1/p62 (Tfb1PH/p62PH) subunit of TFIIH (26, 27). In the case of the p53, the interaction is specific for p53TAD2 (27). Given the sequence homology between the TADs of p53 and EKLF (Fig. 1), we examined whether or not EKLFTAD1 and EKLFTAD2 interact with Tfb1PH/p62PH. To quantitatively determine the apparent dissociation constants (Kd), isothermal titration calorimetry (ITC) studies were performed. The ITC experiments demonstrate that both p62PH and Tfb1PH are able to bind EKLFTAD2 with similar Kd values (1.1 ± 0.1 μM and 1.0 ± 0.2 μM, respectively) (Table 1). A weaker affinity is observed for Tfb1PH with EKLFTAD1 (11 ± 1 μM) by ITC. The Kd values for EKLFTAD2 are comparable to those previously determined for the interaction of p62PH and Tfb1PH with p53TAD2 (3.2 ± 0.6 μM and 0.39 ± 0.07 μM, respectively) (27).
Table 1.
Kd (μM) values determined by ITC
| Kd, μM | ||
| Tfb1PH | EKLFTAD2 | 1.0 ± 0.2 |
| p62PH | EKLFTAD2 | 1.1 ± 0.1 |
| CBP KIX | EKLFTAD2 | 1.2 ± 0.7 |
| CBP IBiD | EKLFTAD2 | 4.2 ± 2.0 |
| CBP TAZ1 | EKLFTAD2 | 3.0 ± 1.0 |
| CBP TAZ2 | EKLFTAD2 | 0.74 ± 0.08 |
| p62PH | EKLF TAD2 W73P | n.b. |
| CBP IBiD | EKLF TAD2 W73P | n.b. |
n.b., no binding detected.
EKLDTAD2 and p53TAD2 Share a Common Binding Site.
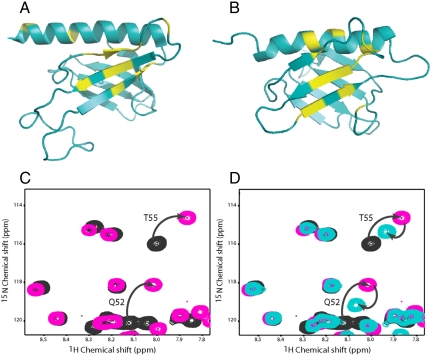
To help define the binding site for EKLFTAD2 on both Tfb1PH and p62PH, we performed NMR chemical shift titration studies. Additions of unlabeled-EKLFTAD2 to either 15N-Tfb1PH or 15N-p62PH resulted in significant changes in 1H and 15N chemical shifts for several signals in both 1H-15N heteronuclear single quantum coherence (HSQC) spectra (Fig. S2). When mapped onto the structure of either Tfb1PH (Fig. 2A) or p62PH (Fig. 2B), the residues exhibiting significant changes upon addition of EKLFTAD2 are located in strands β5, β6, β7 and the H1 helix. The chemical shift perturbations induced by EKLFTAD2 in both Tfb1PH and p62PH are very similar to those observed for p53 binding to Tfb1PH and p62PH (27). In addition, NMR displacement experiments demonstrate that p53TAD2 and EKLFTAD2 compete for a common binding site on Tfb1PH (Fig. 2 C and D).
Fig. 2.
EKLFTAD2 and p53TAD2 share a common binding site on Tfb1PH/p62PH. Ribbon models of the 3D structures of Tfb1PH (A) and p62PH (B). The amino acids showing a significant chemical shift change (Δδ(ppm) > 0.15; Δδ = [(0.17ΔNH)2 + (ΔHN)2]1/2) upon formation of a complex with EKLFTAD2 are shown in yellow (A and B). (C) Overlay of a selected region from the two-dimensional 1H-15N HSQC spectra for a 0.5-mM sample of 15N-labeled p53TAD2 in the free form (black) and in the presence of 0.4 mM unlabeled Tfb1PH (pink). (D) Overlay of a selected region from the two-dimensional 1H-15N HSQC spectra for a 0.5-mM sample of 15N-labeled p53TAD2 in the free form (black), in the presence of 0.4 mM unlabeled Tfb1PH (pink), and after addition of 0.5 mM unlabeled EKLFTAD2 (aqua). Signals that undergo significant changes in 1H and 15N chemical shifts upon formation of the complex with Tfb1PH (C) and that return toward their original position following the addition of EKLFTAD2 (D) are indicated by arrows.
EKLFTAD2 Binds to CBP/p300.
The p53TAD also interacts with the histone acetyltransferases CBP and p300 (CBP/p300) and acetylation of p53 by CBP/p300 is essential for p53-dependent activation (31, 32). Distinct interactions occur with both subdomains of the p53TAD and p53TAD2 interacts with at least four domains (TAZ1/CH1, KIX, the TAZ2/CH3, and IBID) of CBP/p300 (24, 25). Given the fact that EKLF has been shown to interact with CBP/p300 and acetylation of EKLF by CBP is crucial for activation of ß-globin gene expression (12–14), we tested if EKLFTAD2 also binds to the same four domains of CBP/p300 as p53TAD2 (24, 25). As with p53TAD2 (24, 25), ITC studies show that all four domains of CBP/p300 bind to EKLFTAD2 with Kd values ranging between 0.74 ± 0.08 μM (TAZ2 domain) and 4.2 ± 2.0 μM (IBiD domain) (Table 1).
Structure Determination of the Tfb1PH/EKLFTAD2 Complex.
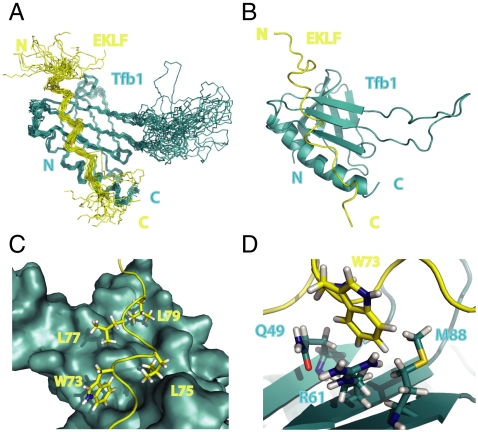
Next, we pursued NMR structural studies of a complex containing Tfb1PH and EKLFTAD2. Tfb1PH (yeast) was chosen as a model system for NMR studies, because it is more stable then the homologous p62PH (human) in solution. The three-dimensional structures of the Tfb1PH/EKLFTAD2 complex were calculated using 1686 NOE-derived distance restraints, 47 intermolecular NOEs (Table S1), 36 hydrogen-bond restraints, and 116 dihedral angle restraints. The structure of the Tfb1PH/EKLFTAD2 complex is well defined by the NMR data. The 20 lowest-energy structures (Fig. 3A) are characterized by good backbone geometry, no significant restraint violation, and low pairwise rmsd values (Table S2).
Fig. 3.
Structure of the Tfb1PH/EKLFTAD2 Complex. (A) Overlay of the 20 lowest-energy structures of the complex between Tfb1PH (blue) and EKLFTAD2 (yellow). The structures were superimposed using the backbone atoms C′, Cα, and N of residues 4–65 and 85–112 of Tfb1PH and residues 59–84 of EKLFTAD2. (B) Ribbon model of the lowest-energy conformer of the complex between Tfb1PH (blue) and EKLFTAD2 (yellow). (C) Three-dimensional structure of Tfb1PH is shown as molecular surface (blue), and EKLFTAD2 is represented as a tube (yellow). The side chains of several hydrophobic residues of EKLFTAD2 (Trp73, Leu75, Leu77, and Leu79) that form the interface with Tfb1PH are shown as sticks. (D) Three-dimensional structure of Tfb1PH is shown as ribbon (blue), and EKLFTAD2 is represented as a tube (yellow). The side chain of Trp73 (EKLFTAD2) is positioned in a pocket formed by Gln49, Arg61, and M88 of Tfb1.
The structure of Tfb1PH in complex with EKLFTAD2 is comparable to that of free Tfb1PH (33), indicating that Tfb1PH does not undergo significant structural changes between the free and complex form (Fig. 3 A and B). Tfb1PH consists of two perpendicular antiparallel-β sheets arranged in a β-sandwich (β1–β7) and flanked on one side by a long α-helix (H1). EKLFTAD2 binds Tfb1PH in an elongated conformation devoid of any regular secondary structural elements (Fig. 3 A and B). β-strands β5–β7 and the H1 α-helix of Tfb1PH form an extensive interface with residues between Ala69 and Thr80 of EKLFTAD2 (Fig. 3 C and D), and this is consistent with chemical shift changes of 15N-EKLFTAD2 upon addition of Tfb1PH (Fig. S2).
Tfb1PH/EKLFTAD2 Interface.
EKLFTAD2 binds to a contiguous shallow groove formed by β-strands β5, β6, and β7 and the H1 helix of Tfb1PH. The shallow groove of Tfb1PH is surrounded by positively charged residues (Lys47, Lys57, Arg61, Arg86, Lys97, Lys101, and Lys112) that position the negatively charged EKLFTAD2 (Fig. S3) so that it can participate in a series of van deer Waals contacts with hydrophobic pockets dispersed along the entire length of the groove (Fig. 3C). Numerous contacts are formed between Tfb1PH and Ala69, Ala71, Thr72, Trp73, Leu75, Leu77, Leu79, and Thr80 of EKLFTAD2 (Fig. 3 C and D). The key residue of EKLFTAD2 along the interface with Tfb1PH appears to be Trp73, which inserts into a pocket formed by Gln49, Ala50, Thr51, Met59, Leu60, Arg61, and Met88 of Tfb1PH. This pocket is centrally located within the groove, and in this pocket Trp73 forms a cation-π interaction with Arg61 and an amino-π interaction with Gln49 (Fig. 3D). Other important van der Waals contacts are generated by Ala71 of EKLFTAD2 inserting into a cleft formed by residues in strand β5 (Thr51), strand β6 (Met59), and the loop between strands β5 and β6 (Pro52 and Lys57) of Tfb1PH at one end of the groove and Leu79 of EKLFTAD2 inserting into a pocket formed by residues in strand β5 (Leu48) and in helix H1 (Gln105, Ile108, and Ser109) of Tfb1PH at the other end. In addition to the interactions involving hydrophobic residues, there are two potential salt bridges between acidic residues of EKLFTAD2 and Tfb1PH (Fig. S3). One is formed between Asp76 of EKLFTAD2 and Lys101 of Tfb1PH, and the second one is formed between Asp70 of EKLFTAD2 and Lys57 of Tfb1PH.
Trp73 Is Crucial for EKLFTAD Functions.
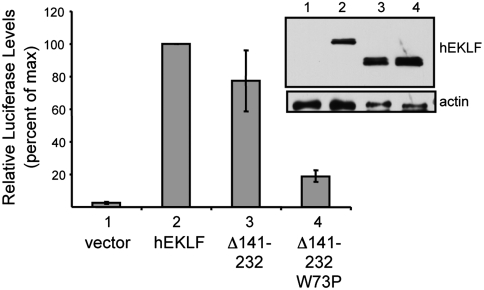
To examine the functional role of Trp73 of EKLF, we generated a proline mutant (W73P) and tested EKLF function in an in vivo β-globin activation assay. Because EKLF contains a second activation domain (residues 140–232 in hEKLF) that is also sufficient for β-globin gene expression (34, 35), the mutation was tested in an EKLF construct deleted for the redundant activation domain (EKLFW73PΔ140–232). In this assay, EKLFΔ140–232 activates β-globin expression in K562 erythroid cells to similar levels as full-length EKLF as expected. In contrast, the EKLFW73PΔ140–232 mutant shows significantly lower activity (approximately 5-fold) (Fig. 4), indicating that Trp73 is important for the β-globin activation associated with the minimal activation domain of EKLF (EKLFTAD). Likewise, a W73P mutant generated in the context of a 104 amino acid mEKLF TAD segment fused to the GAL4 DBD significantly reduces its ability to activate a GAL4-TK-β-globin-reporter gene in K562 cells (17) (Fig. S4). These in vivo results are also consistent with ITC studies demonstrating that an EKLFTAD2 (W73P) mutant is unable to bind to either p62PH or the IBiD domain of CBP/p300 (Table 1). Together, these results indicate that Trp73 of EKLF forms important interactions with CBP/p300 and TFIIH that may be required for EKLF functions in vivo.
Fig. 4.
Trp73 influences in vivo activation of β-globin gene expression. K562 cells were cotransfected with plasmids expressing the luciferase reporter gene under control of ß-globin promoter along with the EKLF constructs (EKLF, EKLFΔ140–232, and EKLFW73PΔ140–232). A plasmid expressing Renilla luciferase was included as a control for normalization of transfection efficiency. Transcriptional activities were normalized to that of full-length hEKLF, which was set at 100%. Error bars represent standard error of the mean of three independent experiments, each performed in triplicate. (Inset) The level of expression of the proteins after transfection into Cos7 cells as monitored by Western blot with an antibody against EKLF. Actin levels are shown as a loading control.
Discussion
The structures of EKLFTAD2, p53TAD2 (27), and VP16C (26) bound to Tfb1PH allow us to make comparisons between the three acidic TADs bound to a common target. In both the p53TAD2/TFb1PH and the VP16C/Tfb1PH complexes (26, 27), three hydrophobic residues on one face of the α-helix make crucial contacts with Tfb1PH and in both cases one of the hydrophobic residues (Phe54 of p53TAD2 and Phe479 of VP16C) is inserted into the same pocket on Tfb1PH as Trp73 of EKLFTAD2 (Fig. S5 A and B). In this pocket, the phenylalanine residues of p53TAD2 and VP16C participate in virtually identical interactions with Tfb1PH as observed for Trp73 in EKLFTAD2. Surprisingly, this is the only real similarity between the EKLFTAD2/Tfb1PH complex and either the p53TAD2/Tfb1PH or VP16C/Tfb1PH complex along the binding interface. However, by binding in an elongated form, EKLFTAD2 contributes a significantly larger interface (≈700 Å2) than either p53TAD2 (≈380 Å2) or VP16C (≈500 Å2) in complex with Tfb1PH.
Previous structural studies indicate that acidic TADs have the capability to interact with multiple partners due to their ability to couple folding with binding (23, 36–38). The transition of p53TAD2 and VP16C from a disordered state to a helical conformation when bound to Tfb1PH is consistent with this idea (26, 27). In contrast, EKLFTAD2 binds to Tfb1PH in a nonhelical form, and this indicates that acidic TADs can recognize identical target proteins using different structural conformations. The structures of p53TAD2, VP16C, and EKLFTAD2 in complex with Tfb1PH/p62PH also suggest that coupling folding and binding is common, but not a strict requirement for TADs interacting with their partner proteins. This is further supported by the structure of a complex containing the acidic carboxyl-terminal domain of the alpha subunit of the general transcription factor IIE (TFIIEαCTD) bound to p62PH. In this structure, an acidic region of TFIIEαCTD binds in a virtually identical conformation as EKLFTAD2 along the same binding interface and inserts a phenylalanine side chain (Phe387) in the same pocket of Tfb1PH as Trp73 (Figs. S5C and Fig. S6) (39, 40).
A similar situation is observed with the structures of the TADs of STAT2, CITED2, and HIF-1α bound to the TAZ1 domain of CBP (36, 38, 41). These TADs bind to the same surface of TAZ1, but they bind using two different orientations (N to C and C to N) and by utilizing different combinations of helical segments. Despite binding through distinct mechanisms they differ from EKLFTAD2 in that all three of these acidic TADs require the formation of an alpha helical conformation through a coupled folding and binding mechanism (36, 38, 41). However, they clearly demonstrate that the target (TAZ1) is able to bind acidic TADs containing different structural conformations.
The interaction of EKLFTAD with Tfb1PH/p62PH is consistent with the fact that TFIIH is required for β-globin expression and a mutation in TFIIH leads to β-thalassemia (28–30). The structure of the Tfb1PH/EKLFTAD2 complex demonstrates that Trp73 is crucial for this interaction and is supported by in vivo results demonstrating that mutations of Trp73 leads to significantly lower levels of β-globin gene activation in K562 cells. The results demonstrating that EKLFTAD2 binds the same four subdomains of CBP as p53TAD2 are in agreement with studies demonstrating that acetylation of EKLF by CBP is required for activation of β-globin gene expression (12–14). As was the case for binding to Tfb1PH/p62PH, Trp73 is important for EKLFTAD2 interaction with CBP. These results also suggest that p53TAD2 and EKLFTAD2 may share other common binding partners, but they may bind these targets using different conformations.
Sequence comparisons indicate that a domain similar to EKLFTAD2 is also present in KLF2, KLF4, KLF5, and KLF15 (Fig. 1) (1). This is supported by studies demonstrating that these four KLF proteins all interact with CBP/p300 through the region that is homologous to EKLFTAD2 (42–45). This strongly suggests that KLF2, KLF4, KLF5, and KLF15 will bind to a number of the same regulatory factors as EKLFTAD2. Additional structural and functional studies are needed to determine whether these homologous regions of the other KLF proteins bind to their target proteins in an elongated form like EKLFTAD2 or if they form helices as typically seen with acidic TADs such as p53 and VP16. In addition to the minimal EKLFTAD, EKLF contains a second domain (residues 140–232) that is sufficient for activation of β-globin gene expression (34, 35). Mutational analysis demonstrated that these domains appear to serve redundant functions (34), although the mechanistic details by which this second region of EKLF functions is not understood (35). The primary sequences of the two domains are very different, with EKLFTAD being very acidic (pI = 3.8) and the region between residues 140 and 232 being very basic (pI = 9.7) with only two acidic residues. However, the redundant functions suggest the two domains recruit the same transcriptional regulatory factors and future studies are needed to determine if this second domain of EKLF also binds TFIIH and CBP/p300.
Experimental Procedures
Cloning and Purification of Recombinant Proteins.
EKLFTAD1 (residues 1–40) and EKLFTAD2 (residues 51–90) were cloned into the pGEX-2T vector generating GST-fusion proteins starting with the human EKLF cDNA. The KIX domain (residues 586–672) of human CBP was provided by Alanna Schepartz (Yale University, New Haven, CT). The IBiD domain (residues 2065–2115) of CBP was cloned in the pGEX-2T vector generating GST-fusion protein starting with the human CBP cDNA. The TAZ1 domain (residues 345–439) of CBP was provided by Timothy Osborne (University of California, Irvine, CA). The TAZ2 domain (residues 1723–1812)/C1738A, C1746A, C1789A, C1790A) of p300 was provided by Ettore Appella (National Institutes of Health, Bethesda, MD). The GST-Tfb1PH (residues 1–115) and GST-p62PH (residues 1–108) were cloned as previously described (33). QuickChange II site-directed mutagenesis kit (Startagene) was used to carry out site-directed mutagenesis. For details of protein purifications, see SI Experimental Procedures.
Isothermal Titration Calorimetry Binding Experiments.
ITC titrations were performed at 25 °C as previously described (27). Proteins were dialyzed into 20 mM Tris buffer pH 7.5 except for KIX, TAZ1, and TAZ2, which were dialyzed into 20 mM sodium phosphate buffer pH 6.4. The protein concentrations were determined from A280. All injections fit a single-binding site mechanism with 1∶1 stoichiometry.
NMR Spectroscopy.
For details of NMR samples, see SI Experimental Procedures. NMR spectra were collected at 300 K on Varian Unity Inova 500-, 600-, and 800-MHz spectrometers. Interproton distance restraints were derived from 3D 15N-edited NOESY-HSQC and 13C-edited HMQC-NOESY spectra (τm = 90 ms). Intermolecular distance restraints were obtained from 3D 15N/13C {F1}-filtered, {F3}-edited NOESY experiments (τm = 90 ms). Hydrogen-bond restraints were derived from slowly exchanging amide protons determined in H/D exchange studies.
Structures Calculation.
The NOE-derived distance restraints were defined as strong (1.8–2.8 Å), medium (1.8–3.4 Å), weak (1.8–5.0 Å), and very weak (3.3–6.0 Å). Backbone dihedral angles were derived with the program TALOS (46). The EKLFTAD2/Tfb1PH structures were calculated using the program CNS (47). The figures were generated with PyMol (http://www.pymol.org).
Transactivation Assays in K562 Blood Cells.
The EKLFΔ140–232 and EKLFW73PΔ140–232 constructs were prepared from full-length hEKLF. K562 cells were cotransfected with plasmids expressing the luciferase reporter gene under control of the ß-globin promoter along with the EKLF constructs as described previously (17, 48). Expression levels of the proteins were determined with an antibody directed against EKLF.
Supplementary Material
Acknowledgments.
NMR experiments (800 MHz) were recorded at the Québec/Eastern Canada NMR Facility. This work was supported by Canadian Institutes of Health Research Grant MOP-209826 (to J.G.O.) and by United States Public Health Service Grant NIH DK46865 (to J.J.B.). The microcalorimeter was purchased with funds from the National Sciences and Engineering Research Council of Canada.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The NMR, atomic coordinates, chemical shifts, and restraints have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2L2I).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017029108/-/DCSupplemental.
References
- 1.Bieker JJ. Kruppel-like factors: Three fingers in many pies. J Biol Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- 2.Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4(2):206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodge D, et al. A global role for EKLF in definitive and primitive erythropoiesis. Blood. 2006;107:3359–3370. doi: 10.1182/blood-2005-07-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perkins AC, Sharpe AH, Orkin SH. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 6.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375(6529):316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 7.Wijgerde M, et al. The role of EKLF in human beta-globin gene competition. Genes Dev. 1996;10:2894–2902. doi: 10.1101/gad.10.22.2894. [DOI] [PubMed] [Google Scholar]
- 8.Borg J, et al. Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nat Genet. 2010;42(9):801–U100. doi: 10.1038/ng.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou DW, Liu KM, Sun CW, Pawlik KM, Townes TM. KLF1 regulates BCL11A expression and gamma- to beta-globin gene switching. Nat Genet. 2010;42(9):742–744. doi: 10.1038/ng.637. [DOI] [PubMed] [Google Scholar]
- 10.Feng WC, Southwood CM, Bieker JJ. Analyses of beta-thalassemia mutant DNA interactions with erythroid Kruppel-like factor (EKLF), an erythroid cell-specific transcription factor. J Biol Chem. 1994;269:1493–1500. [PubMed] [Google Scholar]
- 11.Chen XY, Bieker JJ. Stage-specific repression by the EKLF transcriptional activator. Mol Cell Biol. 2004;24:10416–10424. doi: 10.1128/MCB.24.23.10416-10424.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W, Bieker JJ. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc Natl Acad Sci USA. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang W, Kadam S, Emerson BM, Bieker JJ. Site-specific acetylation by p300 or CREB binding protein regulates erythroid Kruppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol Cell Biol. 2001;21:2413–2422. doi: 10.1128/MCB.21.7.2413-2422.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sengupta T, Chen K, Milot E, Bieker JJ. Acetylation of EKLF is essential for epigenetic modification and transcriptional activation of the beta-globin locus. Mol Cell Biol. 2008;28:6160–6170. doi: 10.1128/MCB.00919-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bottardi S, Ross J, Pierre-Charles N, Blank V, Milot E. Lineage-specific activators affect beta-globin locus chromatin in multipotent hematopoietic progenitors. EMBO J. 2006;25:3586–3595. doi: 10.1038/sj.emboj.7601232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sengupta T, Cohet N, Morle F, Bieker JJ. Distinct modes of gene regulation by a cell-specific transcriptional activator. Proc Natl Acad Sci USA. 2009;106:4213–4218. doi: 10.1073/pnas.0808347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Bieker JJ. Erythroid Krueppel-like factor (EKLF) contains a multifunctional transcriptional activation domain important for inter- and intramolecular interactions. EMBO J. 1996;15:5888–5896. [PMC free article] [PubMed] [Google Scholar]
- 18.Ouyang L, Chen X, Bieker JJ. Regulation of erythroid Kruppel-like factor (EKLF) transcriptional activity by phosphorylation of a protein kinase casein kinase II site within its interaction domain. J Biol Chem. 1998;273:23019–23025. doi: 10.1074/jbc.273.36.23019. [DOI] [PubMed] [Google Scholar]
- 19.Fields S, Jang SK. Presence of a potent transcription activating sequence in the p53 protein. Science. 1990;249:1046–1049. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- 20.Cress WD, Triezenberg SJ. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan SM, et al. Mutational analysis of a transcriptional activation region of the VP16 protein of herpes simplex virus. Nucleic Acids Res. 1998;26:4487–4496. doi: 10.1093/nar/26.19.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Candau R, et al. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene. 1997;15:807–816. doi: 10.1038/sj.onc.1201244. [DOI] [PubMed] [Google Scholar]
- 23.Kussie PH, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor supressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 24.Ferreon JC, et al. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proc Natl Acad Sci USA. 2009;106:6591–6596. doi: 10.1073/pnas.0811023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teufel DP, Freund SM, Bycroft M, Fersht AR. Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proc Natl Acad Sci USA. 2007;104:7009–7014. doi: 10.1073/pnas.0702010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langlois C, et al. NMR structure of the complex between the Tfb1 subunit of TFIIH and the activation domain of VP16: Structural similarities between VP16 and p53. J Am Chem Soc. 2008;130:10596–10604. doi: 10.1021/ja800975h. [DOI] [PubMed] [Google Scholar]
- 27.Di Lello P, et al. Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol Cell. 2006;22:731–740. doi: 10.1016/j.molcel.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Glover-Cutter K, et al. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol. 2009;29:5455–5464. doi: 10.1128/MCB.00637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bird G, Zorio DAR, Bentley DL. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol Cell Biol. 2004;24:8963–8969. doi: 10.1128/MCB.24.20.8963-8969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viprakasit V, et al. Mutations in the general transcription factor TFIIH result in beta-thalassaemia in individuals with trichothiodystrophy. Hum Mol Genet. 2001;10:2797–2802. doi: 10.1093/hmg/10.24.2797. [DOI] [PubMed] [Google Scholar]
- 31.Avantaggiati ML, et al. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 32.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 33.Di Lello P, et al. NMR structure of the amino-terminal domain from the Tfb1 subunit of TFIIH and characterization of its phosphoinositide and VP16 binding sites. Biochemistry. 2005;44:7678–7686. doi: 10.1021/bi050099s. [DOI] [PubMed] [Google Scholar]
- 34.Pandya K, Donze D, Townes TM. Novel transactivation domain in erythroid Kruppel-like factor (EKLF) J Biol Chem. 2001;276:8239–8243. doi: 10.1074/jbc.M008457200. [DOI] [PubMed] [Google Scholar]
- 35.Brown RC, et al. Distinct domains of erythroid Kruppel-like factor modulate chromatin remodeling and transactivation at the endogenous beta-globin gene promoter. Mol Cell Biol. 2002;22:161–170. doi: 10.1128/MCB.22.1.161-170.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wojciak JM, Martinez-Yamout MA, Dyson HJ, Wright PE. Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains. EMBO J. 2009;28:948–958. doi: 10.1038/emboj.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferreon JC, Martinez-Yamout MA, Dyson HJ, Wright PE. Structural basis for subversion of cellular control mechanisms by the adenoviral E1A oncoprotein. Proc Natl Acad Sci USA. 2009;106:13260–13265. doi: 10.1073/pnas.0906770106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Guzman RN, Martinez-Yamout MA, Dyson HJ, Wright PE. Interaction of the TAZ1 domain of the CREB-binding protein with the activation domain of CITED2: Regulation by competition between intrinsically unstructured ligands for non-identical binding sites. J Biol Chem. 2004;279:3042–3049. doi: 10.1074/jbc.M310348200. [DOI] [PubMed] [Google Scholar]
- 39.Okuda M, et al. Structural insight into the TFIIE-TFIIH interaction: TFIIE and p53 share the binding region on TFIIH. EMBO J. 2008;27:1161–1171. doi: 10.1038/emboj.2008.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Lello P, et al. p53 and TFIIEalpha share a common binding site on the Tfb1/p62 subunit of TFIIH. Proc Natl Acad Sci USA. 2008;105:106–111. doi: 10.1073/pnas.0707892105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dames SA, Martinez-Yamout M, De Guzman RN, Dyson HJ, Wright PE. Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response. Proc Natl Acad Sci USA. 2002;99:5271–5276. doi: 10.1073/pnas.082121399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.SenBanerjee S, et al. KLF2 is a novel transcriptional regulator of endothelial proinflammatory activation. J Exp Med. 2004;199:1305–1315. doi: 10.1084/jem.20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evans PM, et al. Krurppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J Biol Chem. 2007;282:33994–34002. doi: 10.1074/jbc.M701847200. [DOI] [PubMed] [Google Scholar]
- 44.Zhang ZP, Teng CT. Phosphorylation of Kruppel-like factor 5 (KLF5/IKLF) at the CBP interaction region enhances its transactivation function. Nucleic Acids Res. 2003;31:2196–2208. doi: 10.1093/nar/gkg310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hussain S, Lara-Pezzi E, Brand N. Role of the KLF15 transcription factor in an anti-hypertrophic pathway involving GSK3 beta. Heart. 2008;94:A53–A54. [Google Scholar]
- 46.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 47.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 48.Siatecka M, Xue L, Bieker JJ. Sumoylation of EKLF promotes transcriptional repression and is involved in inhibition of megakaryopoiesis. Mol Cell Biol. 2007;27:8547–8560. doi: 10.1128/MCB.00589-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.