Abstract
The circadian clock in mammalian organisms is generated by a transcription–translation feedback loop that controls many biochemical pathways at the cellular level and physiology and behavior at the organismal level. Cryptochrome (Cry) is a key protein in the negative arm of the transcription–translation feedback loop. It has been found that Cry mutation in cells with p53-null genotype increased their sensitivity to apoptosis by genotoxic agents. Here we show that this increased sensitivity is due to up-regulation of the p53 gene family member p73 in response to DNA damage. As a consequence, when tumors arising from oncogenic Ras-transformed p53−/− and p53−/−Cry1−/−Cry2−/− cells are treated with the anticancer drug oxaliplatin, p53−/− tumors continue to grow whereas p53−/−Cry1−/−Cry2−/− tumors exhibit extensive apoptosis and stop growing. This finding provides a mechanistic foundation for overcoming the resistance of p53-deficient tumor cells to apoptosis induced by DNA-damaging agents and suggests that disruption of cryptochrome function may increase the sensitivity of tumors with p53 mutation to chemotherapy.
The circadian clock is a global regulatory system that generates rhythmic changes with ∼24-h periodicity in many cellular and organismal functions. At the molecular level the clock is composed of four gene groups, CLOCK/NPAS2, BMAL1, Cry1/2, and Per1/2, that generate a transcription–translation feedback loop (TTFL): The CLOCK/NPAS2-BMAL1 heterodimer transactivates the transcription of cognate genes including Cry1/2 and Per1/2; the Cry and Per proteins, in turn, inhibit CLOCK-BMAL1 after a time lag to close the regulatory loop and generate rhythmic expression or subcellular location of the clock proteins (1–5). The circadian clock directly controls the expression of ∼10% of the genes in a given tissue (6). In addition, the clock network is integrated within all of the major cellular signaling and metabolic pathways (2–4). As a consequence, disruption of the clock is expected to affect cellular and organismal behavior in multiple ways.
We found that circadian clock disruption by cryptochrome (Cry) mutation (7) or Cry down-regulation in a p53-null (8, 9) background renders cells more sensitive to genotoxic agents, such as UV, by making them more prone to apoptosis (Figs. S1 and S2) (7). To gain some insight into the mechanism by which Cry mutation sensitizes cells to apoptosis, we considered the p53-independent pathways for intrinsic apoptosis. Specifically, we focused on contribution of the p73 gene to intrinsic apoptosis. The p73 protein along with p63 and p53 constitute the p53 family (10–12). Of these, p53 is the primary tumor suppressor, which prevents cancer mainly by promoting apoptosis in response to DNA damage or oncogenic transformation. In the absence of p53, the other two members of the family, in particular p73, a structural and functional homolog of p53 (10–12), can substitute for p53 as the proapoptotic tumor suppressor (13–17). Hence, when we discovered that p53−/−Cry1−/−Cry2−/− (hereafter p53KOCryDKO) cells have increased sensitivity to apoptosis compared with p53−/− (p53KO) cells (7), we reasoned that this might have been caused by an elevated level of p73 in p53KOCryDKO, in particular after DNA-damaging treatment, as it is known that p73 is a damage-inducible gene (18). To test this idea, we compared p73 levels and apoptosis in p53KO and p53KOCryDKO cells after UV irradiation or oxaliplatin treatment. We found that these genotoxic treatments induce high levels of p73 in cryptochrome mutant cells and significantly enhance apoptosis and clonogenic cell death in p53KOCryDKO cells relative to p53KO cells. This finding suggests that disruption of cryptochrome function may be used as an adjuvant in cancer chemotherapy with platinum-based drugs.
Results
Effect of Cryptochrome Mutation on p73 Expression.
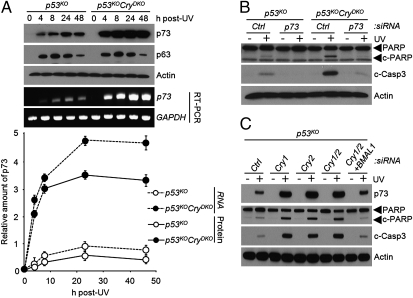
We found that under our assay conditions neither p73 nor p63 is detectable before DNA damage but both are strongly induced after UV irradiation (Fig. 1A). Importantly, however, whereas the Cry mutation in p53-null background had no effect on p63 induction by UV, it enhanced the rates of both p73 mRNA and protein synthesis by a factor of about 10 (Fig. 1A and Figs. S3 and S4). We should note, however, that the enhanced induction of p73 by UV in p53KOCryDKO cells is not unique to cells with p53-null genotype. We find essentially the same dose–response and enhanced induction of p73 in Cry1−/−Cry2−/− (CryDKO) cells as in p53KOCryDKO cells (Fig. S3), indicating that enhanced expression of p73 in Cry-null cells is independent of the p53 status of the cell.
Fig. 1.
Regulation of p73-mediated apoptosis by the circadian clock. (A) Time course of p73 induction in p53KO and p53KOCryDKO cells analyzed by immunoblotting and RT-PCR. The quantitative results of the levels of p73 protein and mRNA (Bottom) are means ± SD (n = 3). The UV dose was 10 Jm−2. (B) Immunoblots showing the role of p73 in UV-induced apoptosis in p53KOCryDKO cells. Cells were transfected with the indicated siRNAs, irradiated with 10 Jm−2, and then analyzed for apoptosis by probing for cleaved PARP (c-PARP) and cleaved caspase 3 (c-Casp3). (C) Enhanced apoptosis observed in cryptochrome-deficient cells is reversed by down-regulation of BMAL1. p53KO mouse embryonic fibroblasts were transfected with the indicated siRNAs and UV-irradiated where indicated with 10 Jm−2, and apoptosis was probed by immunoblotting for cleaved PARP and cleaved caspase 3.
To ascertain that the enhanced apoptosis in p53KOCryDKO was in fact dependent upon p73, we compared the UV-induced apoptosis in p53KOCryDKO and p53KO cells before and after siRNA knockdown of p73. The results indicate that both the residual apoptosis observed in p53KO and the enhanced apoptosis seen in p53KOCryDKO cells are drastically reduced when p73 is down-regulated (Fig. 1B and Fig. S5), suggesting that the enhanced induction of p73 in p53KOCryDKO is responsible for the increased apoptosis seen in these cells.
Regulation of p73 by the Circadian Clock.
These findings suggest a unique regulatory mechanism that is both circadian clock- and DNA damage-dependent. To test this model, we analyzed p73 levels and apoptotic response to DNA damage in p53KO cells after down-regulation of the negative (Cry1/2 or Per1/2) and positive (BMAL1) elements of the core circadian TTFL. The results are shown in Fig. 1C and Figs. S6 and S7. Down-regulation of either Cry1/2 or Per1/2 enhances UV-induced p73 synthesis and apoptosis. Interestingly, down-regulation of either Cry1 or Cry2 yields results similar to down-regulation of both genes with regard to UV-induced p73 levels and the extent of apoptosis as probed by caspase activity (Fig. 1C) and clonogenic survival (Fig. S6). Importantly, both of these effects are reversed by down-regulation of BMAL1 (Fig. 1C and Fig. S6), supporting the conclusion that the expression of p73 is controlled both by DNA damage and by the circadian clock, a unique and unprecedented regulatory mechanism.
Control of p73 Expression by DNA Damage and the Circadian Clock.
Next, we wished to understand how clock disruption by Cry mutation leads to elevated p73 levels. As this increase was dependent on both the circadian TTFL and DNA damage, we investigated the p73 promoter for clock-related sequence elements. We found that the p73 promoter region up to 4 kbp preceding the transcription initiation site did not reveal a canonical E box or the variant E′-box elements, which are key sequence elements in the promoters of most first-order clock-controlled genes. Then, we considered the possibility that p73 may be a second-order clock-controlled gene. The transcriptional regulation of p73 has been investigated in some detail. The gene is negatively controlled by C-EBPα and is activated by Early growth response 1 (Egr1) (18, 19). Treatment of cells with DNA-damaging agents leads to release of the repressive C-EBPα from the promoter and transcriptional up-regulation of p73 in an Egr1-dependent manner. Of particular interest, it was recently reported that Egr1 might be a clock-controlled gene (20, 21). These facts, considered together, lead to certain predictions: First, Egr1 must be up-regulated in CryDKO cells, and this up-regulation must be reversed by down-regulation of BMAL1. Second, it would be expected that BMAL1 will bind to the Egr1 promoter and, finally, both Egr1 and C-EBPα are expected to bind to the p73 promoter where the binding of C-EBPα but not of Egr1 must be affected by UV irradiation.
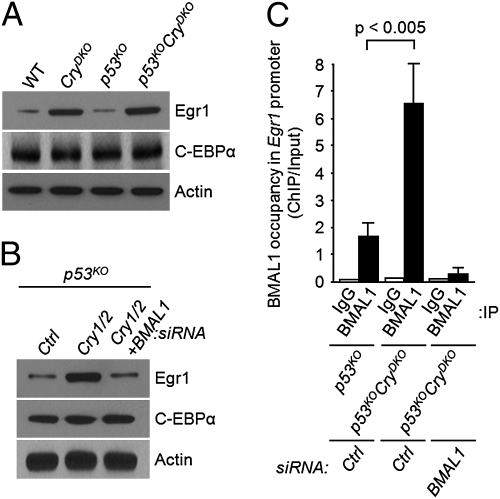
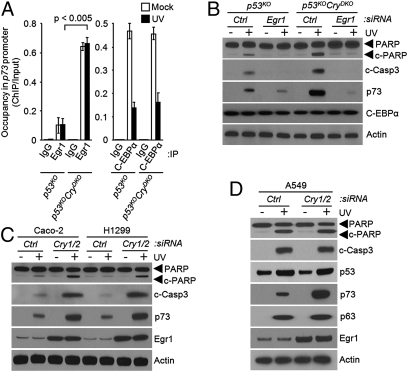
We tested these predictions in experiments summarized in Figs. 2 and 3. As would be predicted for a clock-controlled gene, the level of Egr1 is elevated both in CryDKO and p53KOCryDKO cells (Fig. 2A). Moreover, in accordance with the prediction, the elevation of Egr1 as a consequence of Cry down-regulation can be reversed by BMAL1 knockdown (Fig. 2B), consistent with regulation of Egr1 by the primary circadian feedback loop. In support of the model for positive regulation by BMAL1 and negative regulation by Cry, chromatin immunoprecipitation (ChIP) analysis revealed occupancy of the Egr1 E-box sequence element by BMAL1, which is enhanced in the absence of Cry1/2 and abolished by down-regulation with BMAL1 (Fig. 2C). Finally, when the occupancy of the p73 promoter was analyzed by ChIP, we found greatly enhanced occupancy by Egr1 in p53KOCryDKO cells, which was not affected by UV irradiation (Fig. 3A). In contrast, Cry mutation had no effect on binding of C-EBPα to the p73 promoter, but the occupancy of the promoter by C-EBPα was drastically reduced after UV irradiation (Fig. 3A). Taken together, these data support the model that p73 expression is regulated by the clock-controlled Egr1 and by the DNA damage-controlled C-EBPα, and hence, in Cry-null mutants, increased expression of Egr1 combined with UV damage-induced dissociation of C-EBPα from the p73 promoter leads to a high level of p73 production and the consequent apoptosis. In further support of this model, we find that both in p53KO and in p53KOCryDKO cells, down-regulation of Egr1 by siRNA significantly reduces UV induction of p73 and abolishes apoptosis (Fig. 3B).
Fig. 2.
Egr1 is a clock-controlled gene. (A) Overexpression of Egr1 in cryptochrome-deficient cells. Mouse fibroblasts of the indicated genotypes were analyzed for expression of Egr1 and C-EBPα, the two main regulators of p73. Actin was used as a loading control. (B) Egr1 expression is regulated by the circadian clock. p53KO fibroblasts were transfected with the indicated siRNAs and the levels of Egr1 and C-EBPα were probed by immunoblotting. (C) Binding of BMAL1 (positive arm of the core circadian clock) to the Egr1 promoter. Fibroblasts of the indicated genotypes were transfected with control (Ctrl) or BMAL1 siRNA and then the occupancy of the Egr1 promoter by BMAL1 was analyzed by ChIP. Note the elevated ChIP values in p53KOCryDKO cells relative to p53KO cells, indicating that Cry reduces the occupancy of target promoters by BMAL1 (P < 0.05, n = 3).
Fig. 3.
Requirement for Egr1 in clock-mediated p73-dependent apoptosis. (A) Effect of UV on occupancy of the p73 promoter by Egr1 and C-EBPα in the absence and presence of cryptochrome. Mouse fibroblasts of the indicated genotypes were irradiated with 10 Jm−2 where indicated and the promoter binding of Egr1 and C-EBPα was determined by ChIP. Note that Cry mutation enhances Egr1 binding (P < 0.05, n = 3), which is not affected by UV. In contrast, Cry mutation has no effect on C-EBPα binding, which is significantly reduced by UV irradiation. (B) Elevated Egr1 in Cry mutant cells is responsible for enhanced UV-induced apoptosis. Mouse fibroblasts of the indicated genotypes were transfected with the indicated siRNAs, irradiated with UV, and then analyzed for C-EBPα and p73 expression directly by immunoblotting and for apoptosis by immunoblotting for cleaved PARP and caspase 3. (C) Effect of cryptochrome down-regulation in p53-null human cell lines on Egr1 and p73 expression and UV-induced apoptosis. The cell lines were transfected with the indicated siRNAs, irradiated with 10 Jm−2 of UV where indicated, and probed for apoptosis by immunoblotting. (D) Effect of cryptochrome down-regulation on Egr1 and p73 expression and UV-induced apoptosis in a human tumor cell line with wild-type p53. Whole-cell lysates were analyzed by immunoblotting.
Regulation of p73 by the Circadian Clock in Human Cancer Cell Lines.
The apoptosis-enhancing effect of Cry mutation seems to be unique to both mouse and human p53 mutant cells, because in cells with wild-type p53 the p53-driven apoptosis is expected to be dominant and thus obscure any contribution of the p73 pathway (22). This prediction was tested by analyzing apoptosis in two p53 mutant and one p53 wild-type human cancer cell lines. As seen in Fig. 3C in the Caco-2 and H1299 cell lines in which p53 is not functional, down-regulation of Cry enhances p73 expression and increases apoptosis after UV irradiation. In contrast, in the A549 cell line with a functional p53, even though down-regulation of Cry leads to enhanced levels of p73 after UV, this up-regulation of p73 is not accompanied by enhanced apoptosis (Fig. 3D) because apparently the UV-induced p53 is sufficient to cause maximal level of apoptosis, which cannot be further enhanced by increased p73. Taken together, the data in Figs. 2 and 3 show that Egr1 is a clock-controlled gene and that p73 is a second-order clock-controlled gene that plays a prominent role in promoting apoptosis and clonogenic death (Figs. S6 and S8) in p53-null cells lacking cryptochrome.
Enhancement of the Efficacy of Oxaliplatin by Cryptochrome Disruption.
Next, we wished to determine whether the enhancement of apoptosis induced by genotoxic agents (intrinsic apoptosis) (23, 24) by Cry mutation in p53 mutant cells can be used for therapeutic purposes. Cisplatin and its second- and third-generation derivatives are UV-mimetic agents that kill cells mainly by producing intrastrand diadducts in DNA (25, 26). Hence, we reasoned that cryptochrome disruption might enhance the sensitivity of p53-null cells and tumors to killing by platinum derivatives. We performed our chemotherapy experiments with oxaliplatin, which is the drug of choice for treating colorectal cancers, among others (26). First, we established that Cry mutation sensitizes transformed p53-null cells to oxaliplatin-induced p73 up-regulation and to killing by apoptosis (Fig. S9). This was to be expected, because the major DNA lesions induced by this drug, the GG, AG, and GXG diadducts, can only be repaired by nucleotide excision repair, as are the cyclobutane pyrimidine dimer and the (6-4) photoproduct (27). Then, we used a mouse tumor model to test the efficacy of oxaliplatin in treating clock-normal and clock-disrupted cancers by oxaliplatin.
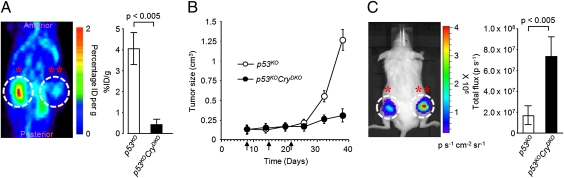
Tumor xenografts were established in 6-wk-old female immunodeficient mice by injection of oncogenically transformed p53KO and p53KOCryDKO cells (7). Each animal received, by subcutaneous injection, p53KO cells in one flank and p53KOCryDKO cells in the other. The tumors of both genotypes grew at the same rate (Fig. S10), and when the tumor volumes reached 0.1 cm3, oxaliplatin treatment was initiated. The mice received 10 mg/kg oxaliplatin once a week for 3 wk. Fig. 4A shows that administration of oxaliplatin drastically inhibits 2-[18F]deoxyglucose ([18F]FDG) uptake in the p53KOCryDKO tumor compared with the p53KO tumor. In parallel with this effect on metabolism, oxaliplatin suppressed tumor growth in p53KOCryDKO but had no measurable effect on the growth rate of the p53KO tumor (Fig. 4B). In vivo measurements of caspase-3 activity of the two groups of tumors after the drug treatment revealed that there was massive apoptosis in the tumor with the p53KOCryDKO genotype (Fig. 4C), which provides a mechanistic explanation of the favorable response to oxaliplatin of tumors of this genotype. Thus, the tumor xenograft study supports the prediction based on apoptosis and toxicity experiments with cultured cells that cryptochrome disruption in p53-null cells makes them more sensitive to chemotherapy by oxaliplatin.
Fig. 4.
Effect of cryptochrome mutation on treating cancer with p53-null mutation. NOD/SCID mice bearing a p53KO tumor xenograft on the left flank (*) and a p53KOCryDKO tumor xenograft on the right flank (**) received oxaliplatin intraperitoneally at 10 mg/kg once a week for 21 d and various tumor-related metrics were recorded. (A) Tumor metabolism. MicroPET scan recorded 1 h after [18F]FDG injection on day 38 after xenograft injection. (Right) The quantification of [18F]FDG signal calculated as the ratio of % injected dose (ID) at the ROI (circle) to the % ID at a background region. Error bars represent means ± SD (n = 4). The dorsal aspect is shown. (B) Tumor growth. Arrows indicate days of oxaliplatin injection following the xenograft injection on day 0. The oxaliplatin treatment was initiated after tumor size had reached 0.1 cm3. Error bars represent means ± SD (n = 10). (C) Oxaliplatin-induced apoptosis in xenografts. NOD/SCID mice were injected with p53KO-Luc cells in the left flank (*) and p53KOCryDKO-Luc cells in the right flank (**). One week later, weekly injections with oxaliplatin were initiated. After the third round of oxaliplatin injection, Z-DEVD-aminoluciferin prosubstrate for caspase 3 was administered intraperitoneally and 10 min later, bioluminescence was measured in the ROI (circle) drawn over the tumor. Quantitative analysis (Right). Error bars represent means ± SD (n = 4).
Conclusion
In Fig. 5, we summarize the findings of this study: Egr1 is a clock-controlled gene and p73 is a second-order clock-controlled gene that is regulated through Egr1 by the clock and through C-EBPα by the DNA damage response reaction. In the absence of Cry, Egr1 is elevated, and upon DNA damage the release of C-EBPα from the p73 promoter leads to Egr1-activated up-regulation of p73 and thus p53-independent apoptosis. These findings suggest that in p53 mutant tumors, which constitute about 50% of all human tumors, targeted inhibition of Cry may improve the efficacy of platinum-based chemotherapy or of combination chemotherapy that includes cis-platinum drugs.
Fig. 5.
Model for targeting the clock to improve the efficacy of oxaliplatin treatment of p53-null tumors. Cry regulates Egr1, which transactivates p73. With Cry inactive, Egr1 is up-regulated. Following oxaliplatin-induced removal of C-EBPα from the p73 promoter, Egr1 causes massive production of p73 and enhanced apoptosis.
Materials and Methods
Establishment of p53−/− and p53−/−Cry1−/−Cry2−/− Tumor Cell Lines.
Cell lines were generated from the p53−/− and p53−/−Cry1−/−Cry2−/− mouse strains as described previously (7, 27). To obtain cell lines, skin patches from the backs of mice at the age of 3 mo were taken under sterile conditions, cut into small pieces, placed in four- or six-well plates, and incubated for 1 wk to let the fibroblasts migrate into the wells. The fibroblasts were passaged at least three times before using them in any experiments. Primary wild-type skin fibroblasts were isolated in the same manner, and passaged eight times without the appearance of spontaneously immortalized clones. Wild-type mouse embryonic fibroblasts were kindly provided by Norman Sharpless (University of North Carolina, Chapel Hill, NC). For Ras-induced oncogenic transformation, cells were seeded into 10-cm plates and transfected with 250 ng of pEGFP-N1 (Clontech) and/or 5 μg of Ras pT24 vector (7) using FuGENE 6 reagent (Roche). The next day, cells were divided into three plates and incubated for 18 d. The plates were stained with Giemsa and colonies of oncogenically transformed cells were counted visually, and the fraction of transformed cells was normalized for transfection efficiencies calculated by counting GFP-positive cells in each plate. These cells were maintained in DMEM (Life Technologies) supplemented with 10% FBS (Gemini Bio Product), 100 units/mL penicillin, and 100 μg/mL streptomycin (Life Technologies). The cells were maintained in an incubator at 37 °C under 5% CO2. The Cry1−/−Cry2−/− cell line has been described (27).
Human Cell Lines.
H1299 cells were kindly provided by Shlomo Melmed (Cedars-Sinai Medical Center, Los Angeles, CA). Caco-2 and A549 cells were obtained from the American Type Culture Collection. These cells were grown in DMEM with 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin at 37 °C under 5% CO2.
Clonogenic Survival Assay.
UV survival assays were performed as described previously (27) using a fluence in the range of 0–10 Jm−2. For oxaliplatin survival assays, cells were seeded into six-well plates at 105, 5 × 104, and 2 × 104 cells per well in duplicate and incubated in growth medium for 10–14 h. The cells were washed and incubated with 0–20 μM oxaliplatin for 2 h. Then, the cells were washed with PBS twice, growth medium was added, and cells were incubated for 10–14 d. Cells were then fixed for 20 min in 3:1 methanol:acetic acid, rinsed with water, stained with 5% methylene blue, and counted. Colonies containing >50 cells were scored, and survival was calculated from the ratio of the number of colonies in the treated samples to those in the control plates.
Immunoblot Analysis.
Antibodies were from the following sources: Egr1, PARP, cleaved caspase 3, and p53 (Cell Signaling Technology), p63 (Novus Biologicals), p73 (Upstate Biotechnology), C-EBPα (Thermo Scientific), and actin and cyclophilin B (Santa Cruz Biotechnology). Mouse Cry1 (monoclonal) and Cry2 (polyclonal) antibodies were prepared in our laboratory (27). Conventional immunoblotting procedures were used to detect the target proteins: Cells were collected, washed once in cold PBS, and then scraped in TEGN buffer (10 mM Tris, pH 8, 1 mM EDTA, 10% glycerol, 0.5% Nonidet P-40, 400 mM NaCl, 1 mM DTT, 0.5 mM phenylmethylsulfonylfluoride, and protease inhibitor mixture containing 1 M benzamidine, 3 mg/mL leupeptin, 100 mg/mL bacitracin, and 1 mg/mL α2 macroglobulin) and incubated on ice for 15 min. Lysates were then cleared by centrifugation at 20,000 × g for 10 min. Total protein concentration was determined via the Bio-Rad protein assay. Equal amounts of protein were separated on 10% SDS/PAGE and the proteins were transferred to nitrocellulose membranes (Schleicher and Schuell). The membranes were then blocked for 1 h in a PBS solution containing 5% nonfat milk powder and 0.1% Tween-20 and then probed with primary antibody overnight in 1% milk, 0.1% Tween-20 in PBS. After washing, membranes were incubated for 1 h with horseradish peroxidase-linked secondary antibody (Sigma) in 1% milk, 0.1% Tween-20 in PBS. Finally, after three 5-min washes in 0.1% PBS/Tween-20, proteins were visualized by enhanced chemiluminescence (Amersham Biosciences). Band intensities were quantified with ImageQuant 5.2 software (Molecular Dynamics).
Chromatin Immunoprecipitation.
ChIP was performed according to manufacturer's instructions (Sigma) with some exceptions. Protein–DNA complexes incubated with rabbit polyclonal antibodies against BMAL1 (Bethyl Laboratories), rabbit monoclonal antibody directed against Egr1 (44D5; Cell Signaling Technology), rabbit polyclonal antibody against C-EBPα (PA1-825; Thermo Scientific), or equivalent IgG control (Cell Signaling Technology) were precipitated using protein A/G-conjugated agarose beads (Calbiochem). Protein–DNA crosslink was resolved and followed by quantitative real-time PCR using SYBR green master mix (Applied Biosystems) with the primers listed in Table S1.
Chemicals and Treatment.
Cells were treated with 20 μg/mL cycloheximide (Sigma) or 5 μg/mL actinomycin D in DMEM as indicated and then protein stability was assayed by conventional immunoblotting. The caspase-3 inhibitor Z-Asp(OMe)-Glu(OMe)-Val-Asp(OMe)-CH2F (Z-DEVD-FMK) (R&D Systems) was used at a final concentration of 25 μM.
RNA Interference and Transfection.
For RNAi experiments, cells at 70% confluency were transfected using DharmaFECT (Dharmacon Research) according to the manufacturer's directions for transfection of ON-TARGET plus SMARTpool siRNA duplexes specific for each target purchased from Dharmacon Research: mouse Cry1 (L-040485-01-0005), mouse Cry2 (L-040486-00-0005), mouse Per1 (L-040487-00-0005), mouse Per2 (L-040489-01-0005), mouse BMAL1 (L-040483-01-0005), mouse Egr1 (L-040286-00-0005), mouse C-EBPα (L-040561-00-0005), mouse p73 (L-043871-00-0005), mouse cyclophilin B (D-001820-20-05) as a control, human Cry1 (L-015421-00-0005), human Cry2 (L-014151-01-0005), human cyclophilin B (D-001820-10-05) as a control, or nontargeting siRNA (D-001810-10-05) as a control. The siRNA experiment was carried out for 48 h, at which time cells were harvested and the extent of knockdown of the target genes was assessed by immunoblotting or RT-PCR. For the determination of the effect of gene knockdown on clonogenic survival, after treating the cells with siRNA for 24 h, the cells were plated at low density to ensure the formation of 400 colonies per six-well plate in the absence of UV irradiation. Following plating, cells were incubated in growth medium for 10–14 h and treated with UV at a fluence rate of 0.5 Jm−2·s−1 from a germicidal lamp for the appropriate doses. The cells were then incubated for 9–10 d until colonies were readily visible. The specificity of the antibodies and siRNAs used are shown in Fig. S11.
RT-PCR.
RNA was extracted from cell pellets using an RNeasy Mini Kit (Qiagen) and quantified by UV-vis spectrophotometry. The cDNAs were generated by reverse transcription with random hexamers using the ImProm-II Reverse Transcription System (Promega). Then RT-PCR was performed with the primers listed in Table S2. Quantitative real-time PCR was then performed using the Expand High Fidelity PCR System (Roche). Conditions for linear amplification were established through template and cycle curves. PCR products (40% of reaction mixture) were then separated on 2% agarose gels and bands were visualized with ethidium bromide staining. Quantification of p73 induction was performed with ImageQuant 5.2 software (Molecular Dynamics).
Tumor Xenograft Model.
Animal studies were conducted in accordance with the regulations of the National Institutes of Health and the Institutional Animal Care and Use Committee of the University of North Carolina School of Medicine. Tumor xenografts were established in 6-wk-old female immunodeficient NOD/SCID mice by subcutaneous inoculation of 2 × 106 cells mixed with an equal volume of Matrigel (BD Biosciences) into both flanks. The NOD/SCID mice were bred and kept under defined-flora pathogen-free conditions at the AAALAC-approved Animal Facility of the Division of Experimental Radiation Oncology, University of North Carolina. Oxaliplatin at 10 mg/kg was administered intraperitoneally once a week for 21 d. Tumor volume and body weight were recorded every 3 d. Tumor size was measured with a caliper in three mutually perpendicular diameters (a, b, and c) and the volume was calculated as V = (π/6) × a × b × c.
Micro Positron Emission Tomography Imaging.
Whole-body images of the biodistribution of [18F]FDG tracers in mice were obtained by micro positron emission tomography (microPET) scans. Mice were kept warm under gas anesthesia (2% isoflurane) and injected with [18F]FDG intraperitoneally. A 1-h interval for uptake was allowed between probe administration and microPET scanning. Data were acquired using a Siemens Preclinical Solutions MicroPET Focus 220 instrument. MicroPET data were acquired for 10 min and were reconstructed using statistical maximum a posteriori probability algorithms into multiple frames. The spatial resolution of microPET is ∼1.5-mm, 0.4-mm voxel size. Three-dimensional regions of interest (ROI) were drawn using AMIDE software (Andreas Loening, Stanford University, Palo Alto, CA). Color scale is proportional to tissue concentration, with red being the highest and lower values in yellow, green, and blue.
In Vivo Imaging of Caspase-3 Activity.
For preparation of Z-DEVD-aminoluciferin, 10 mg of VivoGlo Caspase-3/7 substrate (Promega) was dissolved in 67 μL of surfactant mix (70 μL Tween-80, 640 μL PEG 400, and 290 μL N,N-dimethylacetamide) and then added to 100 μL of 5% glucose in water. The mixture was vortexed to make a homogeneous suspension and injected intraperitoneally into tumor-bearing mice (100 μL per mouse). Ten minutes after Z-DEVD-aminoluciferin injection, the mice were imaged with IVIS-200 (Xenogen) for in vivo photon counting of bioluminescence. The photon emission intensity (photons/s) of each mouse is indicated in a rainbow bar scale.
Statistical Analysis.
Results are shown as means ± SD, which were performed using a two-tailed t test.
Supplementary Material
Acknowledgments
We thank members of our laboratory, in particular T. H. Kang, C. P. Selby, and S. Gaddameedhi, for critical reading of the manuscript and helpful discussions. We are grateful to S. K. Choe, S. Kwak, and S. H. Lee for critical comments and useful discussions. We acknowledge the services of the Mouse Facility Center and Biomedical Research Imaging Center at the University of North Carolina, Chapel Hill. We appreciate experimental assistance and helpful discussions in microPET and bioluminescence imaging analysis from J. H. Lee and S. H. Kim. This work was supported by National Institutes of Health Grants GM31082 and GM32833.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 10379.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106284108/-/DCSupplemental.
References
- 1.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Hastings MH, Reddy AB, Maywood ES. A clockwork web: Circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahar S, Sassone-Corsi P. Metabolism and cancer: The circadian clock connection. Nat Rev Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 5.Vitaterna MH, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 7.Ozturk N, Lee JH, Gaddameedhi S, Sancar A. Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proc Natl Acad Sci USA. 2009;106:2841–2846. doi: 10.1073/pnas.0813028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 9.Lowe SW, et al. p53 status and the efficacy of cancer therapy in vivo. Science. 1994;266:807–810. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- 10.Yang A, Kaghad M, Caput D, McKeon F. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet. 2002;18:90–95. doi: 10.1016/s0168-9525(02)02595-7. [DOI] [PubMed] [Google Scholar]
- 11.Melino G, Lu X, Gasco M, Crook T, Knight RA. Functional regulation of p73 and p63: Development and cancer. Trends Biochem Sci. 2003;28:663–670. doi: 10.1016/j.tibs.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Stiewe T. The p53 family in differentiation and tumorigenesis. Nat Rev Cancer. 2007;7:165–168. doi: 10.1038/nrc2072. [DOI] [PubMed] [Google Scholar]
- 13.Jost CA, Marin MC, Kaelin WG., Jr p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 14.Gong JG, et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 15.Irwin M, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 16.Irwin MS, et al. Chemosensitivity linked to p73 function. Cancer Cell. 2003;3:403–410. doi: 10.1016/s1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- 17.Flores ER, et al. Tumor predisposition in mice mutant for p63 and p73: Evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 18.Marabese M, Vikhanskaya F, Rainelli C, Sakai T, Broggini M. DNA damage induces transcriptional activation of p73 by removing C-EBPα repression on E2F1. Nucleic Acids Res. 2003;31:6624–6632. doi: 10.1093/nar/gkg869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J, Baron V, Mercola D, Mustelin T, Adamson ED. A network of p73, p53 and Egr1 is required for efficient apoptosis in tumor cells. Cell Death Differ. 2007;14:436–446. doi: 10.1038/sj.cdd.4402029. [DOI] [PubMed] [Google Scholar]
- 20.Bai L, et al. Daily oscillation of gene expression in the retina is phase-advanced with respect to the pineal gland. Brain Res. 2008;1203:89–96. doi: 10.1016/j.brainres.2008.01.073. [DOI] [PubMed] [Google Scholar]
- 21.Humphries A, Carter DA. Circadian dependency of nocturnal immediate-early protein induction in rat retina. Biochem Biophys Res Commun. 2004;320:551–556. doi: 10.1016/j.bbrc.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 22.Flores ER, et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 23.Hotchkiss RS, Strasser A, McDunn JE, Swanson PE. Cell death. N Engl J Med. 2009;361:1570–1583. doi: 10.1056/NEJMra0901217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danial NN, Korsmeyer SJ. Cell death: Critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 25.Jung Y, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 26.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 27.Gauger MA, Sancar A. Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Cancer Res. 2005;65:6828–6834. doi: 10.1158/0008-5472.CAN-05-1119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.