Abstract
The human tumor suppressor protein BRCA2 plays a key role in recombinational DNA repair. BRCA2 recruits RAD51 to sites of DNA damage through interaction with eight conserved motifs of approximately 35 amino acids, the BRC repeats; however, the specific function of each repeat remains unclear. Here, we investigated the function of the individual BRC repeats by systematically analyzing their effects on RAD51 activities. Our results reveal the existence of two categories of BRC repeats that display unique functional characteristics. One group, comprising BRC1, -2, -3, and -4, binds to free RAD51 with high affinity. The second group, comprising BRC5, -6, -7, and -8, binds to free RAD51 with low affinity but binds to the RAD51-ssDNA filament with high affinity. Each member of the first group reduces the ATPase activity of RAD51, whereas none of the BRC repeats of the second group affects this activity. Thus, through different mechanisms, both types of BRC repeats bind to and stabilize the RAD51 nucleoprotein filament on ssDNA. In addition, members of the first group limit binding of RAD51 to duplex DNA, where members of the second group do not. Only the first group enhances DNA strand exchange by RAD51. Our results suggest that the two groups of BRC repeats have differentially evolved to ensure efficient formation of a nascent RAD51 filament on ssDNA by promoting its nucleation and growth, respectively. We propose that the BRC repeats cooperate in a partially redundant but reinforcing manner to ensure a high probability of RAD51 filament formation.
Keywords: breast cancer, cancer, chromosome stability, DNA-break repair, recombination
BRCA2 is a tumor suppressor protein. First identified in humans, mutations in BRCA2 cause predisposition to breast, ovarian, and other types of cancer (1, 2). It soon became apparent that BRCA2 is involved in maintaining genomic stability through its interaction with RAD51, a central component of recombinational DNA repair in humans (3). Homologous recombination serves to maintain genomic integrity in somatic cells by promoting the repair of breaks in DNA strands. BRCA2 regulates RAD51 function in DNA repair by recruiting it to the sites DNA breaks (4). For double-stranded DNA breaks, the dsDNA is resected to produce ssDNA that is rapidly bound by Replication protein A (RPA) (5–7). There, BRCA2 mediates the loading of RAD51 protein onto the RPA-ssDNA that was produced by resection (8, 9).
The 3,418 amino acids (aa) of BRCA2 include several hallmark motifs that are conserved in all BRCA2-like proteins and they are regarded as critical for function. One of these key motifs is a sequence of about 35 aa in length, referred to as the BRC repeat, because it is repeated eight times in the human protein, and is separated by variable size linker regions (10–12) (Fig. S1A). These motifs are highly conserved between mammalian species (11), and they confer upon BRCA2 the ability to bind RAD51 (8). In addition to the BRC repeats, an unrelated sequence capable of binding RAD51 was mapped to the C terminus, and it controls BRCA2 function through the cell cycle (13, 14). Adjacent to the BRC repeats in the primary sequence there is also a binding site for DMC1, the meiotic homolog of RAD51 (15), implicating BRCA2 in meiotic recombination. The C-terminal domain of BRCA2 contains a site that enables it to bind single-stranded DNA (ssDNA) (16) and two nuclear localization signals that allow it to recruit RAD51 to the nucleus (17).
The conspicuous conservation of the BRC repeats between mammalian species contrasts with the relatively poor identity among the repeats within a species, suggesting that multiplication of the initial BRC repeat took place before mammalian radiation (11). The divergence of these repeat units suggests that they may have evolved to achieve different functions (18). Nevertheless, based on the crystal structure of RAD51-BRC4 (19), a sequence interaction fingerprint of the BRC repeats and RAD51 has been identified and its conservation is remarkably good especially for repeats BRC1, -2, -4, -7 and -8 (12).
Depending on the assay, pull-down or yeast two-hybrid, six or eight of the BRC repeats have been reported to bind to the core region of RAD51 (20). This conclusion is consistent with the RAD51-binding capacity of full-length BRCA2, which binds at least six RAD51 monomers (8). Using a semiquantitative yeast two-hybrid assay, a fragment containing BRC1-4 was estimated to bind RAD51 with an affinity equivalent to that of a region containing all the BRC repeats, and fivefold tighter than BRC6-8, (21); other groups have confirmed these findings (15, 22). Again by yeast two-hybrid analysis, BRC4 was shown to interact threefold stronger with RAD51 than BRC1 (23).
Apart from the semiquantitative approaches mentioned above, little is known about the individual functions of these different repeats. Previously, we established that BRC4 and a region containing all eight BRC repeats modulates the DNA-binding preference of RAD51 by kinetically slowing nucleation onto dsDNA, while stabilizing complex formation with ssDNA (24). In addition, we showed the BRC4 stabilizes the RAD51-ssDNA filament by halting ATP hydrolysis by RAD51. Here we have extended those findings to include analysis of each of the eight BRC repeats. We discovered that they comprise two separate classes. We can define the roles of the individual BRC repeats based on their distinctive differences in affinity for RAD51 versus the RAD51-ssDNA filament; also, we establish they have contrasting effects on RAD51 functions. These analyses have revealed an unexpected separation of functions that define two distinct classes of BRC repeats: BRC1, -2, -3, and -4 comprise one class, and BRC5, -6, -7, and -8 comprise the other. Together, these two classes of BRC repeats function in a complementary manner to facilitate formation of the RAD51 nucleoprotein filament that is essential for the DNA strand exchange activity, which is required for recombinational DNA repair. These findings further define the function of BRCA2 in DNA repair.
Results
The Individual BRC Repeats Bind RAD51 with Distinct Affinities.
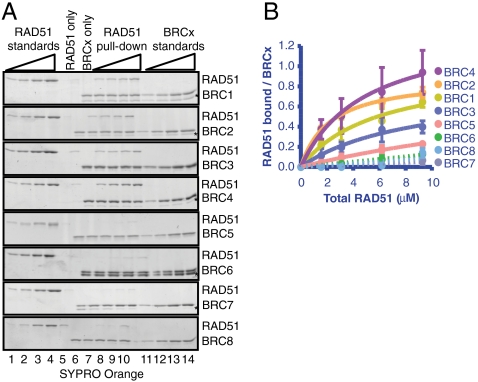
Various studies have established the ability of the BRC repeats to bind RAD51 (20–23); however, a quantitative analysis of the affinity of each BRC repeat for RAD51 has not been reported. We purified BRC1, BRC2, BRC3, BRC4, BRC5, BRC6, BRC7, and BRC8 fused to glutathione-S-transferase (GST) (Fig. S1B). Each GST-BRC fusion was tested individually for interaction with RAD51 in a GST pull-down assay wherein the RAD51 concentration was varied (8, 24). Using known concentrations of purified RAD51 (Fig. 1A, lanes 1–4) and each GST-BRC fusion (Fig. 1A, lanes 11–14) as a standard, RAD51 was incubated with the GST-BRC repeat, captured using glutathione beads, and analyzed by SDS/PAGE (Fig. 1A, lanes 6–10). The gels were stained with SYPRO Orange, and the binding of increasing amounts of RAD51 to a fixed amount of BRC peptide was quantified to generate the binding curves shown in Fig. 1B. Little or no RAD51 was retained by nonspecific binding to the beads (Fig. 1A, lane 5). All of the BRC repeats bound RAD51 to some extent (Fig. 1 A and B). The amount of RAD51 bound to the BRC repeats increased with RAD51 concentration (Fig. 1A, lanes 6–10); for BRC1, -2, -3, and -4, the binding curve approached a plateau, indicating formation of a saturable complex. As expected from prior studies (21–23), BRC1, -2, -3, and -4 bound to RAD51 with higher affinity than either BRC5, -6, -7, or -8, (Fig. 1 A and B and Table 1). BRC2, BRC4, and BRC1 displayed the highest affinity for RAD51 with an apparent Kd of approximately 1 - 2 μM in each case, with BRC3 next at approximately 4 μM. For BRC1, -2, -3, and -4, the stoichiometry reached a plateau at approximately 1 RAD51 monomer per BRC repeat (Table 1).
Fig. 1.
The individual BRC repeats of BRCA2 form complexes with RAD51, with distinct classes of affinities. (A) Each GST-tagged BRC peptide was tested individually for binding to RAD51 in a GST pull-down assay: RAD51 was incubated with the GST-BRC repeats, captured using glutathione beads, washed extensively to remove unbound proteins, analyzed by SDS/PAGE, and stained with SYPRO ORANGE. Lanes 1–4 contain increasing amounts of RAD51 (0.1, 0.2, 04, 0.8 μg) to generate a standard curve. Lane 5 or 6 is a control that contains the highest concentration of RAD51 used in the pull-down (2.4 μg), but in the absence of BRC peptide (for gels with GST-BRC1, -3, -4, and -6, the “RAD51 only” control is lane 6; lane 5 is empty in these gels). The next lane (6 or 7) is a control showing the BRC peptide, “BRCx only”, (0.5 μg) used in the pull-down experiment in the absence of RAD51. Lanes 7–10 (or 8–11) show increasing amounts of RAD51 incubated with each BRC peptide. Lanes 11–14 (or 12–15) contain increasing amounts of each BRC peptide (0.1, 0.2, 0.4, 0.5 μg) to generate a standard curve for the BRC peptide. The gel with GST-BRC8 contains an extra empty lane between lanes 10 and 11. An asterisk denotes the band that corresponds to contaminating GST (the amount of free GST could vary between different preparations of the same peptide). (B) The data from A were fitted to a single-site binding curve (Prism 5.0b), and are shown as solid lines. Error bars represent the standard deviation (SD) for two or more independent experiments. The data for BRC repeats 6–8 could not be fit accurately, and are shown connected by dashed lines.
Table 1.
Affinity and stoichiometry of individual BRC repeat binding to RAD51
| BRC repeat | Kd (μM) | Stoichiometry (Bound RAD51 per BRCx) |
| BRC1 | 2.1 ± 0.9 | 1.0 ± 0.2 |
| BRC2 | 1.1 ± 0.2 | 0.9 ± 0.1 |
| BRC3 | 4.0 ± 2.3 | 0.8 ± 0.3 |
| BRC4 | 1.5 ± 0.7 | 1.4 ± 0.3 |
| BRC5 | 9.0 ± 4.3 | 1.0 ± 0.7 |
| BRC6 | N.A. | N.A. |
| BRC7 | N.A. | N.A. |
| BRC8 | N.A. | N.A. |
The dissociation constant (Kd ± S.D.) and binding stoichiometry (RAD51/BRCx ± S.D.) were derived from fitting of the binding curves to a hyperbola as shown in Fig. 1. For the repeats that bound too weakly to permit reliable analysis, there are no values listed (N.A.); for these, the affinities are estimated to be approximately 100–200 μM.
In comparison, the ability of RAD51 to bind either BRC5, -6, -7, or -8, was greatly reduced. We could only measure with reasonable accuracy the affinity of BRC5 (Kd = 9.0 ± 4.3 μM); due to the lower affinity, the binding stoichiometry derived from the curve fitting shows a large error (1.0 ± 0.7). The binding curves for BRC6, -7, and -8 are represented as dashed lines (Fig. 1B) because they could not be accurately fit to a hyperbolic binding curve; we estimate that their dissociation constants are approximately 100–200 μM. These results are in qualitative accord with our previous study of the full-length human BRCA2 protein revealing two affinities classes for RAD51 binding: one that binds 4.5 ± 1 molecules of RAD51 with high affinity, and a second class that binds at least 2–3 RAD51 molecules with 100- to 1,000-fold lower affinity (8).
Only BRC1, -2, -3, and -4 Inhibit the DNA-Dependent ATPase Activity of RAD51.
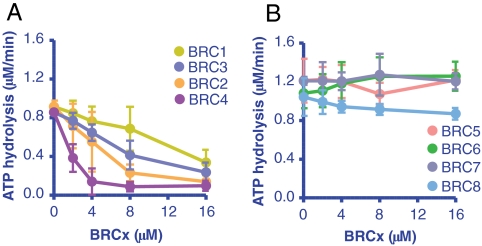
We showed previously (24) that a single BRC repeat (BRC4) can stabilize the RAD51-ssDNA complex by inhibiting the DNA-dependent ATPase activity of RAD51. Using the same rationale, we analyzed the effect of the individual BRC repeats on the ATPase activity of RAD51. Addition of either BRC1, -2, -3, or -4 decreased the ssDNA-dependent ATP hydrolysis by RAD51 in a concentration-dependent manner (Fig. 2A). The reduction in ATP hydrolysis correlated generally with the affinity of the BRC repeats for RAD51 (Table 1), showing the progression: BRC4 > BRC2 > BRC3 > BRC1. As expected from the binding results, BRC5, -6, -7 and -8 did not show any effect on the ATPase activity of RAD51 (Fig. 2B).
Fig. 2.
Only BRC1, -2, -3 and -4 inhibit ssDNA-dependent ATP hydrolysis by RAD51. (A) BRC1, -2, -3, and -4: RAD51 (3 μM) was incubated with increasing concentrations of GST-BRC peptide, as indicated, prior to addition of dT40 ssDNA and was further incubated for 1 h in the presence of 0.5 mM ATP and 4 mM Mg2+. B. Same as A but using BRC5, -6, -7, or -8. Error bars represent the SD for two or more independent experiments; where not seen, the error bars are smaller than the data points.
All of the BRC Repeats Stimulate RAD51-ssDNA Complex Formation, but by Different Mechanisms.
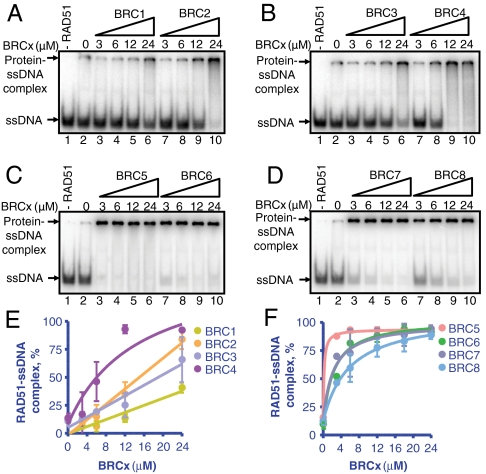
To determine whether the inhibition of ATPase activity results in a stabilization of RAD51-ssDNA complex, we used an electrophoretic mobility-shift assay (EMSA). Briefly, RAD51 was incubated with the individual BRC repeats, and added to ssDNA (dT40) in the presence of ATP, Mg2+, and Ca2+, as done previously for BRC4 (24). BRC1, -2, -3, and -4 stimulated the binding of RAD51 to ssDNA according to their binding affinities (BRC1: Fig. 3A (lanes 3–6); BRC2: Fig. 3A, (lanes 7–10); BRC3: Fig. 3B (lanes 3–6); and BRC4: Fig. 3B (lanes 7–10)). Unexpectedly, BRC5, -6, -7, and -8, despite both very weak binding to RAD51 (Fig. 1) and no effect on the ATPase activity of RAD51 (Fig. 2), greatly stimulated the binding of RAD51 to ssDNA (BRC5: Fig. 3C (lanes 3–6); BRC6: Fig. 3C (lanes 7–10); BRC7: Fig. 3D (lanes 3–6); and BRC8: Fig. 3D (lanes 7–10)). These results are plotted in Fig. 3 E and F. Fitting the data revealed that BRC5, -6, -7, and -8 increased the relative affinity of RAD51 for ssDNA more than either BRC1, -2, -3, or -4 (compare Fig. 3E to 3F; Table 2).
Fig. 3.
Each of the BRC repeats stimulates RAD51-ssDNA complex formation. (A) Autoradiograph of an acrylamide gel showing RAD51 (3 μM) incubated with either GST-BRC1 (lanes 3–6) or GST-BRC2 (lanes 7–10) prior to incubation with 32P-labeled dT40 ssDNA (0.3 μM) for 1 h in the presence of ATP, Mg2+, and Ca2+. Lane 1 contains DNA alone. Lane 2 contains RAD51 and DNA, in the absence of BRC peptide. (B) Same as in A but with GST-BRC3 (lanes 3–6) or GST-BRC4 (lanes 7–10). (C) Same as in A but with GST-BRC5 (lanes 3–6) or GST-BRC6 (lanes 7–10). (D) Same as in A but with GST-BRC7 (lanes 3–6) or GST-BRC8 (lanes 7–10). (E) Quantification of RAD51 binding to ssDNA in the presence of increasing concentrations of BRC1, -2, -3, and -4 calculated from the data shown in A and B. F. Quantification of RAD51 binding to ssDNA in the presence of increasing concentrations of BRC5, -6, -7, and -8 calculated from the data shown in C and D. Error bars represent the SD for two or more independent experiments; where are not seen, the error bars are smaller than the data points.
Table 2.
Apparent affinity of individual BRC repeat binding to the RAD51-ssDNA filament
| BRC repeat | Kd (μM) |
| BRC1 | N.A. |
| BRC2 | N.A. |
| BRC3 | N.A. |
| BRC4 | 14.6 ± 8.9 |
| BRC5 | 0.2 ± 0.1 |
| BRC6 | 2.2 ± 0.6 |
| BRC7 | 2.2 ± 1.1 |
| BRC8 | 5.8 ± 1.8 |
The apparent affinity of the BRC repeats for RAD51-ssDNA nucleoprotein filaments is defined as the concentration of BRCx required for half-maximal RAD51 nucleoprotein filament formation. The apparent dissociation constant (Kd,app ± S.D.) for BRCx binding was obtained from fitting of the EMSA experiments shown in Fig. 3 E and F. For the data that could not be fit reliably, no values are listed (N.A.).
Because BRC5 through 8 cannot bind free RAD51 (Fig. 1) but they do promote binding of RAD51 to ssDNA, we concluded that they must be binding to the RAD51-ssDNA filament rather than to free RAD51. To verify this inference, we performed the same mobility-shift experiment but used BRC5 bound to fluorescently labeled anti-GST as a reporter of the GST-tagged BRC5. Fig. S2 shows two images of the same gel: A shows the radioactive signal coming from the ssDNA and B shows the fluorescent signal coming from the reporter FL-BRC5. As can be seen, the fluorescently labeled BRC5 displayed the same mobility as the radiolabeled RAD51-ssDNA complexes, indicating that it is part of the complex. As expected, BRC5 alone did not shift the mobility of the ssDNA, confirming the absence of direct binding to ssDNA (Fig. S2A, lane 1); however, as seen in Fig. S2B (lane 1), there is some nonspecific retention of the fluorescently labeled BRC5 in the wells due to antibody binding. The fact that the amount of BRC repeat that bound to the filament increased above this background with increasing BRC5 concentration, while the amount of RAD51-ssDNA complex remained constant [compare Fig. S2A (lanes 3–6) and Fig. S2B (3–6)], shows that the BRC5 bound to the RAD51-ssDNA complex, and that it did not need to be at saturating amounts to exert its stabilizing effect. To our knowledge, no function had been previously ascribed to BRC repeats 5 through 8.
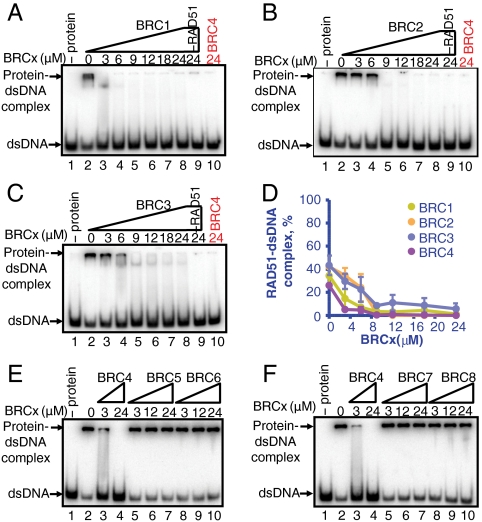
BRC1, -2, -3, and -4 Prevent the Formation of RAD51-dsDNA Nucleoprotein Filaments.
RAD51 binds with similar affinity to ssDNA and dsDNA (25–27). Previously we showed that BRC4 could inhibit the binding of RAD51 to dsDNA (24). The effect of the remaining BRC repeats on the formation of RAD51-dsDNA complex was examined as described previously (24). In the presence of ATP, Mg+2, and Ca+2, about 40% of the DNA was shifted by RAD51 binding, (Fig. 4A (lane 2) and Fig. 4D). The presence of BRC1, -2, -3, or -4 produced a concentration-dependent decrease in RAD51-dsDNA complexes, each to a similar extent, resulting in nearly complete inhibition of complex formation (Fig. 4 A–C and Fig. S3; quantification in Fig. 4D). In contrast, BRC5, -6, -7, -8 did not affect the formation of RAD51-dsDNA complexes (BRC5: Fig. 4E (lanes 5–7); BRC6: Fig. 4E (lanes 8–10); BRC7: Fig. 4F (lanes 5–7); and BRC8: Fig. 4F (lanes 8–10); BRC4 was used as a control in all panels). These results demonstrate that BRC1, -2, -3, and -4 share the common property of inhibiting the binding of RAD51 to dsDNA. Based on our previous work with BRC4, it is reasonable to expect that the mechanism of inhibition by these repeats would be the same: Namely, rather than disrupting the nucleoprotein filaments formed on dsDNA, they slow the nucleation of RAD51 onto dsDNA (24).
Fig. 4.
BRC1, -2, -3, and -4 prevent formation of RAD51-dsDNA complexes. (A) EMSA analysis showing RAD51 (3 μM) incubated with GST-BRC1 prior to incubation with 32P-labeled dA40·dT40 dsDNA (0.3 μM, bp) and further incubation for 1 h in the presence of ATP, Mg2+, and Ca2+. Protein-DNA complexes were resolved in 6% PAGE and analyzed by autoradiography. Lane 1 contains DNA alone; lane 2 contains RAD51 incubated with DNA in the absence of BRC peptide. Lanes 3–8 contain the indicated concentration of BRC1 peptide and 3 μM RAD51. Lane 9 contains the maximum concentration of BRC1 peptide and DNA in the absence of RAD51. Lane 10 contains the indicated concentration of BRC4 and RAD51 incubated with DNA under the same conditions used with BRC1. (B) Same as in A but using BRC2 instead of BRC1. (C) Same as in A but using BRC3 instead of BRC1. (D) Quantification of data in A, B, and C: BRC1 (green), BRC2 (orange), BRC3 (blue), and BRC4 (purple; gel shown in Fig. S3). Error bars indicate SD from at least three independent experiments.
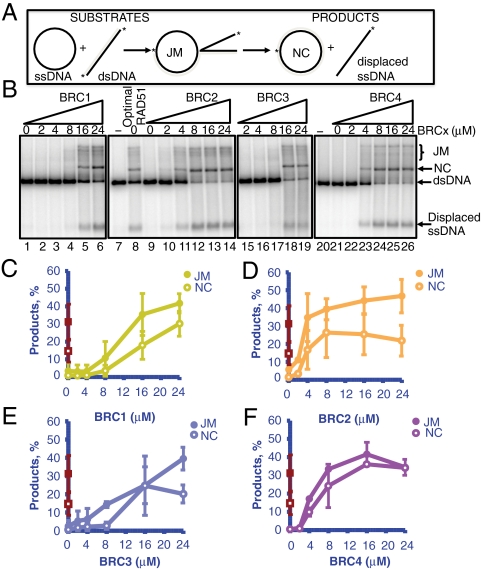
The Individual BRC Repeats Differ in their Ability to Stimulate RAD51-Mediated DNA Strand Exchange.
It is evident that the BRC repeats comprise two classes of regulatory elements that differentially modulate the DNA-binding selectivity of RAD51. To ascertain the consequences of this regulation, we examined their effect on the DNA strand exchange activity of RAD51 (24). Briefly, RAD51 was incubated with each individual BRC repeat in the presence of ϕX174 ssDNA. The mixture was then incubated with the ssDNA-binding protein, RPA, and the reaction was started by addition of 32P-labeled linearized ϕX174 dsDNA (Fig. 5A). At a two-fold higher concentration of RAD51 than the optimal amount required to saturate the ssDNA (Fig. 5B, lane 8), RAD51 binds to the dsDNA partner and decreases product formation to background levels [Fig. 5B (lanes 1, 9, and 21)] (25, 28). As previously shown (24), under these conditions, BRC4 stimulated RAD51-mediated DNA strand exchange (Fig. 5B, lanes 22–26) and the product yields surpassed those of the optimal RAD51 reaction (Fig. 5F, compare red squares versus purple circles). A similar stimulation was observed for BRC1 (Fig. 5B (lanes 2–6) and Fig. 5C), BRC2 (Fig. 5B (lanes 10–14) and Fig. 5D) and BRC3 (Fig. 5B (lanes 15–19) and Fig. 5E). This stimulation is reflected as an increase in the amount of both the intermediates of DNA strand exchange, joint molecules (JM) (Fig. 5 C–F, closed symbols), and the product, nicked circular dsDNA (NC) (Fig. 5 C–F, open symbols). The extent of stimulation was the greatest for BRC2 and BRC4 (Fig. 5 D and F), consistent with their higher affinity for RAD51. In contrast, BRC5, -6, -7, -8 did not stimulate RAD51-mediated DNA strand exchange reaction at the maximum concentrations attainable (Fig. S4). These results strongly suggest that BRC1, -2, -3, and -4 use a similar mechanism to stimulate RAD51-mediated DNA strand exchange activity. In contrast, the capabilities of BRC5, -6, -7, and -8 are restricted to the stimulation of RAD51 nucleoprotein filament formation on ssDNA.
Fig. 5.
Only BRC-1, -2, -3 and -4 can stimulate the DNA strand exchange activity of RAD51. (A) Scheme of DNA strand exchange between ϕX174 circular ssDNA and homologous linear dsDNA to produce joint molecules (JM) and nicked circular dsDNA (NC). The asterisk shows the 32P-label on each strand. (B) Effect of BRC1 (lanes 2–6), BRC2 (lanes 10–14), BRC3 (lanes 15–19), or BRC4 (lanes 22–26) on DNA strand exchange with excess RAD51 (7.5 μM); lane 8 shows DNA strand exchange at the optimal RAD51 concentration (3.75 μM). (C–F) Quantification of the joint molecules (JM, filled symbols) and nicked circular dsDNA (NC, open symbols) products using 3.75 μM RAD51 (red squares, same in every graph) or 7.5 μM RAD51 (circles) and GST-BRC1 (C), BRC2 (D), BRC3 (E), or BRC4 (F). Error bars indicate SD from at least three independent experiments.
Discussion
In spite of the many studies devoted to analysis of the BRC repeats of BRCA2, an unanswered fundamental question is why mammalian organisms carry a BRCA2 protein with eight BRC repeats, whereas BRCA2 homologs from organisms such as Caenorhabditis elegans BRC-2 or Ustilago maydis Brh2 have only a single BRC repeat. A related question concerns the function of each repeat in human BRCA2. In an attempt to answer these questions we studied the effects of the individual BRC repeats on RAD51 function.
Previous studies used semiquantitative methods, such as yeast two-hybrid or GST pull-downs from cell extracts, to estimate the affinity of several BRC repeats for RAD51 (20, 21). Using purified GST-tagged BRC repeats and purified RAD51, we quantified the affinity of each BRC repeat for both RAD51 and RAD51-ssDNA complexes. Our results show an unambiguous difference in the affinities of BRC repeats 1, -2, -3, and -4 versus -5, -6, -7, and -8 for RAD51. The first four repeats showed the strongest interaction with RAD51, and this is in accord with previous estimates (20, 21). BRC repeats 1, -2, -3, and -4 bound to RAD51 with a stoichiometry of approximately 1 to 1, whereas BRC repeats 5, -6, -7, and -8 bound very weakly; this dichotomy is in agreement with our recent work using full-length BRCA2 where two RAD51 binding modes were observed (8). Interestingly, BRC2, which contains several nonconserved residues compared to other BRC repeats (11), was remarkably similar in behavior to BRC4. The sequence of BRC2, however, is very well conserved in the BRC2 sequences of other mammalian species suggesting that residues outside of the canonical BRC repeat consensus sequence also contribute to the modulation of RAD51 DNA-binding affinity (12).
Previously, we demonstrated that either BRC4 or a region comprising BRC repeats 1–8 stabilized the formation of RAD51-ssDNA complex by inhibiting its DNA-dependent ATPase activity (24); this behavior was also manifest by the full-length protein, BRCA2 (8). Interestingly, the analysis reported here reveals that only the first four BRC repeats block ATP hydrolysis by RAD51. These results point to a similar mechanism for stabilization of RAD51-ssDNA complexes by BRC1, -2, -3, and -4. Unexpectedly, despite displaying a low affinity for free RAD51 protein (Fig. 1B, Table 1), BRC5, -6, -7, and -8 stimulated the binding of RAD51 to ssDNA in a BRCx concentration-dependent manner, as do the other repeats (Fig. 3 E and F). These results suggested that the affinity of these repeats for RAD51 increases significantly when RAD51 is assembled into nucleoprotein filaments on ssDNA; we have indeed verified this hypothesis as shown in Fig. 3 and Table 2. In addition, our results show that BRC1, -2 and -3 also inhibit RAD51-dsDNA complex formation to a similar extent as BRC4 (24); however, BRC5, -6, -7, and -8 lack this activity. Lastly, as a consequence of these properties, BRC1, -2, -3, and -4 promote DNA strand exchange when RAD51 is present in an amount excess of that needed to saturate the ssDNA, but BRC5, -6, -7, and -8 do not.
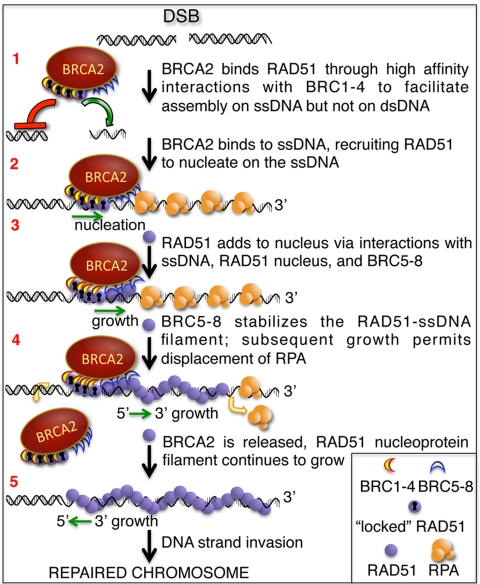
Together, these data suggest that the BRC repeats of human BRCA2 have evolved into two separate groups to ensure RAD51 nucleoprotein filament formation on ssDNA, rather than dsDNA, as indicated in our model (Fig. 6). After resection of a double-strand break (DSB), BRCA2 binds to the free form of RAD51 via BRC1, -2, -3, and -4 to facilitate nucleation of the RAD51 filament on the ssDNA (step 1). The high-affinity binding mode of this BRC group facilitates the rate-limiting nucleation of RAD51 on ssDNA by delivering it to the ssDNA, locally inhibiting ATP hydrolysis, and avoiding nonproductive association with dsDNA (step 2). We propose that the second group, formed by repeats BRC5, -6, -7, and -8, binds to the RAD51 molecules that have extended in a filamentous form from the nucleus on ssDNA (step 3). These repeats stabilize the association of RAD51 with ssDNA without affecting the ATP hydrolysis and, in a sense, serve as a chaperone to guide nascent filament formation. Once the filament starts to grow beyond the region physically occupied by BRCA2 (step 4), the BRCA2 would dissociate, having facilitated both nucleation and nascent growth. The BRCA2 could either dissociate without the RAD51 molecules that were originally bound to the BRCA2 leaving them bound to the ssDNA or, in principle, it could dissociate as a complex with the 4–5 tightly bound RAD51 molecules, which bind to BRCA2 with nanomolar affinity (step 4) (8), to engage another DSB and start the process again. The details of RAD51 filament growth are largely undefined but, similar to RecA (29), it likely can grow both directions (step 5) (30), to fill the ssDNA gap on either side of the nascent RAD51 filament thus enabling DNA strand exchange to proceed in either direction (8, 30, 31), although with a bias opposite to that of RecA (25, 32). Although the disposition of the BRC5-BRC8 relative to BRC1-BRC4 in the tertiary structure of BRCA2 is unknown, in principle, they can be organized either to one side of the BRC1-4 nucleation center in which case they would direct a unidirectional assembly of the RAD51 filament, or they could be located on either side of the nucleation core, in which case they would promote bidirectional growth that is needed to fill ssDNA gaps. The resulting nucleoprotein filament would then promote the homology-search, DNA-pairing, and strand-invasion steps of the recombination process.
Fig. 6.
Proposed model for the role of the BRC repeats in the context of the entire BRCA2 protein in DSB repair (see text for details). Upon formation of a DSB, the dsDNA is resected to generate ssDNA tails. RAD51 binds to BRCA2 through the high-affinity BRC repeats, BRC1-4 (1); via this interaction, the BRC repeats alter the conformation of RAD51, enhancing ssDNA binding and slowing dsDNA binding. The binding of BRCA2 to ssDNA directs RAD51 onto the ssDNA of a processed DSB and restricts assembly onto the dsDNA nearby; the new conformation imposed by BRC1-4 on RAD51 (“locked” RAD51) allows nucleation onto ssDNA and stabilizes the nascent nucleoprotein filament by limiting the ATP hydrolysis by RAD51 (2). After nucleation, BRC repeats 5–8 bind the nascent RAD51 nucleoprotein filament and further stabilize filament extension locally (3). The subsequent BRCA2-independent growth of the RAD51 nucleoprotein filament displaces RPA from the ssDNA. At this point, BRCA2 can be released from the DNA to promote RAD51 nucleation at another DSB (4). The RAD51 filament continues to grow from the BRCA2-stablized nucleus to form the ATP-bound nucleoprotein filament capable of homologous DNA pairing (5).
Many aspects of the BRC repeats remain to be elucidated. For example, recently, two modules within the BRC repeats have been described, adding another layer of complexity to their function (33). Our study of the individual BRC repeats has allowed us to ascribe a function to four of the previously unassigned BRC repeats and to unveil a separation of function between the eight BRC repeats of BRCA2. Within the context of the full protein, it is clear that the BRC repeats act synergistically to bind RAD51 because the affinity for RAD51 is approximately 1,000-fold higher than for an isolated BRC repeat (8, 24). The reinforcing activities of the BRC repeats described here can explain why mutation of a single BRC repeat can increase the risk of breast cancer, but paradoxically all of the BRC repeats are not essential for at least partial BRCA2 function: The redundancy and cooperativity of BRC function within the full protein ensure a high probability of nucleation and growth of RAD51, but clearly a single repeat can suffice as a minimal platform for assembly (34–36), as is the case for bacterial analogs (37, 38). For humans, however, most or all of the repeats are needed to ensure the efficient loading of RAD51 on ssDNA required for ongoing recombinational DNA repair of somatic cells.
Materials and Methods
Construction of GST-BRC Expression Plasmids.
Details are found in SI Text.
Protein Expression and Purification.
RAD51, RPA, and GST-tagged BRC repeats were purified as described (24). A GST contaminant copurifies with some of the GST-BRC preparations as a consequence of leaky expression from the bacterial host; the GST contaminant, however, had no effect because the same the results were obtained when the GST tag was cleaved (24). See SI Text for additional details.
GST Pull-Down Assay.
Glutathione-Sepharose 4B beads (GE) were equilibrated with binding buffer ‘B’: 20 mM TrisHCl (pH 7.5), 100 mM NaCl, 1 mM EDTA, and 1 mM DTT, 10% glycerol, 0.01% Igepal CA-630. Each purified GST-BRC peptide (0.5 μg) was incubated with 0.4–2.4 μg of purified RAD51, for 15 min at 37 °C and then batch bound to glutathione beads in a final volume of 30 μL for 30 min at 37 °C. The complexes were then washed with buffer B containing 0.1% Igepal CA-630 and loaded onto a 12% SDS- polyacrylamide gel. The gel was run for 1 h at 180 Volts and stained with SYPRO Orange (Invitrogen). The protein bands were quantified by ImageQuant software on a Storm 860 PhosphorImager (Molecular Dynamics). The amount of RAD51 pulled down with each GST-BRC peptide in Fig. 1A was determined using standard curves generated from known concentrations of RAD51 and GST-BRC peptides run in the same gel. When present, RAD51 retained by nonspecific binding to the beads was subtracted in the quantification. The input concentration of GST-BRC peptide in each pull-down reaction was 2.3 μM; when present, contaminating free GST in the BRC peptide preparation was accounted for so that the concentrations reported are those for the GST-BRCx peptide. The total input for RAD51 ranged from 1.6 μM to 9.6 μM. The binding data were fit to a hyperbola.
Preparation of GST-BRC Peptide Bound to Fluorescently Labeled Anti-GST.
Details are found in SI Text.
ATP Hydrolysis, DNA Strand Exchange, and Electrophoretic Mobility-Shift (EMSA) Assays.
The procedures and reactions were essentially as described (24); see SI Text for details.
Analysis.
In all graphs, error bars represent the standard deviation derived from at least two independent experiments and, in some cases, error bars are smaller than the symbol; all analyses used GraphPad Prism (version Mac 5.0b).
Supplementary Material
Acknowledgments.
We thank Dr. Ryan Jensen for providing the full-length BRCA2 construct. Thanks to all members of the Kowalczykowski laboratory for helpful comments and insights on the manuscript. This work was supported by grants from the National Institutes of Health (GM41347) and DOD-Breast Cancer Research Program (BC085223) to S.C.K., and a Postdoctoral Fellowship from Ministerio de Educación y Ciencia (Spain) to A.C.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106971108/-/DCSupplemental.
References
- 1.Lancaster JM, et al. BRCA2 mutations in primary breast and ovarian cancers. Nat Genet. 1996;13:238–240. doi: 10.1038/ng0696-238. [DOI] [PubMed] [Google Scholar]
- 2.Wooster R, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 3.Sharan SK, et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 4.Yuan SS, et al. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 5.Cejka P, et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niu H, et al. Mechanism of the ATP-dependent DNA end resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nimonkar AV, et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bork P, Blomberg N, Nilges M. Internal repeats in the BRCA2 protein sequence. Nat Genet. 1996;13:22–23. doi: 10.1038/ng0596-22. [DOI] [PubMed] [Google Scholar]
- 11.Bignell G, Micklem G, Stratton MR, Ashworth A, Wooster R. The BRC repeats are conserved in mammalian BRCA2 proteins. Hum Mol Genet. 1997;6:53–58. doi: 10.1093/hmg/6.1.53. [DOI] [PubMed] [Google Scholar]
- 12.Lo T, Pellegrini L, Venkitaraman AR, Blundell TL. Sequence fingerprints in BRCA2 and RAD51: Implications for DNA repair and cancer. DNA Repair (Amst) 2003;2:1015–1028. doi: 10.1016/s1568-7864(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 13.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 14.Davies OR, Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat Struct Mol Biol. 2007;14:475–483. doi: 10.1038/nsmb1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thorslund T, Esashi F, West SC. Interactions between human BRCA2 protein and the meiosis-specific recombinase DMC1. EMBO J. 2007;26:2915–2922. doi: 10.1038/sj.emboj.7601739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang H, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 17.Spain BH, Larson CJ, Shihabuddin LS, Gage FH, Verma IM. Truncated BRCA2 is cytoplasmic: Implications for cancer-linked mutations. Proc Natl Acad Sci USA. 1999;96:13920–13925. doi: 10.1073/pnas.96.24.13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pellegrini L, Venkitaraman A. Emerging functions of BRCA2 in DNA recombination. Trends Biochem Sci. 2004;29:310–316. doi: 10.1016/j.tibs.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Pellegrini L, et al. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 20.Wong AKC, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem. 1997;272:31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 21.Chen PL, et al. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc Natl Acad Sci USA. 1998;95:5287–5292. doi: 10.1073/pnas.95.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esashi F, et al. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 23.Chen CF, Chen PL, Zhong Q, Sharp ZD, Lee WH. Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J Biol Chem. 1999;274:32931–32935. doi: 10.1074/jbc.274.46.32931. [DOI] [PubMed] [Google Scholar]
- 24.Carreira A, et al. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 26.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 27.Mazin AV, Zaitseva E, Sung P, Kowalczykowski SC. Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing. EMBO J. 2000;19:1148–1156. doi: 10.1093/emboj/19.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 30.Namsaraev EA, Berg P. Rad51 uses one mechanism to drive DNA strand exchange in both directions. J Biol Chem. 2000;275:3970–3976. doi: 10.1074/jbc.275.6.3970. [DOI] [PubMed] [Google Scholar]
- 31.Namsaraev EA, Berg P. Branch migration during Rad51-promoted strand exchange proceeds in either direction. Proc Natl Acad Sci USA. 1998;95:10477–10481. doi: 10.1073/pnas.95.18.10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baumann P, West SC. The human Rad51 protein: polarity of strand transfer and stimulation by hRP-A. EMBO J. 1997;16:5198–5206. doi: 10.1093/emboj/16.17.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajendra E, Venkitaraman AR. Two modules in the BRC repeats of BRCA2 mediate structural and functional interactions with the RAD51 recombinase. Nucleic Acids Res. 2010;38:82–96. doi: 10.1093/nar/gkp873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin JS, Winkelmann N, Petalcorin MI, McIlwraith MJ, Boulton SJ. RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol Cell Biol. 2005;25:3127–3139. doi: 10.1128/MCB.25.8.3127-3139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 36.Saeki H, et al. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc Natl Acad Sci USA. 2006;103:8768–8773. doi: 10.1073/pnas.0600298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morimatsu K, Kowalczykowski SC. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: A universal step of recombinational repair. Mol Cell. 2003;11:1337–1347. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 38.Spies M, Kowalczykowski SC. The RecA binding locus of RecBCD is a general domain for recruitment of DNA strand exchange proteins. Mol Cell. 2006;21:573–580. doi: 10.1016/j.molcel.2006.01.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.