Abstract
The emergence of Vertebrata was accompanied by two rounds of whole-genome duplications. This enabled paralogous genes to acquire novel functions with high evolutionary potential, a process suggested to occur mostly by changes in gene regulation, rather than in protein sequences. In the case of Hox gene clusters, such duplications favored the appearance of distinct global regulations. To assess the impact of such “regulatory evolution” upon neo-functionalization, we developed PANTHERE (PAN-genomic Translocation for Heterologous Enhancer RE-shuffling) to bring the entire megabase-scale HoxD regulatory landscape in front of the HoxC gene cluster via a targeted translocation in vivo. At this chimeric locus, Hoxc genes could both interpret this foreign regulation and functionally substitute for their Hoxd counterparts. Our results emphasize the importance of evolving regulatory modules rather than their target genes in the process of neo-functionalization and offer a genetic tool to study the complexity of the vertebrate regulatory genome.
Keywords: genetics, limb development, global gene regulation, long-range enhancers, mouse genetics
The transition from early chordates to vertebrates was associated with two rounds of whole-genome duplications (2R hypothesis) (1, 2). Duplicated genes could thus acquire novel functions distinct from their ancestral roles during vertebrate evolution (3). This process could have involved modifications of coding sequences, albeit constrained by the original protein structure. Alternatively, the duplication of genomic loci provided an increased flexibility to generate new expression patterns, critical for the emergence of morphological novelties (4, 5). Interestingly, such “regulatory evolution” in vertebrates often involved enhancer elements located at considerable distances from their target promoters, where potential interferences with ancestral control modules are minimized (6–8) (Fig. 1A). Therefore, in contrast to invertebrate systems, megabase-scale genomic contexts have to be considered when attempting to understand the evolution of the vertebrate regulatory genome.
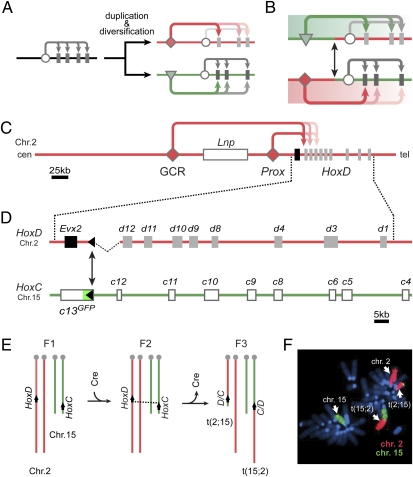
Fig. 1.
Functional diversification of vertebrate paralogous gene clusters via the evolution of specific global regulations. (A) After duplication of a gene cluster, which was structurally constrained by an ancestral regulatory module (circles), functional specializations of twin clusters occurred through the emergence of remote global regulatory controls (red and green arrows). (B) The conservative exchange of such regulatory landscapes in vivo reveals the respective importance of regulatory versus structural evolution of the gene clusters in the acquisition of vertebrate-related innovations. (C) On chromosome 2 (Chr. 2), multiple global enhancer sequences (e.g., GCR and Prox) drive the expression of several posterior Hoxd genes in the distal limb bud. This large region, located centromeric to the HoxD cluster, is referred to as a regulatory landscape (16). (D) Our PANTHERE (PAN-Genomic Translocation for Heterologous Enhancer RE-shuffling) strategy whereby any regulatory landscape containing a loxP site can be associated to a heterologous target locus. Here, a balanced translocation between chromosomes 2 (Chr. 2) and 15 (Chr. 15) exchanges the centromeric landscapes between the murine HoxD and HoxC loci, bringing the latter under control of the HoxD digit-regulatory landscape. (E) Crossing scheme of the PANTHERE approach. Transgenic Cre-expression induces recombination events between nonhomologous chromosomes harboring single loxP sites at the 5′ extremities of the HoxD and HoxC clusters. (F) Metaphase-spread nuclei of cells obtained from a mouse carrying the translocation, stained for chromosomes 2 (Chr. 2, red) and 15 (Chr. 15, green), showing the presence of both translocated chromosomes t(2;15) and t(15;2).
Such extended regulatory landscapes have been found to be associated with the expression of many genes essential for developmental processes. Among these are the four Hox gene clusters, which, after duplications, acquired novel and specific expression patterns due to the recruitment of distinct global enhancer elements (9, 10). For example, the HoxC and HoxD loci, which originated from the same ancestral complex via the second duplication event (11), diversified their functional territories rather significantly such that murine Hoxc genes are now required for tegument development (12), whereas Hoxd genes are involved in patterning of hands and feet (13). Accordingly, deletions of posterior Hoxd, but not Hoxc, genes cause severe digit malformations (14, 15). In the case of Hoxd genes, their transcription in the distal limb bud, the future digits, is controlled by multiple enhancer sequences located far upstream of the HoxD cluster (16, 17) (GCR and Prox in Fig. 1C). These enhancer sequences can act on several Hox genes, as well as on Evx2 and Lnp, two non-Hox transcription units located within this genomic context. As a result, these latter genes show similar expression in the developing hands and feet by means of a bystander effect, with only minor functional relevance for these structures (16, 18).
Recently, we reported that these enhancer sequences located centromeric to HoxD and driving these distal limb expression patterns, are absolutely necessary, as shown by an inversion taking these elements away from an otherwise intact HoxD cluster. This inverted allele phenocopied the digit defects observed after a deletion of all posterior Hoxd genes (19). However, it remained unclear as to whether such a regulatory landscape in itself would be sufficient to fully support the specific Hoxd gene expression in digits, or whether it also required additional structural changes either in the general topology of the cluster or in the gene coding regions. To address this question, we swapped the HoxD and HoxC regulatory landscapes, first to study whether the architecture of the HoxC cluster would be permissive for the HoxD global digit enhancers to take control of Hoxc genes and, second, to see if the resulting ectopic Hoxc expression could functionally substitute for Hoxd genes during digit development.
Results
Reshuffling Genomic Landscapes.
Because of the size and complexity of these regulatory landscapes, we exchanged the entire upstream chromosomal regions of the HoxC and HoxD gene clusters by engineering a targeted translocation (20) (Fig. 1 D and E). To this end, we developed PANTHERE (PAN-genomic Translocation for Heterologous Enhancer RE-shuffling) that enabled us to reshuffle the entire HoxD digit regulatory landscape in vivo by simple means of mouse breeding and genotyping procedures. As parental alleles, we used two stocks of mice harboring single loxP sites at the 5′ extremities of both gene clusters. Because the introduction of the loxP site at the Hoxc13 locus concomitantly inactivated the gene with an in-frame GFP reporter (21), we balanced the posterior Hoxd gene dose by using an allele where the Hoxd13 locus was deleted. This deletion allele induces an important up-regulation of Hoxd12 in the distal limb bud, which in turn is able to rescue to a large extent the digit defects normally associated with the inactivation of Hoxd13 (22). This particular setup would allow us to place the HoxC cluster under the HoxD centromeric regulatory landscape and thus to comparatively evaluate the effect of similar gene doses emanating either from the HoxD or the HoxC locus upon digit patterning. Mice double-heterozygous for both loxP sites were crossed with HprtCre transgenic animals, and F3 offspring were screened for the desired recombination event, giving rise to a balanced translocation in vivo. Animals who had not retained the Cre transgene were selected for further analysis (Fig. 1 E and F).
Hoxc Genes Under the Control of the HoxD Digit Regulatory Landscape Can Substantially Rescue the Distal Limb Skeleton.
As male founder animals were sterile, we crossed females heterozygous for the translocation with males carrying a balancer chromosome that lacks the three posterior-most Hoxd genes (15). In this manner, a full loss of posterior Hoxd gene function occurred in digits. However, Hoxc genes could potentially rescue this deficiency, should they be able to respond to the translocated regulatory landscape and be functional to support digit development.
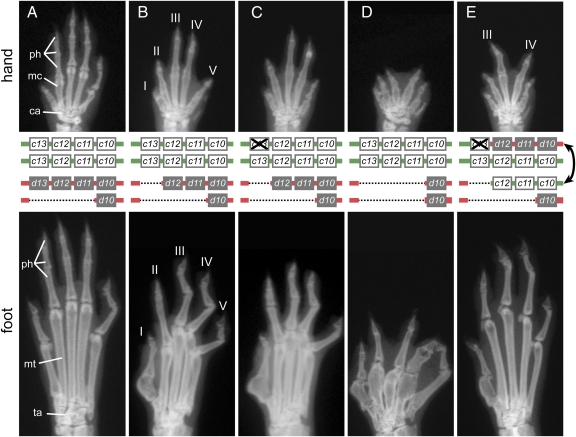
X-ray images of adult specimens confirmed that, whereas mice lacking one dose of posterior Hoxd genes had a wild-type limb skeleton, the deletion of both alleles of Hoxd13 resulted in abnormal distal limb development with shortening and loss of phalanges, a malformed digit I, and metatarsal truncations (Fig. 2 A and B). This phenotype was not further aggravated when, in addition, Hoxc13 was inactivated along with the introduction of the GFP (Fig. 2: compare B and C). The deletion of all posterior Hoxd genes caused severe reductions and malformations of digits, metacarpals, and metatarsals, as well as synpolydactyly in forelimbs and hind limbs (15) (Fig. 2D). Mice trans-heterozygous for both the deletion and the translocation also lacked all Hoxd gene function in distal limbs because one set of Hoxd genes was deleted, whereas the other had been disconnected from its regulatory landscape by the translocation. However, animals of this genotype displayed a clearly improved phenotype. In the forelimb, only digits III and IV were partially rescued (Fig. 2E), whereas the entire foot skeleton was restored to close-to-normal morphology although some phalanges were still missing (Fig. 2E).
Fig. 2.
Phenotypic rescue of Hoxd loss of function in limbs by Hoxc genes under the control of the HoxD regulatory landscape. X-ray images of distal forelimbs (hand, Top) and hind limbs (foot, Bottom) with a schematic of the genotypes (Middle) are shown. Whereas mice heterozygous for a deletion of Hoxd11 to Hoxd13 display an almost wild-type limb (A), removing the second dose of Hoxd13 leads to distal malformations (B). (C) The additional inactivation of one allele of Hoxc13 has no effect. (D) Mice homozygous for a deletion of the three posterior Hoxd genes display a severe distal limb phenotype. (E) Exchanging the 5′ regulatory landscapes of the HoxC and HoxD clusters leads to a significant rescue of the hand skeleton. Digits III and IV of the forelimb are markedly increased in size, with their metacarpals now close to normal in size and morphology. A spectacular rescue is observed for the foot, in particular in the overall aspect of metatarsals and in the number of phalanges. However, the big toe is not equally rescued and still shows the same mutant aspect as in D. ph, phalanges; mc, metacarpals; ca, carpals; mt, metatarsals; ta, tarsals.
Hoxc Genes Respond to HoxD Digit Enhancers and Can Functionally Substitute for Hoxd Gene Functions During Digit Development.
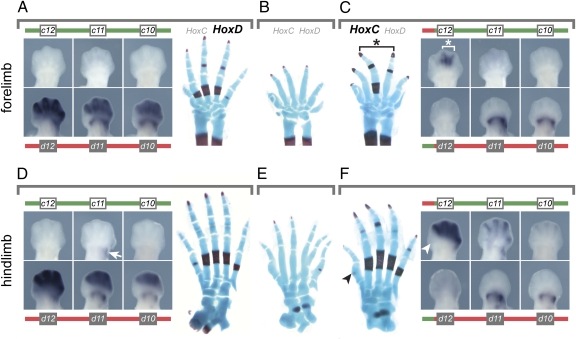
We concluded that one copy of the HoxC cluster placed under the 5′ regulatory landscape of HoxD was sufficient to compensate, to a large extent, for the absence of Hoxd gene function during digit development. To see more precisely which Hoxc genes were instrumental in this functional rescue, we monitored gene expression in vivo using balancer alleles removing the entire set of either Hoxc or Hoxd genes, such that transcription was scored either from a single wild-type or from a single translocated chromosome. In wild-type animals, Hoxd genes were expressed in both proximal and distal parts of the forelimb, with Hoxd12 being stronger distally than Hoxd11 or Hoxd10 (23) (Fig. 3A). In contrast, Hoxc genes were not transcribed at all (Fig. 3A). From the translocated chromosome, expression of Hoxd genes was completely abolished distally, whereas Hoxd11 and Hoxd10 remained expressed in their proximal domain (Fig. 3C), as expected from the telomeric location of this particular enhancer (24). However, Hoxc12 was now expressed in the distal limb bud, although restricted to the areas of presumptive digits III and IV (Fig. 3C).
Fig. 3.
Hoxc genes can functionally substitute for Hoxd genes during digit development. (A) In E12.5 wild-type embryos, posterior Hoxd genes are expressed in both the proximal and the distal part of the growing forelimb. No transcript is detected for Hoxc12, Hoxc11, or Hoxc10. (B) In newborns, the absence of Hoxd gene function in digits leads to a reduction in size and malformation of the cartilage rods, as well as a delay in their ossification. (C) After translocation, Hoxd gene expression is lost distally, whereas Hoxc12 becomes expressed in digits III and IV, i.e., precisely where a rescue in both the size and the ossification pattern is observed (asterisk). (D) As for the forelimb, only Hoxd genes are expressed in the distal domain of the developing hind limb. Proximally, however, transcripts for Hoxc11 are detected (arrow). (E) The loss of distal Hoxd function induces similar effects on foot development, as observed in forelimbs. (F) In distal hind limbs at E12.5, Hoxc12, Hoxc11, and Hoxc10 are expressed in a quantitative collinear manner when controlled by the HoxD regulatory landscape, much like Hoxd genes in their wild-type context. These Hoxc transcripts substantially rescue the cortical ossification pattern in the foot, which is now almost identical to wild type. The big toe is not rescued, as explained by the lack of transcription of Hoxc genes in this presumptive digit (arrowheads).
In hind-limb buds, the situation was comparable, yet the expression of Hoxc12 from the translocated chromosome was much stronger and more widespread than in forelimb buds. Moreover, weak ectopic distal signals were scored for Hoxc11 and Hoxc10 (Fig. 3F), thus following the collinear expression strategy observed at the Hoxd locus under normal conditions. Therefore, from these redistributed expression patterns, it appeared that the rescue in digit morphology mostly originated from the ectopic expression of Hoxc12. The direct morphological effects of these transcriptional reallocations were best demonstrated in newborn postnatal day 0 (P0) skeletons. Although the loss of Hoxd gene expression led to an important delay in primary ossification in digits (Fig. 3 B and E), bone patterns were restored at the exact positions of ectopic Hoxc12 expression. In the forelimb, expression was largely restricted to the areas of presumptive digit III and, to some extent, digit IV. Accordingly, digit III was largely rescued in its length and ossification process, whereas digit IV was moderately rescued (Fig. 3C, asterisk). In the hind limb, Hoxc12 was expressed similarly to Hoxd12 in wild-type limb buds, i.e., in all developing digits with the exception of the thumb. Consequently, digits II–IV were significantly restored, whereas digit I conserved its mutant morphology (Fig. 3 E and F, arrowheads).
Discussion
The rise of the vertebrate lineage was likely facilitated by two genome duplications, which took place early on during chordate evolution (1). They endowed vertebrates with a particular genetic flexibility, in particular for paralogous genes to diverge in functions via sub- or neo-functionalization processes (25). A substantial part of this flexibility was achieved through the evolution of regulatory modules, rather than of the proteins themselves (regulatory evolution). In this view, the functional territories of genes were enlarged and/or modified by changing their associated expression patterns in space and time, whereas the original protein structures were largely preserved. However, the respective contributions of regulatory evolution versus structural modifications of coding regions in vertebrate evolution are controversial, due in part to the difficulty of assessing these various parameters in physiological conditions in vivo. Hox gene clusters, with their associated dense transcriptional landscapes and their global neo-functionalizations, are of particular interest in this respect.
Hox Cluster Architecture and Global Transcriptional Regulations.
In the bilaterian lineage, various degrees of Hox gene clustering are observed (26). Although this structural diversity in itself has important implications for the transcriptional regulation of the resident genes, the topology of the gene cluster may in turn be modified and consolidated through the appearance of novel regulatory controls. The recurrent deployment of global regulatory elements at vertebrate Hox loci might thus have contributed to their exceptionally tight organization (11).
Interestingly, the posterior half of the murine HoxD complex, which is the target of several global regulations involved either in limb or in external genitalia development (10), is more compacted than its HoxC counterpart (Fig. 1D). This may account for the differences in the quantitative collinear responses of posterior Hoxc genes to the HoxD centromeric regulatory landscape, as evidenced by the sharper decrease in transcriptional efficiencies in the translocated chromosome, compared with Hoxd genes in the control condition (Fig. 3F). Moreover, the striking difference in the distal expression between forelimb and hind-limb buds of posterior Hoxc genes when controlled by the HoxD landscape likely reflects the wild-type patterns of Hoxc gene expression, which are indeed normally restricted to a proximal region of hind-limb buds only (27, 28) (Fig. 3). As such, it suggests that the control of Hoxc genes by global enhancers in the translocated chromosome is modulated by cluster-specific regulatory modalities that may be different in forelimb and hind-limb buds.
Hox Genes and the Evolution of Digit Morphologies.
The phenotypic rescue indicates that the HOXC12 protein can substitute for HOXD12, even though the former is not required for digit development under normal circumstances. In fact, the restoration of cortical ossification matches the exact places where ectopic HOXC12 is detected, which illustrates the functional potency of a single dose of a posterior Hox gene product. The spatially restricted restoration functionally demonstrates the local and direct effect of HOX protein levels on the morphogenesis of individual digits via their impact upon the stimulation of primary ossification (29). This, added to the dose-dependent effects of various paralogous Hox groups on digit morphologies (30), may provide part of the explanation of the amazing range of variations observed in tetrapod distal limb skeletons.
In this context, slight modifications in transcriptional regulation, for example, through alterations in cluster architecture or enhancer activities, could have modulated the phenotype and thus participated in the morphological diversification of digits. In particular, we show here that a very localized dosage effect can impact upon a single digit independently from the neighboring digits, a phenomenon that may underlie the development of those autopods where the shape and/or the length of digits are notoriously heterogeneous within the same handplate, as seen, e.g., in bat wings (31, 32). Our work also suggests that any group 12 or 13 HOX protein may contribute to these processes, thus increasing the realm of possibilities for morphological variation to occur.
Evolution at Two Levels.
Our engineered translocation shows that posterior Hoxc genes can in part interpret the HoxD regulatory landscape and adopt the associated transcriptional responses. This indicates that the recruitment of Hoxd gene function during limb development primarily involved the evolution of a complex cis-regulatory landscape located centromeric to the cluster, rather than changes in the protein structure. Therefore, HOX proteins appear rather generic with respect to their functions (33), perhaps due to the functional constraints imposed by the ancestral collinear mechanism acting on all four Hox clusters during axial elongation. Although the regulatory subfunctionalization of their ancestral patterning functions had been demonstrated previously for a pair of paralogous Hox genes (34), we provide evidence that this potential also exists at the level of entire Hox gene clusters, which seem to be equally responsive to emergent global regulations. Clearly, Hoxc genes could theoretically have been recruited during the emergence of tetrapod digits, instead of the HoxD cluster. Nevertheless, following the initial duplication, the HoxC cluster underwent its own neo-functionalization and acquired a different set of regulations, for example, driving gene expression in hair follicles.
The presence of gene-poor regions flanking all four Hox loci (35), a feature generally associated with complex regulatory landscapes, suggests that all Hox gene clusters were equally co-opted by new global regulations, leading to a division of labor in terms of neo-functionalization. It is nevertheless likely that these novel regulatory modalities subsequently led to minor adaptations in protein structure to optimize any new task, while being constrained by the ancestral collinear mechanism occurring earlier in development. Such a scenario may explain why we do not see a complete rescue in the hind-limb digit skeleton after ectopic expression of Hoxc genes.
Studying Regulatory Evolution.
Understanding the vertebrate regulatory genome will require the analysis of very large and complex genomic segments and their interactions in time and space with one or several target genes. Although the genome-wide analyses of potential enhancer regions, chromatin domains, or chromosomal architectures will be essential in this challenge (36–38), the PANTHERE strategy described here to engineer chimeric loci adds a unique genetic tool to these studies. Using this approach, entire cis-regulatory landscapes can be shuffled around using targeted translocations to assess the respective importance of “regulatory” versus “structural” evolutionary mechanisms under physiological conditions in vivo.
Methods
Mouse Strains and PANTHERE.
The translocation allele was generated using the PANTHERE approach (Fig.1 D and E). For parental strains, we used loxP sites from a Hoxc13GFP allele (21) and a deletion of the Hoxd13 locus that also removes the regulatory region RXI (22). Trans-heterozygous animals were bred to HprtCre mice to induce a targeted translocation between nonhomologous chromosomes (20, 39). The allele was maintained on a B6/CBA F1/CD1 hybrid background. The different balancer alleles were described before: Del(11-13)d11lac (15), Del(11-13) (22), Del(1-13)d11lac (6), and HoxCnull (16). For embryo crosses, noon on the day of the vaginal plug was considered as embryonic day 0.5 (E0.5). Embryos were dissected in ice-cold PBS and fixed overnight in 4% paraformaldehyde.
Genotyping.
Genotyping was performed on isolated ear punch or yolk sac DNA using standard PCR protocols. Oligonucleotide sequences used to genotype both breakpoints were as follows: HoxD-fw: 5′-AGGCATTTTCCTACCCTTCA-3′ and HoxC-rv: 5′-GAATGACTGGGCTCTCCAGGC-3′ and GFP-fw: 5′-GTCCTGCTGGAGTTCGTGAC-3′ and HoxD-rv: 5′-TGGGCAAAACAGGCAACGCTCC-3′. Both resulting PCR products were sequenced to check for breakpoint integrity.
Metaphase Fluorescent in Situ hybridization.
Fluorescent in situ hybridization (FISH) on metaphase spreads of ear fibroblasts from translocation-heterozygous mice was performed as modified from ref. 40. Whole-chromosome paint FISH probes for chromosomes 2 and 15 (StarFISH; Cambio) were used according to the manufacturer's instructions. Additionally, to detect the distal 500 kb of chromosome 15, 1 μg of DNA of BAC RP23-220E23 (CHORI BACPAC Resources) was labeled by nick-translation with Spectrum Green (Vysis Inc.) according to the manufacturer's protocol and cohybridized.
X-Ray Imaging, Skeletal Preparations, and In Situ Hybridization.
Two-month-old males were killed and X-ray images of hands and feet were collected. For analyses of newborn skeletons, P0 animals were killed, eviscerated, and stained for cartilage and bone using standard alcian blue/alizarin red protocols (41). For X-rays and skeletal preparations, a minimum of five specimens per genotype was analyzed, with an indistinguishable degree of rescue in the hind limb and only minor fluctuations in the forelimb. Whole-mount in situ hybridization was performed according to standard protocols, with both mutant and control embryos processed in the same well to maintain identical conditions throughout the procedure. All stainings were reproduced in at least two independent experiments. Hoxd probes were as described elsewhere: Hoxd10 and Hoxd11 (42) and Hoxd12 (43). For Hoxc genes, probe templates were PCR-cloned into pGEM-T easy (Promega) from embryonic cDNA pools following the designs outlined in ref. 28. To assess the expression of Hoxd genes from the translocated chromosome, t(2;15)/HoxDΔ embryos were used. Conversely, Hoxc expression from the translocated counterpart chromosome was assessed in t(2;15)/HoxCnull embryos.
Acknowledgments
We thank Mario Capecchi for the generous gift of the Hoxc13GFP stock and Rolf Zeller for comments and suggestions on the manuscript. We also acknowledge Fabienne Chabaud for help with mouse fibroblasts and members of the Duboule laboratories for discussions and reagents. HoxCnull mice were purchased from the RIKEN BioResource Center. This work was supported by funds from the University of Geneva, the Ecole Polytechnique Fédérale, Lausanne, the Swiss National Research Fund, the National Research Center Frontiers in Genetics, the EU program ‘Crescendo’ and the European Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Ohno S. Evolution by Gene Duplication. Heidelberg, Germany: Springer-Verlag; 1970. [Google Scholar]
- 2.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holland PW, Garcia-Fernàndez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl. 1994:125–133. [PubMed] [Google Scholar]
- 4.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 5.Carroll SB. Evolution at two levels: On genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spitz F, et al. Large scale transgenic and cluster deletion analysis of the HoxD complex separate an ancestral regulatory module from evolutionary innovations. Genes Dev. 2001;15:2209–2214. doi: 10.1101/gad.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 8.Zuniga A, et al. Mouse limb deformity mutations disrupt a global control region within the large regulatory landscape required for Gremlin expression. Genes Dev. 2004;18:1553–1564. doi: 10.1101/gad.299904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deschamps J. Ancestral and recently recruited global control of the Hox genes in development. Curr Opin Genet Dev. 2007;17:422–427. doi: 10.1016/j.gde.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Duboule D. The rise and fall of Hox gene clusters. Development. 2007;134:2549–2560. doi: 10.1242/dev.001065. [DOI] [PubMed] [Google Scholar]
- 11.Ravi V, et al. Elephant shark (Callorhinchus milii) provides insights into the evolution of Hox gene clusters in gnathostomes. Proc Natl Acad Sci USA. 2009;106:16327–16332. doi: 10.1073/pnas.0907914106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Godwin AR, Capecchi MR. Hoxc13 mutant mice lack external hair. Genes Dev. 1998;12:11–20. doi: 10.1101/gad.12.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dollé P, Izpisúa-Belmonte JC, Falkenstein H, Renucci A, Duboule D. Coordinate expression of the murine Hox-5 complex homoeobox-containing genes during limb pattern formation. Nature. 1989;342:767–772. doi: 10.1038/342767a0. [DOI] [PubMed] [Google Scholar]
- 14.Zákány J, Duboule D. Synpolydactyly in mice with a targeted deficiency in the HoxD complex. Nature. 1996;384:69–71. doi: 10.1038/384069a0. [DOI] [PubMed] [Google Scholar]
- 15.Suemori H, Noguchi S. Hox C cluster genes are dispensable for overall body plan of mouse embryonic development. Dev Biol. 2000;220:333–342. doi: 10.1006/dbio.2000.9651. [DOI] [PubMed] [Google Scholar]
- 16.Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez F, Duboule D, Spitz F. Transgenic analysis of Hoxd gene regulation during digit development. Dev Biol. 2007;306:847–859. doi: 10.1016/j.ydbio.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 18.Hérault Y, Hraba-Renevey S, van der Hoeven F, Duboule D. Function of the Evx-2 gene in the morphogenesis of vertebrate limbs. EMBO J. 1996;15:6727–6738. [PMC free article] [PubMed] [Google Scholar]
- 19.Tschopp P, Duboule D. A regulatory ‘landscape effect’ over the HoxD cluster. Dev Biol. 2011;351:288–296. doi: 10.1016/j.ydbio.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 20.Wu S, Ying G, Wu Q, Capecchi MR. Toward simpler and faster genome-wide mutagenesis in mice. Nat Genet. 2007;39:922–930. doi: 10.1038/ng2060. [DOI] [PubMed] [Google Scholar]
- 21.Godwin AR, Stadler HS, Nakamura K, Capecchi MR. Detection of targeted GFP-Hox gene fusions during mouse embryogenesis. Proc Natl Acad Sci USA. 1998;95:13042–13047. doi: 10.1073/pnas.95.22.13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kmita M, Fraudeau N, Hérault Y, Duboule D. Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs. Nature. 2002;420:145–150. doi: 10.1038/nature01189. [DOI] [PubMed] [Google Scholar]
- 23.Montavon T, Le Garrec JF, Kerszberg M, Duboule D. Modeling Hox gene regulation in digits: Reverse collinearity and the molecular origin of thumbness. Genes Dev. 2008;22:346–359. doi: 10.1101/gad.1631708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zákány J, Kmita M, Duboule D. A dual role for Hox genes in limb anterior-posterior asymmetry. Science. 2004;304:1669–1672. doi: 10.1126/science.1096049. [DOI] [PubMed] [Google Scholar]
- 25.De Robertis EM. Evo-devo: Variations on ancestral themes. Cell. 2008;132:185–195. doi: 10.1016/j.cell.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Fernàndez J. The genesis and evolution of homeobox gene clusters. Nat Rev Genet. 2005;6:881–892. doi: 10.1038/nrg1723. [DOI] [PubMed] [Google Scholar]
- 27.Oliver G, Wright CV, Hardwicke J, De Robertis EM. Differential antero-posterior expression of two proteins encoded by a homeobox gene in Xenopus and mouse embryos. EMBO J. 1988;7:3199–3209. doi: 10.1002/j.1460-2075.1988.tb03187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson RL, Papenbrock T, Davda MM, Awgulewitsch A. The murine Hoxc cluster contains five neighboring AbdB-related Hox genes that show unique spatially coordinated expression in posterior embryonic subregions. Mech Dev. 1994;47:253–260. doi: 10.1016/0925-4773(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 29.Villavicencio-Lorini P, et al. Homeobox genes d11-d13 and a13 control mouse autopod cortical bone and joint formation. J Clin Invest. 2010;120:1994–2004. doi: 10.1172/JCI41554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zákány J, Fromental-Ramain C, Warot X, Duboule D. Regulation of number and size of digits by posterior Hox genes: A dose-dependent mechanism with potential evolutionary implications. Proc Natl Acad Sci USA. 1997;94:13695–13700. doi: 10.1073/pnas.94.25.13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen CH, Cretekos CJ, Rasweiler JJ, IV, Behringer RR. Hoxd13 expression in the developing limbs of the short-tailed fruit bat, Carollia perspicillata. Evol Dev. 2005;7:130–141. doi: 10.1111/j.1525-142X.2005.05015.x. [DOI] [PubMed] [Google Scholar]
- 32.Ray R, Capecchi M. An examination of the Chiropteran HoxD locus from an evolutionary perspective. Evol Dev. 2008;10:657–670. doi: 10.1111/j.1525-142X.2008.00279.x. [DOI] [PubMed] [Google Scholar]
- 33.Greer JM, Puetz J, Thomas KR, Capecchi MR. Maintenance of functional equivalence during paralogous Hox gene evolution. Nature. 2000;403:661–665. doi: 10.1038/35001077. [DOI] [PubMed] [Google Scholar]
- 34.Tvrdik P, Capecchi MR. Reversal of Hox1 gene subfunctionalization in the mouse. Dev Cell. 2006;11:239–250. doi: 10.1016/j.devcel.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 35.Lee AP, Koh EG, Tay A, Brenner S, Venkatesh B. Highly conserved syntenic blocks at the vertebrate Hox loci and conserved regulatory elements within and outside Hox gene clusters. Proc Natl Acad Sci USA. 2006;103:6994–6999. doi: 10.1073/pnas.0601492103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 37.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Steensel B, Dekker J. Genomics tools for unraveling chromosome architecture. Nat Biotechnol. 2010;28:1089–1095. doi: 10.1038/nbt.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang SH, Silva FJ, Tsark WM, Mann JR. A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis. 2002;32:199–202. doi: 10.1002/gene.10030. [DOI] [PubMed] [Google Scholar]
- 40.Pinkel D, et al. Fluorescence in situ hybridization with human chromosome-specific libraries: Detection of trisomy 21 and translocations of chromosome 4. Proc Natl Acad Sci USA. 1988;85:9138–9142. doi: 10.1073/pnas.85.23.9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inouye M. Differential staining of cartilage and bone in fetal mouse skeleton by alcian blue and alizarin red. S Cong Anom. 1976;16:171–173. [Google Scholar]
- 42.Gérard M, et al. In vivo targeted mutagenesis of a regulatory element required for positioning the Hoxd-11 and Hoxd-10 expression boundaries. Genes Dev. 1996;10:2326–2334. doi: 10.1101/gad.10.18.2326. [DOI] [PubMed] [Google Scholar]
- 43.Izpisúa-Belmonte JC, Falkenstein H, Dollé P, Renucci A, Duboule D. Murine genes related to the Drosophila AbdB homeotic genes are sequentially expressed during development of the posterior part of the body. EMBO J. 1991;10:2279–2289. doi: 10.1002/j.1460-2075.1991.tb07764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]