Abstract
In the male germline in mammals, chromatoid bodies, a specialized assembly of cytoplasmic ribonucleoprotein (RNP), are structurally evident during meiosis and haploidgenesis, but their developmental origin and regulation remain elusive. The tudor domain containing proteins constitute a conserved class of chromatoid body components. We show that tudor domain containing 7 (Tdrd7), the deficiency of which causes male sterility and age-related cataract (as well as glaucoma), is essential for haploid spermatid development and defines, in concert with Tdrd6, key biogenesis processes of chromatoid bodies. Single and double knockouts of Tdrd7 and Tdrd6 demonstrated that these spermiogenic tudor genes orchestrate developmental programs for ordered remodeling of chromatoid bodies, including the initial establishment, subsequent RNP fusion with ubiquitous processing bodies/GW bodies and later structural maintenance. Tdrd7 suppresses LINE1 retrotransposons independently of piwi-interacting RNA (piRNA) biogenesis wherein Tdrd1 and Tdrd9 operate, indicating that distinct Tdrd pathways act against retrotransposons in the male germline. Tdrd6, in contrast, does not affect retrotransposons but functions at a later stage of spermiogenesis when chromatoid bodies exhibit aggresome-like properties. Our results delineate that chromatoid bodies assemble as an integrated compartment incorporating both germline and ubiquitous features as spermatogenesis proceeds and that the conserved tudor family genes act as master regulators of this unique RNP remodeling, which is genetically linked to the male germline integrity in mammals.
Keywords: germ cells, germinal granules, nuage
Germinal granules/nuage are a conserved feature of germ cells in many animals and are ribonucleoprotein (RNP)-rich cytoplasmic compartments with an amorphous architecture (1). In model animals such as Drosophila and Caenorhabditis elegans, prospective germ cells are fate-determined by maternal factors, several of which are concentrated in germinal granules in the germ plasm (2). These germinal granules are asymmetrically partitioned to the germ cell lineage, leading to a notion that they act as an assembly site of germ cell determinants. In contrast, prospective germ cells in mammals are induced among pluripotent cells, and the presence of germinal granules in early mammalian embryos remains obscure. However, germinal granules similar to those observed in other species are discerned at later stages of the mammalian germline, spermatogenesis and oogenesis (1, 3). These mammalian germinal granules are classified into two types. One is intermitochondrial cement [intermitochondrial material/bar; also termed “pi-body” (4)], which is assembled among clusters of mitochondria in fetal prospermatogonia, postnatal spermatogonia, and meiotic spermatocytes in the male and in developing oocytes in the female. Another more conspicuous form of mammalian germinal granules is chromatoid bodies, which are massive RNP aggregates that develop in mid-to-late meiotic spermatocytes and haploid spermatids (1, 3). Germinal granules of diverse animals, including mammals, conserve not only morphological similarities but also key molecular components, suggesting that they have a common and essential role(s), which, however, remains enigmatic.
As mentioned, maternal gene products in Drosophila essential for the germline are localized in germinal granules (2). Mammals possess homologs of such genes, including vasa, tudor, piwi, and mael, and these gene products are also localized in mammalian germinal granules (3–13). Such germline-specific and conserved composition suggests that germinal granules are a bona fide feature that characterizes the germline. However, germinal granules are enriched with another class of components, which in somatic cells are assembled as processing bodies/GW bodies, a form of RNP compartment widely conserved in eukaryotes and the presumed function of which is RNA decay and/or translational control. Chromatoid bodies share several components with somatic processing bodies, such as RNA decapping enzymes and argonaute proteins (14). Similarly, germinal granules in Drosophila and C. elegans exhibit compositional overlap with somatic processing bodies (15, 16), leading to a controversy over whether germinal granules are a modified analog of ubiquitous processing bodies or a germline-specific feature.
We previously showed that tudor domain-containing (TDRD) proteins, including TDRD1, -6, -7, and -9, constitute conserved components of chromatoid bodies in mice (5, 6, 9, 10). TDRD1 binds to arginine dimethylated MILI, a mouse piwi protein; TDRD9 associates with MIWI2, another essential piwi protein; and the two Tdrd members cooperate in the piwi-interacting RNA (piRNA) pathway to establish retrotransposon silencing in fetal prospermatogonia (4, 9, 12, 17, 18). Remarkably, TDRD1 and MILI are localized in intermitochondrial cement (pi bodies), whereas TDRD9 and MIWI2 are enriched at processing bodies (piP bodies) in prospermatogonia (4, 9, 12), demonstrating that germinal granules and processing bodies are distinct RNP assemblies at the fetal stage of male germ cells.
In this study, we show that Tdrd7 and Tdrd6, which sequentially act in spermiogenesis, define key developmental programs for the unique and ordered RNP remodeling of chromatoid bodies. Our data demonstrate that chromatoid bodies are not a static RNP aggregation but undergo dynamic structural and compositional reorganization during spermatogenesis and provide genetic evidence that the conserved tudor family genes closely link male haploid development to chromatoid bodies, a long-recognized but enigmatic RNP architecture in mammalian spermatogenesis.
Results
Early Spermiogenesis Phenotype in Tdrd7−/− Gene-Targeted Males.
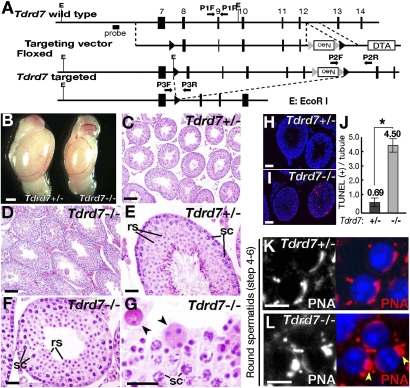
We previously showed that TDRD7 is a tandem tudor domain protein that is expressed in male germ cells and localizes to chromatoid bodies (10). The expression and localization of TDRD7 in spermatogenesis is summarized in Fig. S1. Tdrd7 is abundantly expressed in the testis, but is also detectable in several somatic tissues, including the nervous system and the eye (Fig. S1D). The role of Tdrd7 in eye development was recently described (19). In this study, we made a gene-targeted null mutation of Tdrd7 (Fig. 1A and Fig. S2 A–G) and investigated its spermiogenesis phenotype in detail. Tdrd7−/− mice were viable and obtained at a Mendelian ratio (+/+:+/−:−/− = 42:80:45). However, male homozygotes were sterile (8.8 ± 0.7 pups/litter for Tdrd7+/− males and no pups for Tdrd7−/− males; n = 12) whereas females were fertile (8.1 ± 0.4 pups/litter for Tdrd7+/− females and 7.7 ± 0.8 for Tdrd7−/− females; n = 12). Tdrd7−/− testes were small (Fig. 1B) with spermatogenesis being arrested at a round spermatid stage (Fig. 1 C–G and Fig. S2 H and I). This early spermiogenesis arrest was consistent with increased apoptosis among round spermatids (Fig. 1 H–J) and decreased levels of late spermatid markers (Fig. S2J), which was likely due to the differentiation defect and/or cell death in Tdrd7−/− testes. The spermiogenesis defect by Tdrd7−/− mutation was comparable to that by Tdrd6 mutations (Fig. S2K–Z) (11). However, the Tdrd7−/− phenotype preceded that of Tdrd6−/−. Tdrd7−/− spermatids were impaired before nuclear condensation commenced (steps 4–6) with initial acrosome development being defective (Fig. 1 K and L), whereas Tdrd6−/− spermiogenesis proceeded to an elongating stage (steps 13–14) (Fig. S2 AA and AB) where transition proteins and protamines replaced nuclear histones (Fig. S3 A–J). In both Tdrd7−/− and Tdrd6−/− mutants, spermatogonia and spermatocytes were unaffected (Fig. S3 K–R), which contrasted to the meiotic catastrophe of Tdrd1−/− and Tdrd9−/− spermatocytes (Fig. S3 S–Z) (5, 9). Thus, Tdrd7 and Tdrd6 constitute a spermiogenic class of tudor family genes, which function in a sequential and nonredundant manner. A recent study unveiled that Tdrd7 has an additional somatic function in the eye and controls the development of cataracts as well as glaucoma (19), which we also observed in our aged Tdrd7−/− mice (Fig. S3 AA and AB). Such disorder was not seen in other Tdrd mutants examined (Tdrd1, -6, and -9), suggesting that the age-related function in lens development is likely preferential to Tdrd7.
Fig. 1.
Tdrd7−/− mutation causes haploid spermatid defect. (A) A gene-targeting construct for Tdrd7. (B) Tdrd7+/− and Tdrd7−/− testes at 10 wk. Tdrd7−/− testes (67 ± 5 mg, n = 12) are smaller than Tdrd7+/− testes (109 ± 10 mg, n = 12) (P < 0.01). (C–G) Hematoxylin and eosin H&E-stained sections of Tdrd7+/− (C and E) and Tdrd7−/− (D, F, and G) testes at 2 mo. The arrowheads (G) mark Tdrd7−/− spermatids with aberrant nuclear morphologies. rs, round spermatids; sc, spermatocytes. (H and I) TUNEL staining (red) of apoptotic cells in Tdrd7+/− (H) and Tdrd7−/− (I) testis sections counterstained with a Hoechst dye (blue). (J) Quantification of apoptotic cells in Tdrd7+/− and Tdrd7−/− testes (means and SE from 50 seminiferous tubule cross-sections, P < 0.01, Student's t test). (K and L) Acrosome staining [peanut agglutinin (PNA), red] of Tdrd7+/− (K) and Tdrd7−/− (L) spermatids counterstained with a Hoechst dye (blue). In Tdrd7−/− spermatids (L), the acrosomes were severely fragmented (arrowheads). (Scale bars: B, 1 mm; C and D, 50 μm; E–I, 25 μm; K and L, 10 μm.)
Tdrd7 Regulates Dynamic RNP Remodeling of Chromatoid Bodies.
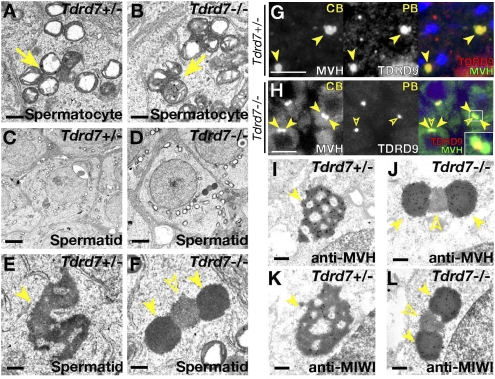
We next examined germinal granules in Tdrd7−/− spermatogenesis in detail by electron microscopy. In Tdrd7−/− testes, intermitochondrial cement was normally seen (Fig. 2 A and B). In contrast, chromatoid bodies showed a peculiar architecture in Tdrd7−/− mutants (Fig. 2 C–F), which was never seen in other Tdrd mutants examined (Tdrd1, -4, -5, -6, -9) (5, 9, 11, 13, 20). Normally in wild type, chromatoid bodies are observed as massive amorphous aggregates in the cytoplasm (for wild type, see Fig. S4A; for Tdrd7+/− spermatids, see Fig. 2 C and E). However, in Tdrd7−/− mutants, two unusual types of round structures were observed (Fig. 2 D and F) in early spermatids (steps 1–3), which were never seen in wild types. These two forms of aggregates had different electron densities and were closely adjoined with each other with a central body of electron-medium density (Fig. 2F, open arrowhead) being interposed between two outside electron-dense aggregates (Fig. 2F, arrowheads).
Fig. 2.
Tdrd7 regulates RNP remodeling of chromatoid bodies. (A–F) Electron microscopy of Tdrd7+/− (A, C, and E) and Tdrd7−/− (B, D, and F) spermatocytes (A and B) and spermatids (C–F). (E and F) Higher magnification views of C and D. Arrows, intermitochondrial cement; arrowheads, chromatoid bodies; open arrowhead, processing body. (G and H) Double immunostain of Tdrd7+/− (G) and Tdrd7−/− (H) spermatids for chromatoid bodies (MVH) and processing bodies (TDRD9). (I–L) Immunoelectron microscopy of Tdrd7+/− (I and K) and Tdrd7−/− spermatids (J and L) with anti-MVH (I and J) and anti-MIWI (K and L) antibodies. The arrowheads indicate chromatoid bodies and the open arrowheads mark processing bodies. (Scale bars: A, B, E, F, and I–L, 500 nm; C and D, 2 μm; G and H, 10 μm.)
To elucidate the developmental and molecular origin of these peculiar structures in Tdrd7−/− mutants, we went back to the wild type and reinvestigated the spatiotemporal assembly of chromatoid bodies in detail. Chromatoid bodies in wild-type spermatogenesis first became discernible in pachytene spermatocytes (Fig S4A, Center). At this stage, intermitochondrial cement was also present, and these two forms of germinal granules shared most of their components, such as TDRD1, TDRD7 (Fig. S4 B, E, F, and G), and MILI (3, 4, 9, 12). Processing bodies, identified by AGO2 and GWB, were observed as distinct assemblies (Fig. S4 H and I), showing that “early” chromatoid bodies, intermitochondrial cement, and processing bodies are different RNP complexes that are independent of each other in meiotic spermatocytes (Fig. S4X). In haploid spermatids, chromatoid bodies were increased in mass and observed mostly as solitary aggregates in the cytoplasm (Fig. S4A, Right, and J–O). Intermitochondrial cement was no longer seen. At this haploid stage, processing body proteins were enriched in chromatoid bodies, as previously reported (Fig. S4 P–R) (14). Thus, “late” chromatoid bodies in spermatids contained both the authentic components of “early” chromatoid bodies and the ubiquitous proteins of processing bodies, resulting in a hybrid composition. Chromatoid bodies also contained RNA, as exemplified by Tnp2 mRNA (Fig. S4 S–W). Together, these observations showed that, in wild type, chromatoid bodies undergo dynamic change in their composition with processing body components being incorporated as spermatogenesis proceeds (Fig. S4X).
In Tdrd7−/− mutants, the marker proteins of “early” chromatoid bodies were observed as pairs of round aggregates in round spermatids (steps 1–3), which sandwiched a single assembly of processing body proteins (Fig. 2 G and H). Immunoelectron microscopy corroborated that the outer pairs of electron-dense aggregates corresponded to chromatoid bodies (Fig. 2 I–L). These findings indicated that chromatoid bodies and processing bodies are indeed assembled as structurally distinct compartments, after which the two assemblies are adjoined, followed by RNP merger under the genetic control of Tdrd7. The unusual structures of chromatoid bodies in Tdrd7−/− mutants were fragmented and dispersed at later stages of spermiogenesis, when the spermatid degeneration proceeded.
The chromatoid body phenotype in Tdrd7−/− mutants was quite unique. In other mutants of chromatoid bodies identified so far, such as Tdrd6−/−, Miwi−/−, and Grth/Ddx25−/− (Fig. S5 A and B for Tdrd6−/−) (11, 21, 22), chromatoid bodies were fragmented and reduced in amount, but the basic architecture and molecular composition were maintained (Fig. S5 C–F for Tdrd6−/− chromatoid bodies). These data indicated that the assembly of chromatoid bodies is sequentially programmed and that the two spermiogenic tudor family genes, Tdrd7 and Tdrd6, play key roles in the early RNP remodeling and later structural maintenance, respectively.
We also found that chromatoid bodies possess another class of components, mRNA cap-binding EIF4E (Fig. S5 G and J), poly(A)-binding PABP (Fig. S5 H and K), and translation repressor TIAR (Fig. S5 I and L), which, together with RNA-binding HuR (23), are all characteristics of stress granules. Stress granules are RNP aggregates induced by stress conditions, including DNA damage and viral infection, and are supposed to sequester translation initiation complexes (24). In somatic cells, stress granules intimately associate with processing bodies, and such a close association was also observed between chromatoid bodies and processing bodies in spermatogenesis (Fig. 2H and Fig. S4 H and I), suggesting that chromatoid bodies may have a feature analogous to somatic stress granules. Notably, in Tdrd7−/− mutants, EIF4E, PABP, and TIAR, which are essential regulators of translation initiation, were all delocalized from chromatoid bodies and instead were cytoplasmically diffuse (Fig. S5 M–R), suggesting that Tdrd7 may affect the translation profile of spermiogenesis (see below).
Tdrd7−/− Deficiency Causes LINE1 Deregulation in Spermatocytes.
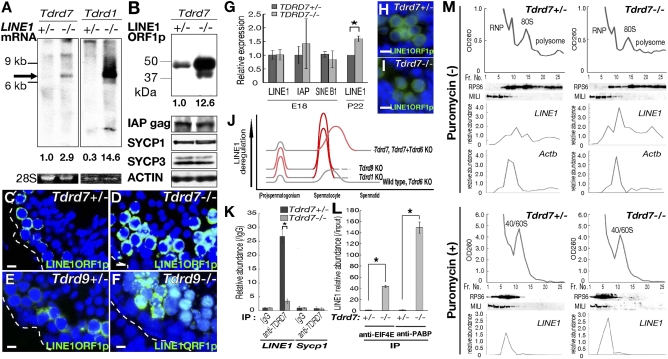
We sought to identify molecular changes underlying Tdrd7−/− mutation. A microarray analysis between Tdrd7−/− and Tdrd7+/− testes at postnatal day 22 (P22) revealed that the expression of several spermiogenesis genes was decreased (more than twofold), including transition protein and protamine genes. However, we could not distinguish whether these changes were under the direct control of Tdrd7 or if they were a consequence of the spermatid defect. In contrast, when retrotransposons (LINE1, IAP, and SINEB1) were individually examined, LINE1 mRNA was moderately but detectably up-regulated in Tdrd7−/− testes (Fig. 3A), although the increase was smaller compared with other mutants of retrotransposon suppressors, Tdrd1, Tdrd9, Mili Miwi2, Mael, and Gasz, etc. (Fig. 3A for Tdrd1−/−) (4, 8, 9, 12, 18, 25–27). LINE1 protein ORF1, in contrast, was more highly elevated in Tdrd7−/− testes (Fig. 3B) compared with LINE1 RNA. By immunofluorescence, LINE1 ORF1 was elevated at the mid-pachytene stage in Tdrd7−/− testes (Fig. 3 C and D), which was at a later stage than LINE1 derepression in other retrotransposon suppressor mutants (Fig. 3 E and F for Tdrd9−/−). Also, LINE1 increase was not observed in Tdrd7−/− fetal prospermatogonia (Fig. 3 G–I). Thus, the kinetics of LINE1 deregulation by Tdrd7−/− deficiency was different from those by previously reported mutations, including Tdrd1−/− and Tdrd9−/− (Fig. 3J).
Fig. 3.
LINE1 deregulation in Tdrd7−/− spermatocytes. (A) Northern blots of total testis RNA at P28 for LINE1 (arrow). Relative band intensities are given below. 28S rRNAs were used as controls. The identity of the upper band (>9 kb) is currently unknown. (B) Western blots of Tdrd7+/− and Tdrd7−/− testes probed for LINE1 ORF1 protein, IAP gag protein, SYCP1, SYCP3, and β-ACTIN. (C–F) Immunostain for LINE1 ORF1 of Tdrd7+/− (C), Tdrd7−/− (D), Tdrd9+/− (E), and Tdrd9−/− (F) testis sections. Dashed lines demarcate seminiferous tubules. (G) Real-time RT-PCR of Tdrd7+/− and Tdrd7−/− embryonic testes (E18.5) for LINE1, IAP, and SINEB1 and of P22 testes for LINE1. Fold changes normalized with β-actin are shown as the means and SE (n = 3). Asterisk, P < 0.05. (H and I) Immunostain of Tdrd7+/− (H) and Tdrd7−/− (I) fetal testis sections (E18.5) for LINE1 ORF1. (J) Developmental kinetics of LINE1 regulation (31) by Tdrd genes. (K) Real-time RT-PCR for LINE1 and Sycp1 (used as a control) from Tdrd7+/− and Tdrd7−/− testis immunoprecipitates with anti-TDRD7 antibody. Fold changes relative to IgG controls are shown as the means and SE (n = 3). Asterisk, P < 0.01. (L) Real-time RT-PCR for LINE1 from Tdrd7+/− and Tdrd7−/− testis immunoprecipitates with anti-EIF4E and PABP antibodies. Fold changes relative to input samples are shown as the means and SE (n = 3). (M) Testis lysates from Tdrd7+/− (Left) and Tdrd7−/− (Right) mice (3 wk) were fractionated on a sucrose gradient in the absence (Top panels) or presence (Bottom panels) of puromycin and analyzed for A260; Western blotting was used to analyze RPS6 and MILI, and the relative abundance of Actb and LINE1 was analyzed by real-time RT-PCR. A260, RPS6, and LINE1 RNA patterns are shifted to upper fractions (Left) in the presence of puromycin. (Scale bars: C–F, H, and I, 10 μm.)
Tdrd1 and Tdrd9 act to suppress retrotransposons through the piRNA pathway (9, 12, 18). We carried out deep sequencing analyses of small RNAs from Tdrd7−/− and control Tdrd7+/− testes at P18 and at 6 mo. However, the piRNA profile of Tdrd7−/− testes was not affected with respect to length distribution, genome sequence annotation, base preferences, sense and antisense patterns, and genome cluster distribution (Fig. S6). DNA methylation at LINE1 loci was also not impaired in Tdrd7−/− testes and FACS-purified germ cells (Fig. S7 A and B). These indicated that LINE1 deregulation in Tdrd7−/− mutants was not due to a primary defect in piRNA biogenesis per se. In accordance, TDRD7 expression and localization was not affected in Mili−/− and Miwi2−/−mutants (Fig. S7 C–J).
We examined LINE1 transcription by primary culture of testicular cells in the presence of uridine analog 5-ethynyluridine, followed by biotinylation by click chemistry (Fig. S7K), and RNA stability by primary culture in the presence of 5,6-dichlororibofuranosylbenzimidazole (DRB), which inhibits RNA polymerase II (Fig. S7L). From these experiments we determined that the relative transcription and RNA stability were not significantly different between Tdrd7−/− mutants and Tdrd7+/− controls. We then analyzed the translation profile of LINE1. By immunoprecipitation, LINE1 RNA was detected in TDRD7 precipitates from control Tdrd7+/− testes, but was abolished in Tdrd7−/− testes (Fig. 3K). Notably, the association of LINE1 RNA with cap-binding EIF4E and poly(A)-binding PABP proteins, key translation initiation factors, was increased in Tdrd7−/− testes (Fig. 3L), suggesting that LINE1 translation was likely elevated. In accordance, sucrose gradient fractionation showed that LINE1 RNA was increased in monosomal and polysomal fractions in Tdrd7−/− mutants compared with Tdrd7+/− controls (Fig. 3M), suggesting that LINE1 translation was increased. LINE1 protein stability, in contrast, was not elevated in Tdrd7−/− mutants (Fig. S7M). Together, these indicated that LINE1 deregulation by Tdrd7−/− deficiency does not accompany transcriptional up-regulation, but rather involves translational regulation at the protein level. This is consistent with the finding that the steady-state level of LINE1 was more highly elevated at the protein level than RNA (Fig. 3 A and B). The moderate RNA increase observed may stem from the increased LINE1 protein, which should protect its RNA by making retrotransposon particles (such effect may not be detectable during the time course of the DRB experiment), although we cannot exclude a possibility that more direct RNA regulation may operate in the mutant. Also, although piRNA biogenesis per se was not disrupted by Tdrd7−/− mutation, Tdrd7 may act downstream of the piRNA pathway and affect translation.
In contrast to Tdrd7, LINE1 deregulation was not detected in Tdrd6−/− testes. The key phenotype of Tdrd6−/− was manifested at a later stage than other Tdrd mutants and thus after the critical period of retrotransposon suppression. Instead, we found that the Tdrd6−/− mutation exerted another phenotypic effect on chromatoid bodies when this RNP structure in wild type exhibited an aggresome-like property (28) with a close spatiotemporal association with the autophagy system (Fig. S8 A–G, the legend of which provides details). In Tdrd6−/− spermatids, the initial contact between chromatoid bodies and autophagosomes was observed (Fig. S8H), but subsequent association with late autophagosomes/endosomes (Fig. S8I) and lysosomes (Fig. S8J) was disrupted, suggesting that the possible clearance of chromatoid body factors may be impaired, which could affect global proteolysis. Indeed, transition protein 2 (TNP2) was aberrantly accumulated in the cytoplasm of Tdrd6−/− spermatids (Fig. S3J, arrowheads), and these cells were mostly degenerating. Although the details of the possible underlying mechanism are currently unclear, these observations suggest that Tdrd6 acts via a distinct cellular pathway compared with Tdrd7 at a later stage of spermatid development.
Tdrd7−/− and Tdrd6−/− Double Mutation Disrupts the Initial Assembly of Chromatoid Bodies.
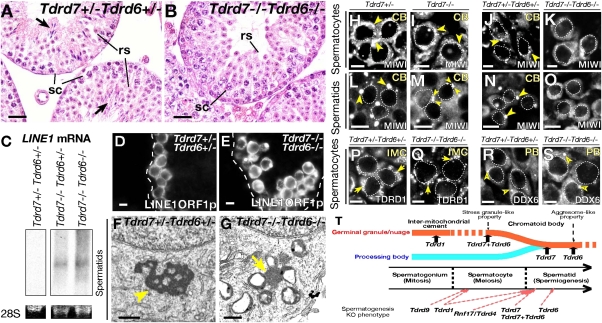
We generated double-null mice of Tdrd7 and Tdrd6. Tdrd7−/− Tdrd6−/− homozygotes were viable, but were male-specific-sterile, showing a round spermatid arrest (Fig. 4 A and B and Fig. S9 A and B) at the same stage as Tdrd7−/− mutants (Fig. 1 F, G, and L) with spermatogonia and spermatocytes being unaffected (Fig. S9 C–F). As in Tdrd7−/− mutants, LINE1 was up-regulated in Tdrd7−/− Tdrd6−/− spermatocytes (Fig. 4 C–E) with cognate DNA methylation being unchanged (Fig. S9G). Thus, Tdrd7−/− Tdrd6−/− double mutation phenocopied Tdrd7−/− single mutation with respect to male sterility, spermatid deficiency and LINE1 deregulation.
Fig. 4.
Tdrd7 and Tdrd6 are key requirements for chromatoid bodies. (A and B) H&E-stained sections of Tdrd7+/− Tdrd6+/− (A) and Tdrd7−/− Tdrd6−/− (B) testes at 2 mo. The arrows indicate mature spermatozoa. (C) Northern blots of total testis RNA at P28 for a LINE1 probe. 28S rRNAs are shown as controls. (D and E) Immunostain for LINE1 ORF1 of Tdrd7+/− Tdrd6+/− (D) and Tdrd7−/− Tdrd6−/− (E) adult testis sections. Dashed lines demarcate seminiferous tubules. (F and G) Electron microscopy of Tdrd7+/− Tdrd6+/− (F) and Tdrd7−/− Tdrd6−/− (G) spermatids. The arrowhead marks a chromatoid body (F), and the arrow points to intermitochondrial cement (G). (H–S) Immunostain of Tdrd7+/− (H and L), Tdrd7−/− (I and M), Tdrd7+/− Tdrd6+/− (J, N, P, and R), and Tdrd7−/− Tdrd6−/− (K, O, Q, and S) spermatocytes (H–K and P–S) and spermatids (L–O) for chromatoid bodies (H–O, MIWI), intermitochondrial cement (P and Q, TDRD1), and processing bodies (R and S, DDX6). Chromatoid bodies (CB), intermitochondrial cement (IMC), and processing bodies (PB) are marked with arrowheads, arrows, and open arrowheads, respectively. (T) Schematic summary for RNP remodeling and male germ-cell development regulated by the Tdrd genes. (Scale bars: A and B, 25 μm; D, E, and H–S, 10 μm; F and G, 500 nm.)
However, Tdrd7−/− Tdrd6−/− testes showed one clear difference compared with each single mutation. In Tdrd7−/− Tdrd6−/− mutants, chromatoid bodies were completely absent from their early assembly stage in spermatocytes to later-stage in haploid spermatids (Fig. 4 F and G). Consistent with this, MIWI, a marker of chromatoid bodies, was diffuse in Tdrd7−/− Tdrd6−/− spermatocytes (Fig. 4 H–K) as well as in spermatids (Fig. 4 L–O). In contrast, intermitochondrial cement was observed in double mutants (Fig. 4G) with TDRD1, an authentic marker of intermitochondrial cement showing fine granular distribution (Fig. 4 P and Q). Processing bodies were normally seen in double mutants (Fig. 4 R and S). Together, Tdrd7 and -6 are critical requirements for the initial establishment of chromatoid bodies.
To examine whether protein methylation, which tudor domains recognize, was essential for chromatoid bodies, we carried out primary culture of testicular cells in the presence or absence of the protein methylation inhibitor, methylthioadenosine (MTA). After treatment with MTA, the disassembly of chromatoid bodies, identified by TDRD6 and TDRD7, was frequently observed (Fig. S9 H–M and P), and such effect was more clearly seen in spermatocytes when early chromatoid bodies had just emerged, rather than in spermatids. Processing bodies were not affected by MTA treatment (Fig. S9 N and O). We also examined several other chemicals, including nocodazole, cycloheximide, and leptomycin B, but specific changes in chromatoid bodies were not observed during the time course of our experiments (3 and 6 h) or only gross cell death was seen. In contrast, MTA induced a relatively fast response before the cell viability was affected (3 h, Fig. S9 P and Q). Together, the active turnover of protein methylation is essential to maintain the integrity of chromatoid bodies, and this is in accordance with the genetic dependence of chromatoid bodies on Tdrd7 and Tdrd6, both of which encode multiple tudor domains that target methylated proteins to assemble macromolecular complexes.
Discussion
The Tdrd gene family constitutes an essential class of spermatogenesis genes with each member having a distinct function at different differentiation stages. In this study, we showed that Tdrd7 is essential for early spermatid differentiation and is a master regulator of the biogenesis of chromatoid bodies. Unlike other Tdrd genes, Tdrd7 mutation causes other somatic phenotypes, cataracts and glaucoma, which become severe with age. These ocular disorders were reported in one N-ethyl-N-nitrosourea point mutant mouse line and in two human cases (a patient and family) (19), and we also confirmed cataracts in our Tdrd7−/− gene-targeted null mice (Fig. S3 AA and AB). Notably, TDRD7 in lens fiber cells was shown to localize to RNP aggregates named TDRD7 granules, suggesting that the RNP control is the common mode of action for TDRD7. Whether other TDRD family members in addition to TDRD7 also constitute somatic RNPs and whether they have relevant physiological function(s) remain undetermined and need future investigation.
In this study, we determined that chromatoid bodies, a long-recognized but enigmatic structure in spermatogenesis, undergo previously unrecognized RNP remodeling, with initially exhibiting a germline-specific/preferential composition similar to intermitochondrial cement and then undergo structural and compositional RNP coalescence with ubiquitous processing bodies, resulting in a hybrid complex composed of both germline and somatic features (Fig. 4T). This process, genetically defined by Tdrd7, settles the previous controversy over the relationship between chromatoid bodies and processing bodies (3, 14). In terms of evolution, such RNP remodeling of germinal granules would be worth examination in other species, and whether similar RNP regulation operates under the control of conserved tudor homologs, such as Drosophila tudor, krimper, and tejas (29), is another interest.
One characteristic of germinal granules is the enrichment of piwi pathway components. In Drosophila, germinal granules are concentrated with a class of piwi pathway proteins (2, 29). In mammals, intermitochondrial cement is also enriched with MILI, TDRD1, and GASZ, which are essential for piRNA biogenesis (3, 4, 9, 12, 25). These and other observations proposed that germinal granules are a subcellular site important for the piRNA operation. Chromatoid bodies also abound with a number of proteins essential for piRNA biogenesis, including MILI, TDRD1, TDRD9, MVH, and MAEL (3–9, 12, 18, 26, 30). However, Tdrd7−/− and Tdrd6−/− mutants, both of which exhibit severe deficiencies in chromatoid bodies, do not affect piRNA biogenesis (12) and relevant DNA methylation. Thus, chromatoid bodies or at least their structural integrity are not a prerequisite for piRNA biogenesis.
However, LINE1 was still deregulated in Tdrd7−/− testes (as well as in Tdrd7−/− Tdrd6−/− testes). LINE1 desuppression by Tdrd7−/− mutation showed a different developmental pattern and distinct mechanism compared with Tdrd1−/− and Tdrd9−/−. This indicates that the germline is equipped with two separate systems that use different Tdrd members to protect against retrotransposons. Tdrd7 may act as a backup pathway to ensure retrotransposon silencing after Tdrd1-Mili and Tdrd9-Miwi2 genes operate.
Because Tdrd7 regulates massive cytoplasmic RNP, chromatoid bodies, it is conceivable that the deficiency affects a posttranscriptional process(es) in spermiogenesis. There may be other endogenous proteins that are also affected in addition to LINE1 protein. We investigated the proteome of Tdrd7−/− testes by 2D fluorescence difference gel electrophoresis, but did not identify specific candidates, except for a presumed posttranslational modification of mitochondrial hydroxyacylglutathione hydrolase. One limitation of studying germ cells is that the germ lineage is quite susceptible to gross cell death. This precludes detailed identification of molecular changes underlying mutant phenotypes of interest. Such difficulty may be circumvented by introducing genetic backgrounds that block cell death pathways in the future.
Remarkably, all loss-of-function mutations of chromatoid body components reported show male fertility defects with severe germ cell degeneration (except for ubiquitous genes whose mutations lead to embryonic lethality) (3, 5–9, 11, 13, 21, 22). This suggests that chromatoid bodies have a critical function(s) in male germline integrity, the molecular basis of which is just starting to be explored. Our study provides seminal information on this enigmatic RNP, elucidating the key sequence of the structural and molecular biogenesis, including the ordered transformation during spermatogenesis. The Tdrd genes are master regulators of such developmental programs of chromatoid bodies, which are genetically closely associated with male haploid differentiation in mice. We propose that similar RNP regulation may be conserved and critical for the germline integrity of divergent species.
Materials and Methods
Generation of Gene-Targeted Mice.
The gene-targeting vectors were constructed using a BAC recombineering system. The vectors were electroporated into V6.5 and KY1.1 ES cells, and then chimeric mice were produced from recombinant ES cell clones by aggregation with C57BL/6 × DBA/2 F1 hybrid morulas or by injection into blastocysts.
Electron Microscopy.
Testes were fixed with 2% glutaraldehyde and postfixed with 1% OsO4 and 0.1 M sucrose in phosphate buffer. For immunoelectron microscopy, 2% paraformaldehyde and 0.02% glutaraldehyde were used. Tissues were dehydrated and embedded in epoxy resin. Sections (70–90 nm) were stained with uranyl acetate and lead citrate or immunostained with specific antibodies. Detailed information is available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank N. G. Copeland for the BAC system, J. Takeda for KY1.1 ES cells, R. Jaenisch for V6.5 ES cells, Sandra L. Martin for the LINE1 ORF1 antibody, Marvin Fritzler for the GWB antibody, S. Kistler for the TNP antibodies, and R. Balhorn for the PRM antibodies. We are grateful to Alex Bortvin, Godfried W. van der Heijden, Mitinori Saitou, Yukihiro Yabuta, and Takashi Shinohara for helpful discussion. This work was supported by the Ministry of Education, Culture, Sports, Science and Technology, Japan (S.C. and N.N.), by the Agence National de la Recherche (R.S.P.), by a kind gift from Kathryn W. Davis, and grants from the National Institutes of Health (to G.J.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015447108/-/DCSupplemental.
References
- 1.Eddy EM. Germ plasm and the differentiation of the germ cell line. Int Rev Cytol. 1975;43:229–280. doi: 10.1016/s0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- 2.Saffman EE, Lasko P. Germline development in vertebrates and invertebrates. Cell Mol Life Sci. 1999;55:1141–1163. doi: 10.1007/s000180050363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chuma S, Hosokawa M, Tanaka T, Nakatsuji N. Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: Germinal granules in mammals. Mol Cell Endocrinol. 2009;306:17–23. doi: 10.1016/j.mce.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Aravin AA, et al. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 2009;5:e1000764. doi: 10.1371/journal.pgen.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chuma S, et al. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci USA. 2006;103:15894–15899. doi: 10.1073/pnas.0601878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chuma S, et al. Mouse Tudor Repeat-1 (MTR-1) is a novel component of chromatoid bodies/nuages in male germ cells and forms a complex with snRNPs. Mech Dev. 2003;120:979–990. doi: 10.1016/s0925-4773(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 7.Kuramochi-Miyagawa S, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 8.Soper SF, et al. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15:285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shoji M, et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17:775–787. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 10.Hosokawa M, et al. Tudor-related proteins TDRD1/MTR-1, TDRD6 and TDRD7/TRAP: Domain composition, intracellular localization, and function in male germ cells in mice. Dev Biol. 2007;301:38–52. doi: 10.1016/j.ydbio.2006.10.046. [DOI] [PubMed] [Google Scholar]
- 11.Vasileva A, Tiedau D, Firooznia A, Müller-Reichert T, Jessberger R. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr Biol. 2009;19:630–639. doi: 10.1016/j.cub.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vagin VV, et al. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yabuta Y, et al. TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J Cell Biol. 2011;192:781–795. doi: 10.1083/jcb.201009043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, Sassone-Corsi P. The chromatoid body of male germ cells: Similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc Natl Acad Sci USA. 2006;103:2647–2652. doi: 10.1073/pnas.0509333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallo CM, Munro E, Rasoloson D, Merritt C, Seydoux G. Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev Biol. 2008;323:76–87. doi: 10.1016/j.ydbio.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 16.Thomson T, Liu N, Arkov A, Lehmann R, Lasko P. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech Dev. 2008;125:865–873. doi: 10.1016/j.mod.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Saxe JP, Tanaka T, Chuma S, Lin H. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr Biol. 2009;19:640–644. doi: 10.1016/j.cub.2009.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reuter M, et al. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol. 2009;16:639–646. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- 19.Lachke SA, et al. Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science. 2011;331:1571–1576. doi: 10.1126/science.1195970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan J, et al. RNF17, a component of the mammalian germ cell nuage, is essential for spermiogenesis. Development. 2005;132:4029–4039. doi: 10.1242/dev.02003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai-Morris CH, Sheng Y, Lee E, Lei KJ, Dufau ML. Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is essential for spermatid development and completion of spermatogenesis. Proc Natl Acad Sci USA. 2004;101:6373–6378. doi: 10.1073/pnas.0401855101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng W, Lin H. Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen Chi M, et al. Temporally regulated traffic of HuR and its associated ARE-containing mRNAs from the chromatoid body to polysomes during mouse spermatogenesis. PLoS ONE. 2009;4:e4900. doi: 10.1371/journal.pone.0004900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kedersha N, et al. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma L, et al. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet. 2009;5:e1000635. doi: 10.1371/journal.pgen.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuramochi-Miyagawa S, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carmell MA, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Haraguchi CM, et al. Chromatoid bodies: Aggresome-like characteristics and degradation sites for organelles of spermiogenic cells. J Histochem Cytochem. 2005;53:455–465. doi: 10.1369/jhc.4A6520.2005. [DOI] [PubMed] [Google Scholar]
- 29.Patil VS, Kai T. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr Biol. 2010;20:724–730. doi: 10.1016/j.cub.2010.02.046. [DOI] [PubMed] [Google Scholar]
- 30.Kuramochi-Miyagawa S, et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 2010;24:887–892. doi: 10.1101/gad.1902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trelogan SA, Martin SL. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc Natl Acad Sci USA. 1995;92:1520–1524. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.