Abstract
Drosophila melanogaster neural-specific protein, ELAV, has been shown to regulate the neural-specific splicing of three genes: neuroglian (nrg), erect wing, and armadillo. Alternative splicing of the nrg transcript involves alternative inclusion of a 3′-terminal exon. Here, using a minigene reporter, we show that the nrg alternatively spliced intron (nASI) has all the determinants required to recreate proper neural-specific RNA processing seen with the endogenous nrg transcript, including regulation by ELAV. An in vitro UV cross-linking assay revealed that ELAV from nuclear extracts cross-links to four distinct sites along the 3200 nucleotide long nASI; one EXS is positioned at the polypyrimidine tract of the default 3′ splice site. ELAV cross-linking sites (EXSs) have in common long tracts of (U)-rich sequence rather than a precise consensus; moreover, each tract has at least two 8/10U elements; their importance is validated by mutant transgene reporter analysis. Further, we propose criteria for ELAV target sequence recognition based on the four EXSs, sites within the nASI that are (U) rich but do not cross-link with ELAV, and predicted EXSs from a phylogenetic comparison with Drosophila virilis nASI. These results suggest that ELAV regulates nrg alternative splicing by direct interaction with the nASI.
Keywords: Alternative splicing, UV cross-linking, RRM, Drosophila, transgenic studies, neuron-specific
Posttranscriptional mechanisms that control differential splicing of nascent transcripts and regulate mRNA stability, transport, and translation are fundamental to regulating gene activity. Members of the ELAV family proteins found in Drosophila, Caenorhabditis elegans, and vertebrates have been implicated in cytoplasmic RNA regulation and nuclear differential splicing (for review, see Antic and Keene 1997). With the exception of one subclass within this family, HuA, their expression is largely restricted to the nervous system (Okano and Darnell 1997). Posttranscriptional regulation is especially critical in the nervous system with its enormous cellular diversity in which neurons differ from each other in morphology, biochemical phenotypes, and function.
Drosophila melanogaster ELAV, the first identified member of the ELAV family, is specifically expressed in all neurons and has a predominantly nuclear localization (Robinow and White 1988, 1991; Yannoni and White 1997). elav gene provides a vital function that is important to differentiation and maintenance of neurons (for review, see Yao et al. 1993). ELAV has been implicated in the nuclear processing of transcripts of three genes: nrg, erect wing, and armadillo (Koushika et al. 2000). All three genes are characterized by broad expression of the transcript and protein and neural-specific alternative splice forms. In each case, the level of the neural-specific spliced transcript correlates with the level of ELAV (Koushika et al. 2000). Although we have shown that ELAV promotes the formation of neural-specific splice forms, the role of ELAV in this process has not been defined.
ELAV proteins are characterized by a minimal structure of two tandem RNA recognition motifs (RRM), followed by a hinge domain and a terminal RRM (Birney et al. 1993). Moreover, RNA binding of each RRM in ELAV serves a nonredundant function (Lisbin et al. 2000). Thus far within the ELAV family, ELAV alone has been implicated in splicing. All other members of ELAV family have been ascribed roles in cytoplasmic RNA processing (Jain et al. 1997; Myer et al. 1997; Fan and Steitz 1998; Levy et al. 1998; Peng et al. 1998; Antic et al. 1999; Ford et al. 1999), including the D. melanogaster RBP9 protein (Park et al. 1998; Kim-Ha et al. 1999). ELAV has also been implicated in autoregulation (Samson 1998). Outside of the ELAV family, the first two tandem RRMs of ELAV are related to the well-studied D. melanogaster Sxl protein (Robinow et al. 1988; Birney et al. 1993; Lisbin et al. 2000). Sxl protein regulates alternative splicing as well as translation of genes in the sex determination cascade (McKeown and Madigan 1992; Bashaw and Baker 1997). The RNA target sequences of Sxl show stretches of U (Sosnowski et al. 1989; Inoue et al. 1990). Reported RNA targets for the vertebrate ELAV proteins are poly(U)-rich sequences of varying lengths interrupted with single (A) residues (for reviews, see Antic and Keene 1997; Wang and Tanaka Hall 2001).
Alternative splicing in neurons was first discovered in the CA/CGRP gene encoding two peptides that differ in the terminal exon: calcitonin in thyroid cells and CGRP in specific neurons (Amara et al. 1982; Crenshaw et al. 1987). Since then, alternative splicing has been found to be a common mechanism used in the generation of isoforms in a variety of functionally important neuronal genes. Although the biological significance of much of the alternative splicing is not always readily evident, in many instances it is thought to be important in the fine tuning of neuronal function. In certain cases, however, the significance of specific isoforms has been shown. For example, isoforms of the potassium channel encoded by the Drosophila Shaker locus show distinct physiological properties (Iverson et al. 1988; Iverson and Rudy 1990), and only the neuronal isoform of agrin has the ability to cluster acetylcholine receptors at the neuromuscular junction (Ferns et al. 1992).
Studies on factors that influence neuronal alternative splicing have revealed that similar to other tissue-specific splicing, certain ubiquitously present RNA-binding proteins are important to splicing regulation (for reviews, see Horowitz and Krainer 1994; Grabowski 1998). Data from several systems suggest that differential concentration of specific RNA-binding proteins, or modulation of their activity by posttranslational modifications as is the case with SR proteins, is crucial to splicing regulation (for review, see Manley and Tacke 1996). These regulatory proteins, along with other proteins that they either recruit or are in complex with, recognize specific enhancer sequences in the pre-mRNA to mediate specific splicing events (Black 1992).
Although most of the neuron-specific splicing model systems have failed to reveal participation of neuron-specific regulatory proteins, it appears that at least in some instances, strictly neuron-specific factor is involved as an essential component of the regulatory complex. Recently, Nova-1, a member of the KH-type RNA-binding protein in mammals with an expression exclusively within specific brain regions, has been shown to be required for alternative splicing of receptor pre-mRNAs for glycine α2 exon 3A and GABAA exon γ2L receptor. Nova-1 binds in vitro to these pre-mRNAs in a sequence-specific manner (Buckanovich et al. 1996; Jensen et al. 2000). Our data suggest that Drosophila ELAV, exclusively present in all neurons, influences splicing (Koushika et al. 2000).
We initiated this study to assess the role of ELAV in nrg neuron-specific splicing. Two nrg spliceforms are generated from the nrg transcript by 3′ splice site (3′SS) choice between the penultimate default exon (D) and the ultimate neural exon (N) (Fig. 1A). This results in Nrg protein isoforms with distinct cytoplasmic domains (Hortsch et al. 1990). Initially we set out to define the sequences within the nrg transcript that were sufficient to mimic the endogenous nrg alternative splicing. A reporter minigene, comprised of the alternative spliced nrg intron (nASI) placed upstream of β-galactosidase gene, was constructed, transformed, and analyzed in vivo in transgenic flies. We show that this minigene reporter produces β-galactosidase in neurons, and that the neural-specific processing is responsive to ELAV levels. Having defined the regulated intron as sufficient for recreating neural-specific splicing, we asked if ELAV bound directly to the intron in vitro. We show that ELAV in embryonic nuclear extracts cross-links to a subset of the poly(U)-rich sequences found throughout the 3.2 kb intron sequence. The ELAV cross-linking sequences (EXS) are strongly poly(U) in character but do not share a precise consensus sequence; rather, the length appears to be critical. The reporter gene simultaneously deleted for the EXS sequences shows reduced signal compared to the parent reporter gene. In addition, to allow an evolutionary comparison, we cloned and sequenced the nASI from D. virilis, which aided in the analysis of sequences essential in the splicing regulation and provided a context for positioning of the ELAV cross-linking sites along the intron. Based on the data presented in this paper, we present a working model for ELAV's role in nrg alternative splicing.
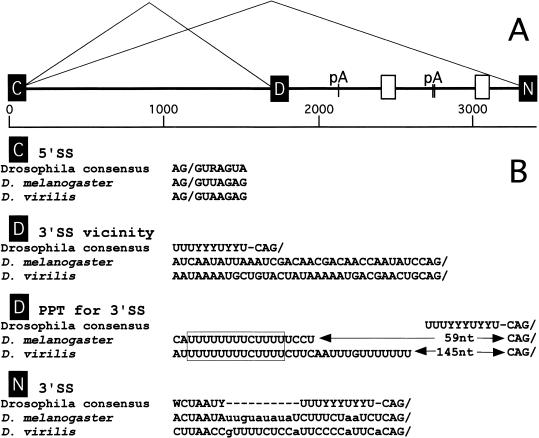
Figure 1.
(A) Schematic of the alternatively spliced intron (nASI) of the nrg gene of D. melanogaster. C, D, and N denote the common, the default, and the neural-specific exons, respectively. The exons C, D, and N correspond to exons VI, VIIa, and VIIb of the published nrg gene (Zhao and Hortsch 1998). The nASI is flanked by the exons C and N. The default 3′SS is at position 1636 and the neural-specific 3′SS is at position 3299 of the sequence. pA indicates the positions of the three consensus polyadenylation signal sequences. The white boxes indicate blocks of homologous sequences between the two species. (B) Comparison of splice site sequences from the nASI of D. melanogaster and D. virilis. Note the conservation of the unconventional distance between the PPT, the 3′ default SS, and the 13 nt poly(U)-rich element between the two species (boxed). The default 3′SS differs considerably more from the Drosophila consensus (Mount et al. 1992) than the neural-specific 3′SS.
Results
nASI from D. melanogaster and D. virilis
The nASI diagrammed in Figure 1 was cloned and sequenced from D. melanogaster (∼3.2 kb). The nASI sequence was also cloned and analyzed from D. virilis (∼4.3 kb), with the expectation that a phylogenetic comparison could yield information about essential intron sequences and potential target sequences for ELAV regulation. An antibody specifically recognizing the neural-specific Nrg180 also recognizes a protein in the D. virilis photoreceptors and larval brain (data not shown); thus, neural-specific splicing is likely to be conserved in D. virilis. The 5′ splice site (5′SS) of the common exon (C) and the 3′SS of the neural exon (N) are close to the Drosophila consensus (Fig. 1B). However, the 3′SS of the default exon (D) differs from the consensus sequence in that the polypyrimidine tract (PPT) is separated from the 3′SS by an (A)-rich sequence, and obvious candidates for a branchpoint sequence are lacking in proximity to the PPT (Fig. 1B). This character of the default 3′SS is conserved in both Drosophila species, suggesting this feature may be functionally significant. A 13-nt block of poly(U)-rich homology (Fig. 1B, outlined box) appears within the default PPT and stands out as a potential ELAV-binding site, as it is reminiscent of the tra target sequence suggested for the ELAV-relative Sxl protein (Sosnowski et al. 1989; Inoue et al. 1990; Handa et al. 1999). Furthermore, two blocks of nonpoly(U) sequences showed significant homology.
D. melanogaster and D. virilis introns have extensive regions of poly(U) sequence, which are thought to be ELAV family member targets (for reviews, see Antic and Keene 1997; Wang and Tanaka Hall 2001). However, outside of the default PPT, no blocks of AU-rich or poly(U)-rich sequence identity were evident.
Reporter transgenes CNV and CNVt express specifically in neurons
To study in vivo the regulated alternative splicing of the nrg transcript, we constructed a minigene reporter from the P-element vector, pCV (Mottes and Iverson 1995) and a 3.3-kb fragment containing the D. melanogaster nASI (Koushika et al. 1996). The resulting construct, termed CNV, consists of the Hsp70 promoter, driving expression of the nASI, followed by the Escherichia coli LacZ open-reading frame and Hsp70 trailer (Fig. 2A). CNVt is identical to CNV except that the tubulin 3′ trailer sequence has replaced those of the Hsp70 trailer. The nrg intron sequences were inserted such that the exon C lies in frame with an open-reading frame that initiates upstream within the transcribed region of the Hsp70 promoter, and the exon N lies in frame with the LacZ open-reading frame. Therefore, only neural-specific splicing of the nASI sequence will generate β-galactosidase.
Figure 2.
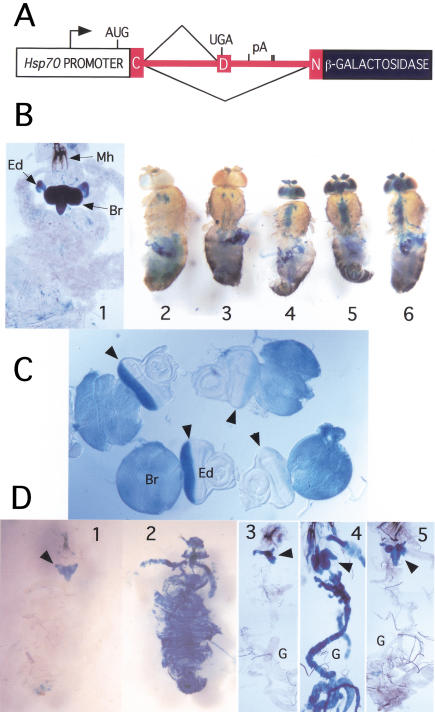
β-galactosidase expression from CNV minigene. (A) CNV minigene construct. The Hsp70 promoter drives the transcription of the nASI (shown in red) in frame with the downstream LacZ reporter gene. Splicing to the proximal 3′SS leads to the translation of a short peptide, whereas splicing to the distal 3′SS leads to the generation of β-galactosidase. (B) CNV expression in adults and larvae is restricted to the nervous system. CNV transgene-carrying flies or larvae were subjected to heat-shock treatment, dissected, and stained at 2 h. Variations in the treatment as noted. (1) The larval cuticle was teased apart and the entire larva was bathed in the staining solution. Note the intense staining of the brain (Br) and in the developing photoreceptors in the eye discs (Ed). Mh, mouth hooks. (2) No CNV. (3) CNV without heat shock. (4) CNV, 35°C pre heat shock only. (5) CNV, heat shock, dissected after 1 h. (6) CNV, heat shock, except 2 h at 38°C. Note that controls 2 and 3 show only the endogenous staining, but in the CNV flies after heat shock, brain, antennae, retina, and the thoracic ganglion also stain. (C) β-Galactosidase expression from the CNV is reduced in photoreceptors with reduced ELAV. Third instar CNV larval brain and eye disc stained for β-galactosidase in two genetic backgrounds for elav: elav+; CNV (wild-type ELAV level) and elave5; elavedr; CNV (reduced ELAV) expression in the photoreceptors. Each eye disc was photographed with attached brain lobe. The two left eye discs with normal ELAV levels show strong staining in the photoreceptor field of the eye disc behind the morphogenetic furrow (arrowhead) as well as staining of the attached brain lobes. Note that the right eye discs with reduced ELAV expression show a significant reduction in the staining compared to the left eye discs with wild-type ELAV expression. (D) Global expression of ELAV leads to global β-galactosidase expression from the CNV transgene. Third instar larvae were heat shocked, dissected, fixed, and stained. In panels 1 and 2, larval cuticle is cut open in the anterior and the whole animal is stained, whereas in panels 3, 4, and 5, larva is dissected and displayed with the brain attached to the mouth hooks and the digestive track (G). CNV larvae (1,3); CNV; Hsp–ELAV larvae (2,4); CNV; Hsp–Sxl (5) larvae. Note that only larvae carrying both CNV and Hsp–ELAV depict global staining (2,4).
CNV transcription was induced by subjecting the animals to an initial 35°C heat shock, followed by a 38°C heat shock. The mild heat shock protects splicing from the effects of the subsequent heat shock (Yost and Lindquist 1986; Mottes and Iverson 1995). Two hours after heat shock, CNV wandering third instar larvae stained intensely in the brain lobes and the ventral ganglion (Fig. 2B, panel 1), as well as in the eye imaginal discs above each brain lobe, but not in nonneuronal tissues. X-gal staining in the developing eye disc is coincident with the photoreceptor neuron field behind the morphogenetic furrow (Fig. 2C, left side).
In control (lacking the CNV transgene) and in non heat shocked CNV adult flies, X-gal staining occurs predominantly in the gut indicative of endogenous β-galactosidase (Fig. 2B, panels 2,3). After heat shock, however, CNV transgenic adults show staining of the nervous system as well (Fig. 2B, panels 4–6). Strong X-gal staining is observed in the antennae, optic lobes, brain, and thoracic ganglion, but not in nonneuronal tissues. Similar results were seen with CNVt (see Fig. 6A, below; data not shown).
Figure 6.
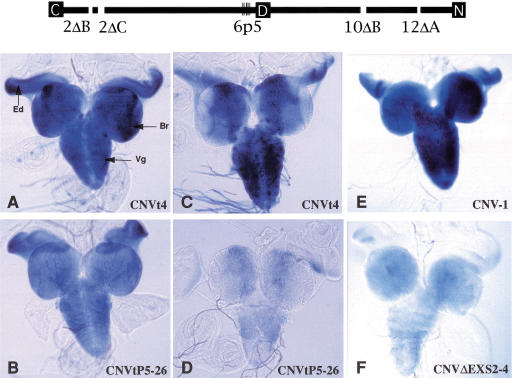
Representative β-galactosidase expression from mutated reporter minigenes. nASI schematic at the top shows the mutated sites in transgene CNVtp5 (6p5) and transgene CNVΔEXS (2ΔB, 2ΔC, 6p5, 10ΔB, and 12ΔA). Refer to Table 1 for sequences. C, common exon; D, default exon; N, neural-specific exon. Third instar larvae-carrying reporter minigenes were heat shocked and central nervous systems with attached eye discs were processed for X-gal staining at 4 h later (A,B,E,F) or 20 h later (C,D). The expression of CNVtp5-26 (B,D) is reduced compared to CNVt-4 (A,C). The expression of CNVΔEXS2-4 (F) is reduced compared to CNV-1 (E). Br, brain lobe; Vg, ventral ganglion; Ed, eye disc. Expression from at least two inserts was tested for each transgene.
ELAV regulates CNV transgene processing
The processing of the CNV transgene indicates that sequences within nASI are competent to generate the neuron-specific isoform similar to that observed for the endogenous nrg. Previously we had shown that reduced expression of ELAV in the eye disc caused a concomitant reduction in the neural-specific protein isoform, Nrg180, but not in the ubiquitous protein isoform, Nrg167 (Koushika et al. 1996). To test if the transcript generated by the CNV transgene is regulated by ELAV, we examined the CNV transgene expression in the third instar larval eye discs with genetically reduced levels of ELAV expression in the photoreceptors (Fig. 2C). Eye discs attached to brain lobes of the genotypes +;CNV/+ (wild-type ELAV) and elave5;elavedr/CNV (reduced ELAV) were fixed concurrently and stained for β-galactosidase activity. In Figure 2C, a pair of wild-type eye disc/brain lobes on the left show robust X-gal staining in the photoreceptor field behind the morphogenetic furrow (solid arrowhead), whereas there is substantial reduction in the staining of the photoreceptors in the pair on the right with reduced ELAV. Thus, the neural processing of the CNV transgene is reduced with reduced levels of ELAV. Note that X-gal staining is not reduced in the elave5;elavedr/CNV brain lobes, where ELAV protein levels are not altered significantly.
Because ectopic expression of ELAV induces the neural-specific Nrg isoform, we tested if it would also induce CNV processing to generate ectopic β-galactosidase. Larvae of the genotype Hsp–ELAV/CNV and control larvae CNV and Hsp–Sxl/CNV were heat shocked, dissected, and stained. On heat shock, Hsp–ELAV, Hsp–Sxl, and CNV are all transcribed globally. Hsp–Sxl is a control for nonspecific effects that could arise from the ectopic expression of a poly(U)-binding protein. Neural-specific staining is seen in the CNV controls (Fig. 2D, panels 1,3), whereas global X-gal staining is observed in Hsp–ELAV/CNV larvae (Fig. 2D, panels 2,4). Control Hsp–Sxl/CNV larvae do not show global X-gal expression (Fig. 2D, panel 5). These data suggest that ELAV is sufficient to alter the processing pathway of the CNV-derived transcript outside the nervous system. This is confirmed at the level of RNA processing in which ectopic ELAV leads to the appearance of the neuron-specific spliced CNV transcript in nonneural tissue (see below).
CNV transgene splicing
RT–PCR was used to analyze the processing of CNV transcripts from heat-shocked CNV transgenic adult flies. Sequence analysis confirmed proper splice site usage for the default and neural-specific spliced transcripts (data not shown).
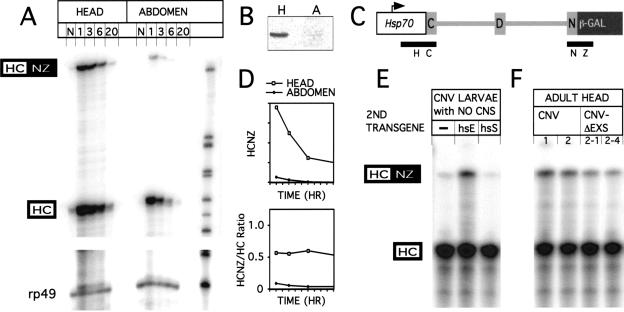
RNAse protection assays were used to analyze RNA processing of the CNV minigene in adult flies. RNA was isolated from CNV fly heads and abdomens after 1-, 3-, 6-, and 20-h post heat shock (Fig. 3A). Heads contain the fly brain and are enriched for neurons, whereas abdomen is neuron poor as it has only sensory and enteric neurons. For RNAse protection assays, antisense probe HCNZ was used (Fig. 3C). Neural-specific spliced CNV transcripts are abundant in heads relative to abdomens. This is consistent with (1) the enrichment of neural tissue in adult heads, and therefore ELAV levels as well, and (2) relatively few neurons in the abdomen (Fig. 3B). Whether default splicing also occurs in neurons is not known. At 1 h post heat shock, the ratio of neural-spliced protected RNA signal to nonneural-spliced protected RNA signal was 0.56 for heads and 0.09 for abdomens (Fig. 3D, lower panel). This translates to a molar ratio of neural spliced CNV RNA to total CNV RNA of 28% for heads and 5.9% for abdomens. In addition, the decay rates for the two CNV RNAs appear to be similar so that the RNA ratio stays relatively constant for a number of hours (Fig. 3D, lower panel). Endogenous nrg transcripts or any transcript from the endogenous β-galactosidase gene were not detected in this experiment.
Figure 3.
RNA processing of the CNV transgene. (A) RNAse protection analysis of CNV transcripts. Heads and abdomens from 2- to 5-day-old CNV flies collected with no heat shock (N) or at 1-, 3-, 6-, or 20-h post 38°C heat shock. RNAse protection assay was performed as described in Materials and Methods with 10 μg of total head or abdomen RNA per lane. The antisense probe used for the protection analysis is termed HCNZ, for Hsp70 C exon/N exon lacZ (C). 321 nt are protected for a neural-specific splice, HCNZ, and 192 nt are protected for the default splice, HC (or unspliced). The rp49 probe controls for total RNA loaded. Size markers are pBR322/MspI ladder. (B) Immunoblot with α-ELAV antibodies to show ELAV level in 10 μg of total protein from adult heads, H, and abdomens, A. (D) The data from A was analyzed and quantitated on a PhosphorImager to show relative amounts of neural-processed CNV RNA. (Top graph) HCNZ-protected RNA amounts for both head and abdomen in arbitrary units. The HCNZ RNA levels decrease in the hour after heat shock. (Bottom graph) The ratio of HCNZ to HC stays relatively constant for several hours as both neural and default RNAs appear to decay with similar rates. Note: In independent experiments using an HCI probe (I, intron), the unspliced CNV, which would be seen as the HC-protected species in the figure above, is present, but in very low amounts (data not shown). (E) Ectopic expression of ELAV induces neural-specific processing of CNV transcripts in nonneuronal tissue. Third instar larvae were heat shocked, and 1 h later, the CNS was dissected out and the remaining tissue subjected to RNA extraction. RNA (5μg) was subjected to RNAse protection analysis with the HCNZ antisense probe (C). Fly heads with CNV transgene alone (−) or with Hsp–Sxl transgene (hsS) have only reduced levels of the neural-specific transcript. However, the ectopic expression of ELAV when CNV is expressed with Hsp–ELAV (hsE) leads to increased neural processing of the CNV transcripts. (F) Deletion/mutation of EXS elements within the nASI leads to a reduction in neural-specific splicing. RNA from heat-shocked CNV and CNV–ΔEXS fly heads was extracted 2 h after heat shock and used in RNAse protection analysis. The ratios of HCNZ/HC from two insertion lines of CNV (1,2) are reduced threefold in two insertion lines of the CNVΔEXS (2-1, 2-4).
To analyze effects of ectopic expression of ELAV on CNV RNA processing in nonneural tissue, RNAse protection assays were performed on third instar larvae carrying just CNV or both CNV and Hsp–ELAV (Fig. 3E). After heat shock, the CNS was dissected out and RNA was extracted from the remaining tissue. In parallel, larvae ectopically expressing Hsp–Sxl were similarly processed. In the absence of ELAV, there is little neural-specific splicing; however, in the presence of ectopic ELAV, neural-specific processing of the CNV transcript is greatly enhanced. Ectopic Sxl had no effect on neural-specific splicing.
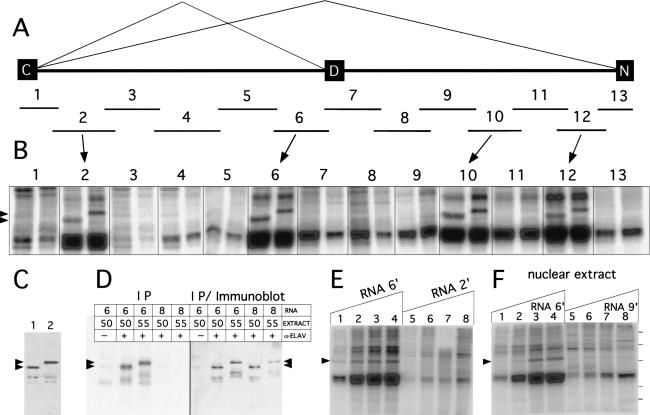
ELAV cross-links to four regions of the nASI
We searched for in vitro ELAV-binding sites within the nASI sequences, as these sequences were sufficient for ELAV regulation. Thirteen overlapping RNAs (1–13, size range 200–380 nt) that span the entire 3200-nt length of the nASI were tested in the UV cross-linking assay with Drosophila embryonic nuclear extracts (Fig. 4A,B). Nuclear extracts were made from 6- to 18-hour-old Canton S embryos. To identify unambiguously cross-linked ELAV, we generated two parallel extracts that differed only in the source of ELAV protein, D. melanogaster ELAV (50 kD) and D. virilis ELAV (55 kD) (Fig. 4C). D. virilis ELAV is fully functional in D. melanogaster in vivo (Yao and White 1991).
Figure 4.
UV cross-linking of ELAV from Drosophila nuclear extracts to the nASI. (A) Schematic diagram of the nASI with the 13 overlapping RNAs (1–13) used in the UV cross-linking experiment. (B) Four of the 13 RNAs UV cross-link to ELAV. [α-32P]UTP RNAs (1.2 nM) were incubated in 2 mg/mL nuclear extract with 50-kD ELAV (left) or 55-kD ELAV (right). Arrowheads indicate the two sized ELAV bands. RNAs 2, 6, 10, and 12 show cross-linking to the ELAV protein. (C) ELAV immunoblot of the two Drosophila embryonic nuclear extracts with ELAV-positive bands at 50 and 55 kD. (D) UV cross-linked ELAV is revealed by immunoprecipitation with α-ELAV in UV cross-linked extracts with RNA 6 but not with RNA 8. The two RNAs were UV cross-linked with the 50- and 55-kD nuclear extracts. Immunoprecipitates were subjected to SDS-PAGE, transferred to nitrocellulose, and exposed to PhosphorImaging plates (left panel). The filter was subsequently subjected to immunoblot analysis with α-ELAV antibodies (right panel). Note that the cross-linking bands observed with RNA 6 (left panel) correspond to the 50- and 55-kD ELAV bands (right panel). (E) UV cross-linking with increasing amounts of positive and negative ELAV cross-linking RNAs 6‘ and 2‘ (see Fig. 5E). 55-kD ELAV nuclear extract was kept constant at 1.5 mg/mL. Arrow indicates the ELAV cross-linking signal. The RNA concentrations are: (lane 1) 0.1 nM; (lane 2) 0.5 nM; (lane 3) 1.0 nM; (lane 4) 1.5 nM; (lane 5) 0.11 nM; (lane 6) 0.57 nM; (lane 7) 1.1 nM; (lane 8) 1.7 nM. (F) Titration of 55-kD ELAV nuclear extract with constant RNA concentrations. RNA 9‘ (see Fig. 5E) does not UV cross-link ELAV in embryonic nuclear extracts. RNA 6‘ (lanes 1–4) is 0.5 nM and RNA 9‘ (lanes 5–8) at 0.87 nM. Nuclear extract concentrations in mg/mL: (lanes 1 and 5) 0.5; (lanes 2 and 6) 1.0; (lanes 3 and 7) 2.0; (lanes 4 and 8) 4.0. Protein markers on the right are 90, 70, 55, 38, and 33 kD.
RNAs 1 to 13 were tested in the UV cross-linking assay with the dual embryonic nuclear extracts (Fig. 4B). ELAV cross-linking, indicated by the 50- and 55-kD bands (arrowheads) for the dual extract lanes, occurs with RNAs 2, 6, 10, and 12, but not with the other nine RNAs. RNA 6 contains the default 3′SS and the U-rich element conserved between the two Drosophila species. Robust non-ELAV signals in the range of 40–45 kD are ubiquitous and appear to coincide roughly in intensity with the poly(U) content of the RNA tested (i.e., RNA 3 has no poly(U) content and no 40–45 kD band). These bands, presently of unknown identity, are most intense in lanes where ELAV cross-linking occurs.
Further confirmation that the cross-linked species is ELAV, comes from the immunoprecipitation of ELAV protein from nuclear extracts, which is radiolabeled only on cross-linking with a radiolabeled RNA. Both the 50- and 55-kD ELAV within the extracts became radiolabeled when immunoprecipitated after cross-linking with RNA 6 but not with non-ELAV cross-linking RNA 8 (Fig. 4D).
To determine the linear range for the cross-linking signal, increasing amounts of the RNA 6‘ or the non-ELAV cross-linking RNA 2‘ (see Fig. 5A) were added to 1.5 mg/mL extract. The ELAV cross-linked species is detected for RNA 6‘ even at the lowest concentration of RNA (0.1 nM). However, the RNA 2‘ cannot be detected even at the highest concentration, although it is also rich in poly(U) sequences (Fig. 4E).
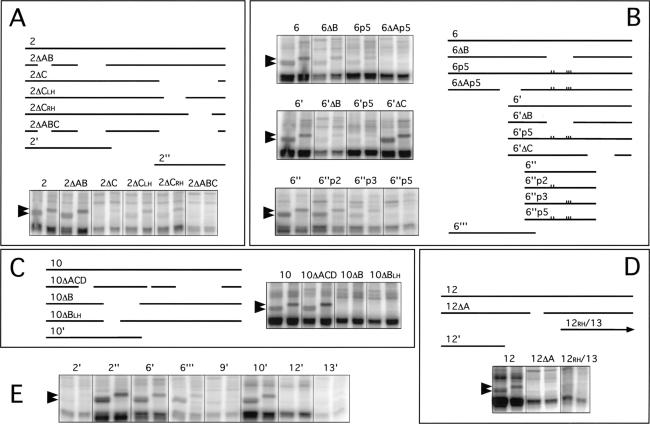
Figure 5.
Deletion analysis to determine sequences responsible for ELAV UV cross-linking. Drosophila embryonic extracts were cross-linked with [α-32P]UTP nASI RNAs and processed as explained in Materials and Methods. For each RNA, deletion map schematic and dual-lane cross-linking data as in Figure 4 are shown. Deletion and point mutant sequences are given in Table 1. Arrowheads indicate the 50-KD ELAV (left lane) and 55-kD ELAV (right lane). (A) The 358 nt RNA 2 has three internal deletions, A, B, and C. RNAs deleted for C, 2ΔC, and 2ΔABC lack the ELAV cross-linking signal. Deletions of the left half 2ΔCLH or right half 2ΔCRH of C reduce but do not eliminate the ELAV signal. (B) The 319-nt RNA 6 has three internal deletions, A, B, and C, and truncated RNA 6‘, 6“‘, and 6“ have large 5′, 3′, and both 5′ and 3′ deletions, respectively. The default PPT region is deleted by ΔB and is disrupted by triple point mutant p3, whereas the nearby AU4AU3AU element is disrupted by double point mutant p2. Point mutant locations are indicated by marks on the line drawings. Data indicate the primary ELAV cross-linking site is the PPT and adjacent 5′ sequence and a suboptimal site 5′ in the ΔA region of 6“‘. (C) Four deletions were made in the 322-nt RNA 10. Deletions A, C, and D, 10ΔACD, have no effect on ELAV cross-linking; however, deletion of B, 10ΔB, and just the left half of B, 10ΔBLH, is sufficient to abolish ELAV cross-linking. (D) Deletion of part of the only significant poly(U) sequence, 12ΔA, eliminated ELAV cross-linking in the 311 nt RNA 12. (E) UV cross-linking with RNAs from A, B, C, and D with large 5′ or 3′ deletions and RNAs 9 and 13. RNA sizes in nt: (2‘) 146; (2“)112; (6‘) 198; (6“‘) 147; (9‘) 144; (10‘) 200; (12‘) 129; (13′) 106.
The concentration of nuclear extract was varied and the input RNAs kept constant (Fig. 4F). RNA 6‘ cross-linking to ELAV is not detectable at 0.5 mg/mL nuclear extract (Fig. 4F, lane 1), barely detectable in the 1 mg/mL nuclear extract (Fig. 4F, lane 2), but strong at 2 mg/mL and above. The ELAV noncross-linking RNA 9‘ is either not detectable or only faintly detectable at all concentrations of nuclear extract tested, even though it also has substantial poly(U) content.
Sequences responsible for the ELAV cross-linking
To further limit the sequences within RNAs 2, 6, 10, and 12, which are responsible for the cross-links to ELAV, a set of deletions was tested for each of the four ELAV cross-linking RNAs (Fig. 5). Deletions were made in the most poly(U)-rich regions within each RNA; their sequences are listed in Table 1.
Table 1.
Sequences of the deletions and point mutations in Figure 5
| RNA #a
|
X- linkb
|
Deletions or point mutantsc
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2ΔA | +++ | U | U | U | U | U | U | A | C | A | C | U | A | U | U | U | U | U | U | U | U | U | U | U | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2ΔB | +++ | U | U | U | U | U | G | U | G | A | U | C | U | U | A | A | C | U | U | A | A | A | A | A | U | A | A | A | A | A | U | G | U | A | U | U | U | U | U | U | U | A | G | U | C | G | C | A | U | G | U | U | U | U | G | U | |||||||||||||||||||
| 2ΔC | +/− | U | U | U | U | U | G | U | U | G | U | C | U | C | U | C | U | U | G | G | U | U | G | U | U | U | U | U | U | U | C | A | C | U | C | U | C | U | U | G | A | U | U | U | C | C | U | U | U | G | C | C | A | C | A | U | U | U | G | U | U | U | U | U | G | G | U | U | U | U | A | ||||
| 2ΔCLH | ++ | A | U | U | U | U | U | G | U | U | G | U | C | U | C | U | C | U | U | U | G | G | U | U | G | U | U | U | U | U | U | U | C | ||||||||||||||||||||||||||||||||||||||||||
| 2ΔCRH | ++ | A | U | U | U | C | C | U | U | U | G | C | C | A | C | A | U | U | U | G | U | U | U | U | U | G | G | U | U | U | U | A | U | U | U | C | |||||||||||||||||||||||||||||||||||||||
| 6ΔA | + | U | U | U | U | G | U | G | U | U | C | C | U | U | G | U | U | A | U | G | U | G | U | U | C | U | U | U | G | U | U | U | |||||||||||||||||||||||||||||||||||||||||||
| 6ΔB | + | A | U | U | U | U | A | U | U | U | A | U | A | U | U | A | U | A | U | U | U | G | U | A | U | C | A | U | U | U | U | U | U | U | U | C | U | U | U | U | U | ||||||||||||||||||||||||||||||||||
| 6p5 | + | A | U | A | U | U | A | A | U | U | A | U | A | U | U | A | U | A | U | U | U | G | U | A | U | C | A | U | C | U | C | U | U | C | U | C | U | U | U | U | U | ||||||||||||||||||||||||||||||||||
| 6p2 | ++ | A | U | A | U | U | A | A | U | U | A | U | A | U | U | A | U | A | U | U | U | G | U | A | U | C | A | U | U | U | U | U | U | U | U | C | U | U | U | U | U | ||||||||||||||||||||||||||||||||||
| 6p3 | ++ | A | U | U | U | U | A | U | U | U | A | U | A | U | U | A | U | A | U | U | U | G | U | A | U | C | A | U | C | U | C | U | U | C | U | C | U | U | U | U | U | ||||||||||||||||||||||||||||||||||
| 6ΔC | +++ | A | A | A | C | A | C | G | U | A | A | A | A | U | A | A | A | C | A | C | A | A | U | C | A | C | A | A | U | C | A | A | U | A | U | U | A | A | A | U | C | G | A | C | A | A | C | G | A | C | A | A | C | C | A | A | U | A | U | C | C | ||||||||||||||
| 10ΔA | +++ | U | U | U | U | A | G | C | U | U | A | C | A | U | G | U | U | U | A | C | U | A | C | U | U | U | A | U | U | U | |||||||||||||||||||||||||||||||||||||||||||||
| 10ΔB | NO | U | U | U | G | U | U | U | U | U | A | U | U | U | C | A | U | U | A | U | U | U | U | A | U | U | U | U | U | U | U | U | A | G | U | U | C | G | U | G | U | A | A | U | G | A | C | U | U | U | G | A | A | U | U | U | U | U | U | U | G | C | U | U | U | ||||||||||
| 10ΔBLH | NO | U | U | U | G | U | U | U | U | U | A | U | U | U | C | A | U | U | A | U | U | U | U | A | U | U | U | U | U | U | U | U | |||||||||||||||||||||||||||||||||||||||||||
| 10ΔC | +++ | U | U | U | G | U | U | U | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10ΔD | +++ | A | U | U | U | U | U | U | C | A | G | U | G | U | A | U | U | U | A | A | U | A | U | U | A | A | A | G | A | A | U | G | A | A | U | G | A | C | G | A | A | A | A | C | A | U | C | A | U | A | A | U | U | A | U | A | U | G | A | A | A | A | U | A | U | A | A | U | U | U | U | U | U | U | A |
| 12ΔA | NO | U | U | U | U | A | U | C | U | U | U | U | A | A | G | U | A | A | U | U | A | U | U | U | G | U | U | U | |||||||||||||||||||||||||||||||||||||||||||||||
| 9‘ | A | U | U | U | U | U | U | U | U | U | A | U | U | U | U | A | A | A | G | A | A | A | U | U | U | A | A | ||||||||||||||||||||||||||||||||||||||||||||||||
| 12‘ | C | A | U | U | U | A | C | U | U | U | C | C | A | U | U | U | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13′ | A | U | U | U | A | A | C | A | U | A | U | C | A | U | U | U | U | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
RNAs as depicted in Figure 5. Sequences for RNAs 9‘, 12‘, and 13′ are primary poly(U)-elements, not deletions. The sequences that carry EXS are bold and underlined.
The behavior of RNAs 2, 6, 10, and 12 carrying deletions in the ELAV cross-linking assay. +++, ++, +/−, and NO are qualitative judgements based on data shown in Figure 5 or by inference in case of 6ΔA.
The point mutants in 6p5, 6p2 and 6p3 are in bold and underlined.
Analyses of the nASI RNA 2
Deletions A, B, and C were made in RNA 2 (Fig. 5A). The ELAV cross-linking signal was unaffected by deletions A and B (2ΔAB), but was abolished by A, B, and C together (2ΔABC) and C alone (2ΔC) (Fig. 5A). Deletion C was examined further by deleting either the left or the right half (2ΔCLH and 2ΔCRH). Both RNAs show reduced ELAV cross-linking, but neither eliminated the signal entirely, suggesting that both halves contain elements that contribute to the interaction with ELAV. The results with two smaller RNAs, 2‘ and 2“ (Fig. 5A), are consistent with the deletion analysis, with only RNA 2“, which contains the ΔC sequence, cross-linking to ELAV (Fig. 5E).
Analyses of the nASI RNA 6
To analyze RNA 6, in addition to generating deletions, we mutated the default PPT with point mutations designed to disrupt the poly(U) character (Table 1; Fig. 5B). p2 and p3 have two and three point mutations in the AU4AU3A element 15 nt upstream of the PPT and in the conserved 13-nt block in the PPT, respectively. p5 has all the five point mutations.
As shown in Figure 5B and E, RNAs 6‘, 6“‘, and 6ΔB that delete terminal 5′, terminal 3′ regions, and the default PPT, respectively, show robust, reduced, and much-reduced ELAV cross-linking, suggesting the presence of two cross-linking sites—the default PPT and a site within 6“‘. The strong cross-linking to the PPT is further supported by robust ELAV signals with RNAs that retain the PPT (6‘, 6‘ΔC, and 6“) and weak signals by RNAs that do not retain the PPT (6ΔB and 6‘ΔB). Further, PPT mutant p5 also greatly diminishes ELAV cross-linking (6p5, 6‘p5, and 6“p5). The second 3′ cross-linking site is within ΔA, as those RNAs in which A is deleted and PPT is mutated (p5) abolish the ELAV signal (6ΔAp5 and 6“p5), whereas deletions that retain the ΔA still show weak cross-linking (6p5 and 6‘p5).
To assess the relative importance of the PPT and the upstream AU4AU3AU, we analyzed cross-linking of RNAs 6“p2, 6“p3, and 6“p5. ELAV cross-linking is slightly diminished in 6“p2 and greatly diminished in 6“p3, but eliminated only in 6“p5. This suggests that disruption of the U8 in the PPT is more deleterious to ELAV binding than disruption of the AU4AU3A element (6“p2), but both elements contribute to binding.
Analyses of the nASI RNA 10
Of the four primary deletions made in RNA 10, only deletion B (10ΔB) eliminated the ELAV cross-linking signal, whereas the other three deletions combined (10ΔACD) had no effect (Fig. 5C; Table 1). Deletion of just the left half of the 10ΔB sequence (10ΔBLH) was sufficient to abolish the ELAV cross-linking signal. RNA 10‘, harboring the ΔB sequence, generates an ELAV cross-linking signal (Fig. 5E).
Analyses of the nASI RNA 12
RNA 12 had the least amount of poly(U) sequence, but the single deletion made (12ΔA) was sufficient to eliminate the ELAV cross-linking signal (Fig. 5D; Table 1). Truncated RNAs that lie either upstream or downstream of ΔA, 12‘, and 12RH/13, respectively, do not cross-link to ELAV (Fig. 5D,E), suggesting that the ΔA sequence as solely responsible for the interaction.
The poly(U)-rich element in nASI RNA 9
nASI RNA 9 has the sequence element AU9AU4A, similar to the nASI RNA 6 sequence, AU8CU5C, a sequence implicated in ELAV cross-linking (Figs. 1B, 5B). RNA 9‘ (Table 1) did not cross-link to ELAV in nuclear extracts (Figs. 4F, 5E), demonstrating that this poly(U) tract is insufficient on its own as an ELAV target in the nuclear extracts.
Deletion/mutation of EXS affects reporter expression
To test if the two blocks of poly(U) in 6“ are important for neural-specific splicing, minigene CNVtp5, which incorporated the same five point mutations as 6“p5 (Table 1), was generated in the parent CNVt vector. The splicing was assayed as before. Larvae of the genotype CNVtp5, along with the control CNVt, were heat shocked and assayed for β-galactosidase staining 4 and 20 h later. The X-gal signal in CNVtp5 brain and eye disc was reduced when assayed 4 h after the heat shock, and the reduction in samples assayed 20 h after the heat shock was significant when compared to the CNVt controls (Fig. 6A–D). The reduction in reporter signal validates the importance of the poly(U)-sequence elements within the polypyrimidine tract of the default 3′SS.
To test if further diminishing of poly(U) content contributes to splicing regulation, we generated a transgene, CNVΔEXS, in the CNV parent background, in which in addition to the 6“p5 mutations, the other three EXSs were deleted. Thus, effectively, all of the EXSs were either deleted or mutated. When assayed, the X-gal staining was reduced even in the 4 h after heat shock sample, consistent with the idea that these sites contribute to ELAV regulation (Fig. 6E–F).
We analyzed RNA processing of the mutant transgenes in adult heads 2 h after heat shock using RNAse protection and the HCNZ antisense probe. Two insertion lines of germ-line-transformed flies were examined for each transgene, along with two lines of CNV. The nervous system processing was reduced in CNVΔEXS lines (Fig. 3F), and quantitation by comparing ratios of protected probes HCNZ:HC showed a threefold reduction in HCNZ in CNVΔEXS mutants. Analysis of two lines of CNVtp5 showed a twofold reduction (Lisbin 1999).
Derivation of putative ELAV target sequence
We analyzed the four EXS sequences as defined by the analysis above. 2ΔC, 6ΔB, 10ΔBLH, and 12ΔA constitute core deletions within EXS2, EXS6 EXS10, and EXS12, respectively (see Table 1), and together with the poly(U)-rich sequences that do not cross-link to ELAV, the sequence patterns that ELAV recognizes were deduced. We did not find precisely conserved sequences. Rather, the common feature in each EXS was the presence of long, predominantly poly(U) tracts (Fig. 7D), ranging from 59% to 66% U content and interrupted by A, C, or G residues at apparently random positions. Tracts with a minimum of 8/10Us, with only single-base interruptions, were found in each EXS (Fig. 7D, green shapes). EXS12, which has the least poly(U) content of the four EXSs, has two blocks of 8/10Us that contain no more than four consecutive Us, whereas all the others have runs of five to eight consecutive Us. Additionally, most 8/10U tracts are preceded by a 5′ A.
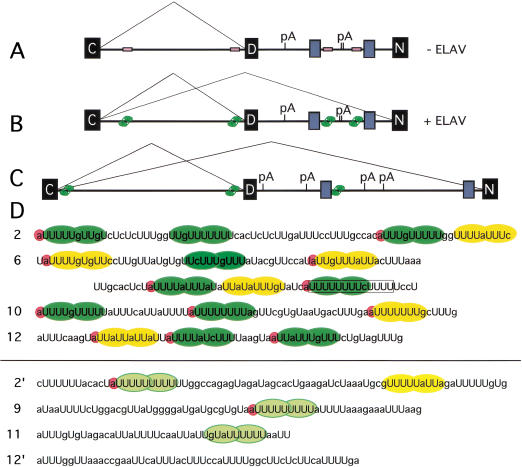
Figure 7.
Model for ELAV regulation of the nASI. D. melanogaster nASI (A,B) and D. virilis (C) nASI in the absence (A) and presence (B,C) of ELAV. Black boxes represent exons and gray boxes represent regions of homology between the two species. EXS sites are shown as unoccupied empty beige boxes (A) or ELAV-occupied green boxes (B,C). EXS sites in C and default splicing in neurons are assumed. pA, polyadenylation sites, are deduced from the sequence. (D) The four D. melanogaster nASI EXSs, above the line, show extended poly(U) tracts that contain multiple stretches of 8/10U (green oval shapes) that potentially accommodate ELAV RRMs 1 and 2. 5′A nucleotide associated with 8/10U stretches are highlighted in pink. The yellow ovals show blocks with only 7/10U poly(U)-rich sequence elements. The sequences below the line show nASI poly(U) elements (light green and yellow) that do not cross-link to ELAV. These contain shorter stretches that lack multiple 8/10U stretches.
Further, the presence of at least two 8/10U tracts in each EXS is important. The inspection of sequences within the nASI that do not cross-link, despite having stretches of poly(U) content very similar to EXS, reveals that the noncross-linkers are shorter and do not contain two 8/10U stretches embedded in them (Fig. 7D). In RNA 2‘, six Us are followed by six bases and then 11 Us, the longest continuous stretch of poly(U) in the nASI; still, it does not show cross-linking to ELAV (Figs. 5E, 7D). Also compare the poly(U) element in RNA 9‘ with the PPT portion of EXS6 (Fig. 7D). This leads us to propose that the specificity of ELAV binding may be in part attributable to the requirement for multiple-binding sites.
Using at least two 8/10U tracts as the minimum criterion, we analyzed the D. virilis nASI sequence for candidate ELAV-binding sites. Three sequences that most fit the EXS model lie in the positions shown in Figure 7C.
Discussion
Our previous studies have indicated that the ELAV, a pan-neural protein, is likely to regulate splicing in neurons. Pan-neural regulators of splicing have not been identified previously. In fact, other factors that, while not tissue-specific, are enriched in some tissues relative to others, are more commonly responsible for regulated splicing (Manley and Tacke 1996). In contrast, only a few examples of tissue-specific splicing factors have been documented. The best studied of tissue-specific splicing factors are Sxl and Tra, the two proteins in the sex determination pathway that are expressed in female but not male tissue (McKeown and Madigan 1992). Other examples are KH-domain RNA-binding proteins PSI and Nova-1. PSI, expressed in somatic but not germ-line tissue, suppresses transposase splicing in the somatic tissue (Siebel et al. 1994, 1995), and Nova-1, expresses specifically in certain brain regions (Buckanovich et al. 1996).
nASI has the determinants for nrg alternative splicing
We set out to determine if the nASI had all of the elements necessary for proper expression and regulation. The expression pattern of the CNV transgene conformed to the expression of neural-specific isoform Nrg180 (Hortsch et al. 1990). This finding was confirmed at the level of the CNV RNA (Fig. 3). Furthermore, ELAV was both necessary and sufficient for the neural-specific expression of the β-galactosidase in the photoreceptor neurons, as previously observed for Nrg180 (Koushika et al. 1996). Two other considerations support the conclusion that the 3′UTR or the reporter coding sequence is not the major regulating factor. First, reporter transgenes that were constructed with either LacZ or GFP (green fluorescent protein) and have HSP 70 trailer, tubulin trailer, or actin trailer show similar expression patterns (data for GFP reporter or actin trailer not shown). Second, the mutated transgenes that show reduced reporter expression have the same 3′UTR (Fig. 6). Thus, we can eliminate from consideration any other sequences from the nrg gene and the non-nASI sequences in the CNV vector as playing an essential role in ELAV regulation, including the 3′ UTR.
ELAV target recognition
In these studies, endogenous ELAV from a crude supernatant of embryonic nuclei was used in UV cross-linking experiments with radiolabeled nrg RNAs. We pursued this strategy for ELAV cross-linking as it presented a more in vivo-like situation in an in vitro assay. The initial studies identified four ELAV cross-linking RNAs: 2 (358 nt), 6 (319 nt), 10 (322 nt), and 12 (311 nt). Deletions coupled with cross-linking assays were used to further limit the EXS within the cross-linking RNAs (Fig. 5).
The deduced ELAV target recognition sequences each have at least two blocks of 8/10Us, with an adjacent 5′ A nucleotide. The finding of 8/10Us, as at least part of the likely ELAV target sequence, is highly relevant in light of ELAV’s homology with Sxl, particularly in the RRM 1 and 2 residues that directly contact RNA. That ELAV is likely to be similar to Sxl in its RNA recognition mechanism has been suggested by Handa et al. (1999) who described the cocrystal structure data of the Sxl RRMs with the target tra RNA, UGUUUUUUU. We propose that ELAV RRMs 1 and 2 recognize these 8/10U tracts. The structure of the ELAV target sequence may help explain, for example, why ELAV and Sxl, expected to have considerable overlap in target recognition, apparently do not interfere with each other's targets.
Whereas much of the ELAV cross-linking data with the nASI RNAs appear to be all or none, the deletion analysis suggests that weak ELAV binding can occur in suboptimal sites, as evidenced by the data from 6“‘, 6ΔA, and 2ΔCRH. These constructs have only one 8/10U but have at least one additional 7/10U element that may allow ELAV to bind, although with reduced affinity.
That each EXS has more poly(U) in addition to the 8/10U tracts suggests that there is more to ELAV–RNA interaction that perhaps involves ELAV RRM3 or auxiliary proteins. The cross-linked protein bands in the range of 40 kD are candidates, because they appear to be most intense in RNAs in which ELAV cross-linking occurs (Fig. 4A), although impaired ELAV binding does not necessarily diminish the 40-kD species (Fig. 5, 6ΔAp5, 6‘p5, and 10ΔB). In the case of 2ΔCRH, in which ELAV cross-linking is impaired but in which two 8/10U still remain, we suggest that the remaining U-tract is insufficient for another component of target recognition.
Within the ELAV family of proteins, homology is highest in the three RRMs (≥70%), particularly in the residues inferred to contact RNA. Therefore, RNA recognition is likely to be very similar among ELAV family members. We recently showed that RRM3 of Rbd9 or HUD can functionally replace the ELAV RRM3 in vivo, but that RRM1 or RRM2 replacements were largely nonfunctional (Lisbin et al. 2000). In vitro RNA selection procedures on vertebrate ELAVs have shown an RNA-binding preference for interrupted stretches of poly(U) (Levine et al. 1993; Gao et al. 1994; Abe et al. 1996). Target sequences have been reported for ELAV relatives HuD, Hel-N1, and HuR, consisting of 3′ UTR AU-rich elements (ARE), which are implicated in mRNA stability. These elements are comprised of predominantly poly(U) tracts with interspersed A residues. The ELAV cross-linking data is consistent with some reports on vertebrate ELAV proteins, such as GLUT-1 3′ UTR (Jain et al. 1995), HPV-1 3′ UTR (Sokolowski et al. 1999), TNF-α ARE (Ford et al. 1999), c-fos ARE and not c-jun ARE (Peng et al. 1998). Other targets reported, for example Myer et al. (1997), which provide in vivo evidence for short AREs interacting with vertebrate ELAV, may represent a different mode of target recognition.
Results from the studies with the nASI minigenes, in which the poly(U) nature of the EXS6 is disrupted, suggest that the default PPT plays a regulatory role and it is likely to occur through the interaction with ELAV. The minigene with deletion/mutation of all four EXSs (CNVΔEXS) also expresses weakly in the nervous system. Many factors could contribute to the residual neural-specific expression from the mutated minigenes. These include additional ELAV-binding sequences not recognized in the UV cross-linking assay, or transcript sequences not included in the minigene, or the deletions themselves could also affect non-ELAV-regulated splicing properties of the nASI. The results from the CNVΔEXS would have to be confirmed further with individual deletions before a definitive conclusion can be drawn. Collectively, these data point to the importance of ELAV’s interaction with the nASI in neural-specific splicing.
Model for splicing regulation
Taken together, the presence of EXS sites within the nASI and its ability to mimic the regulatory aspects of the endogenous nrg, strongly suggests that ELAV influences splicing regulation by direct binding to the nASI. ELAV could be affecting alternative splicing directly and/or affect polyadenylation of the D exon, and as a result, the splicing choice between two terminal exons. For example, in the case of the well-studied Calcitonin gene, regulation of the alternative splicing of the two terminal exons occurs by influencing polyadenylation (Lou et al. 1996). Furthermore, nuclear polyadenylation and splicing of the terminal intron have been shown to be linked events (Niwa et al. 1990; Niwa and Berget 1991).
Our data point to the PPT of the default 3′SS that is included within the EXS6 as playing a pivotal role in splicing regulation. This is similar to the scenario for the alternative splicing of tra, or the Sex-lethal autoregulated intron, in which Sxl targets the default PPT (Sosnowski et al. 1989; Valcarcel et al. 1993). The Sex-lethal autoregulated intron is perhaps a more apt comparison with the nASI because its of similar size and multiple Sxl-binding sites, and Sxl autoregulation also does not occur through a simple blockage model of the default 3′SS (Sakamoto et al. 1992; Horabin and Schedl 1993). Could ELAV additionally regulate polyadenylation of the default transcript? There are three possible polyadenylation sites downstream of the exon D positioned 1959, 2650, and 2673 nt from the 5′SS. None of the sites reside within the conserved elements between D. melanogaster and D. virilis or have conserved elements in close proximity. A CNV minigene with a deletion at site 1959 has no effect on reporter expression (data not shown). Nevertheless, current data does not allow us to rule out regulation at the level of polyadenylation.
We envision a model in which, in the absence of ELAV, the default 3′SS is solely used, despite lacking a consensus branch-point sequence and a proximal PPT. We hypothesize that the conserved intron elements, along with the default exon and polyadenylation site(s) and perhaps other as yet unrecognized sequences, conspire to positively maintain splicing exclusively to an otherwise weak default 3′SS. In the presence of ELAV, however, this positive maintenance is disrupted or partially disrupted, either by directly competing with poly(U)-binding proteins, or by countering the effects of the conserved sequences, leading to 3′SS recognition of the more consensus-like but distal neural-specific 3′SS. This model has predictive value. For example, the exclusive use of the default 3′SS could be compromised by deletion of one or both conserved intron elements. Further studies will be needed to address these questions.
Materials and methods
Cloning and transgene construction
To construct P{w+ = CNV}, nASI was cloned from cosmid 48C2 (Madueno et al. 1995) by PCR using two sets of oligos complimentary to the flanking exon sequences resulting in pBluescript KS+nASI (see Koushika et al. 1996). The 3325 base genomic region of the nrg gene included in CNV extends from the 3′ 59 nt of the C exon to the 5′ 56 nt of the N exon. The nASI was cloned into the CaSpeR transformation vector, pCV, which contains the gene for β-galactosidase under the control of Hsp70 promoter (Mottes and Iverson 1995). To facilitate cloning, site-directed mutagenesis (Kunkel et al. 1991) of KS+nASI was performed to insert a KpnI site into exon C and a NotI site in exon N of nASI. The deletions and point mutants of CNVtp5 and CNVΔEXS were generated by site-directed mutagenesis (Kunkel et al. 1991) of KS+nASI and subsequent cloning into pCV. CNVtp5 was mutated with oligos EXS6p2 and EXS6p3, sequentially, and CNVΔEXS was generated by further mutation with oligos ΔEXS2, ΔEXS10, and ΔEXS12. The sequences are: EXS6p2, CTTTAAATTGCACTCTATATTAATTATATTATA TTTGTATCAT; EXS6p3, TTATATTATATTTGTATCATCT CTTCTCTTTTTCCTAAACACGT; ΔEXS2, CTACAACTGCT CATTACACGCATTGATGACGATTG; ΔEXS10, TTAAGCC TATTGTGTGTAGAAATTGAAACAATGTTTTAG; ΔEXS12, GTATAATTTCAAGTATTATTATTATTACTGTAGTTTGCC TGACAA. To construct P{w+ = hsp–ELAV}, a 1.8 kb elav cDNA fragment was fitted with EcoRI linkers and subcloned into the EcoRI site in pHsp70. Next, the 3.5-kb XbaI fragment, containing the Hsp70–elav cDNA–β-tubulin trailer, was inserted into the XbaI site of CaSpeR (Pirrotta 1988). A D. virilis genomic library (gift of J. Belote, Department of Biology, Syracuse University, NY) was screened with nrg cDNA fragment spanning the nASI as described (Yao and White 1991). The D. virilis clone was then sequenced.
Germ-line transformation and genetic crosses
Flies were raised at room temperature or at 25°C. Below are listed abbreviations of the alleles and transgenes used followed by their descriptions: elave5 is a null allele at the elav locus; elavDVORF (P{w+ = elavDVORF}) is a transgene expressing D. virilis ELAV protein under the control of elav promoter (Yao and White 1991). elavedr is a specific insert of elavDMORF (P{w+ = elavDMORF}), a genomic elav transgene (Yao and White 1991); in the elavedr, elav expression is specifically reduced in photoreceptors (Koushika et al. 1996). Hsp–Sxl (P{w+ = Hsp–Sxl}) and Hsp–ELAV (P{w+ = Hsp70–ELAV}) express Sxl and ELAV cDNAs, respectively, under the hsp–70 promoter control CyOUbGFP is a balancer chromosome CyO with an inserted transgene P{w+ = Ub–GFP}. Ub–GFP expresses green fluorescent protein under the control of the ubiquitin promoter.
Germ-line transformants were generated for the P{w+ = CNV}, P{w+ = Hsp-Sxl} (a gift from L. Bell, Department of Biological Sciences, University of Southern California), and P{w+ = Hsp–ELAV} constructs as described previously (Lisbin et al. 2000). To generate larvae with reduced ELAV expression in the eye and CNV transgene, elave5/elave5; elavedr/+ females were crossed to CNV males. Male larvae of genotype elave5/Y; elavedr/+ were used for further analysis.
To generate flies with CNV and Hsp–ELAV or Hsp–Sxl, CNV females were crossed to Hsp–ELAV/CyOUbGFP or Hsp–Sxl/CyOUbGFP. Larvae of genotype CNV;Hsp–ELAV or CNV;Hsp–Sxl were selected from siblings carrying the GFP balancer by absence of GFP fluorescence.
Heat-shock and β-galactosidase activity assays
Adults were heat shocked according to Mottes and Iverson (1995) for 1 h at 35°C followed by 1 h at room temperature (22°C) and the primary heat shock of 38°C for the stated time. Third instar wandering larvae were heat shocked for 30 min at 35°C , for 40 min at room temperature, and for 40 min at 38°C. At stated times after heat shock, the samples were dissected in 1X PBS on ice, fixed in 10% formalin for 3 min and stained in Fe/NaP with 1/40 of 8% X-gal in DMF (Hiromi et al. 1985; Yao and White 1994). For each transgene, at least two independent inserts were analyzed.
RT–PCR/RNAse protection
Total RNA was isolated from flies or larvae using TRIzol reagent (GIBCO BRL/Life Technologies). For RT–PCR, AMV reverse transcriptase was used to generate cDNA from total CNV RNA with probes complementary to lacZ (LacR, GGCGAT TAAGTTGGGTAACG), the default exon (DR, CTTTGCGTC CATATTGGCC), or the intron (IR, TTCCAACGTAGCAT TACC). The cDNAs were amplified subsequently by a standard PCR with an Hsp70 oligo (HCF, GCTCTAGAAAGTAACCAG CAACCAAG), generating HCNZ, HCD, and HCI, respectively. These PCR products were XbaI cut, and the XbaI/blunt fragments were cloned into pKS+, generating pKS+ HCD, pKS+ HCNZ, and pKS+HCI and sequenced with Prizm (Perkin-Elmer/Cetus).
For RNAse protection assays, pKS+ HCNZ was linearized with XbaI and used as template for antisense probe. The HCNZ antisense probe is 381 nt; 321 nt are protected by neural-spliced RNA and 192 nt are protected by default or nonspliced RNA. rp49 antisense probes control for total RNA (Hardin et al. 1990), with 65 of the 113 nt protected in the processed RNA. In vitro transcriptions were performed with T3 RNA polymerase in the presence of [α-32P]UTP. The transcribed RNA was gel isolated and eluted with standard procedures. RNAse protection assays were performed essentially as reported in Hardin et al. (1990). 0.2 or 1 × 106 cpm probe (∼107 cpm/μg) was added to 5 or 10 μg total RNA in 80% deionized formamide, heated to 85°C for 3 min, and hybridized at 45°C for 16 h. After treatment with RNAse T1 (3 units) and RNAse A (12 μg), and Proteinase K digestion (60 μg), each RNA sample was phenol:chloroform extracted and precipitated with 2.5 volumes of EtOH. Samples were run out on 5% sequencing gels, dried, and analyzed on a PhosphorImager (Molecular Dynamics). Assessment of neural-specific RNA levels from different lines were derived by comparing ratios of the HCNZ:HC signal.
In vitro transcriptions and template construction
All nASI template fragments were generated by PCR; primer sequences are given in Table 2. The “forward” oligos contain a KpnI site at their 5′ end, which facilitated KpnI/blunt cloning into pKS+ vectors. Some templates had subsequent deletions and point mutations introduced by site-directed mutagenesis (Kunkel et al. 1991); for mutated sequences, see Table 1. The template vectors were linearized with XbaI before transcription.
Table 2.
Oligos used to PCR amplify nASI RNAs
| RNA
|
5′–3′
|
|||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1F | G | G | G | G | T | A | C | C | G | C | T | G | G | T | C | G | C | C | A | A | T | C | C | G | T | G | ||||||||||||
| 1R | T | T | C | C | A | A | C | G | T | A | G | C | A | T | T | A | C | C | ||||||||||||||||||||
| 2F | G | G | G | G | T | A | C | C | G | T | A | A | T | G | C | G | T | T | G | G | A | A | A | C | ||||||||||||||
| 2R | A | A | C | A | C | A | A | T | C | G | T | C | A | T | C | A | A | T | G | C | ||||||||||||||||||
| 3F | G | G | G | G | T | A | C | C | A | T | G | A | C | G | A | T | T | G | T | G | T | T | T | G | G | G | ||||||||||||
| 3R | A | G | A | G | C | G | A | A | A | G | G | A | T | C | G | C | T | C | ||||||||||||||||||||
| 4F | G | G | G | G | T | A | C | C | A | G | C | G | A | T | C | C | T | T | T | C | G | C | T | C | T | C | ||||||||||||
| 4R | T | C | G | G | A | A | G | C | C | T | G | C | A | A | G | G | A | G | ||||||||||||||||||||
| 5F | G | G | G | G | T | A | C | C | T | G | C | T | A | T | A | C | A | T | T | T | G | A | C | T | G | C | ||||||||||||
| 5R | G | A | T | G | A | T | C | G | A | T | T | T | C | C | A | T | C | G | ||||||||||||||||||||
| 6F | C | A | C | G | A | G | C | T | C | G | C | T | C | G | T | C | A | A | T | G | A | G | A | T | A | T | T | C | G | |||||||||
| 6R | C | T | T | T | G | C | G | T | C | C | A | T | A | T | T | G | G | C | C | |||||||||||||||||||
| 7F | G | G | G | G | T | A | C | C | A | T | G | A | A | T | G | A | A | G | A | T | G | G | A | T | C | C | ||||||||||||
| 7R | A | G | T | T | G | T | T | T | G | G | C | A | G | T | A | T | T | G | ||||||||||||||||||||
| 8F | G | G | G | G | T | A | C | C | A | A | G | T | G | C | A | A | A | C | A | C | C | A | C | G | A | C | ||||||||||||
| 8R | C | A | T | C | C | C | C | A | T | A | A | C | G | T | C | C | A | G | ||||||||||||||||||||
| 9F | G | G | G | G | T | A | C | C | T | G | C | A | T | G | C | G | A | A | T | C | A | A | A | C | C | G | ||||||||||||
| 9R | C | T | A | A | A | A | C | G | G | A | G | A | T | T | T | G | A | C | ||||||||||||||||||||
| 10F | G | G | G | G | T | A | C | C | G | T | C | A | A | A | T | C | T | C | C | G | T | T | T | T | A | G | ||||||||||||
| 10R | A | T | A | T | G | C | G | A | A | C | G | T | A | C | A | T | C | A | A | C | ||||||||||||||||||
| 11F | G | G | G | G | T | A | C | C | T | T | T | T | C | C | G | G | G | A | A | G | A | G | C | A | A | C | ||||||||||||
| 11R | A | C | C | T | G | G | C | A | A | A | T | G | C | G | T | C | A | G | ||||||||||||||||||||
| 12F | G | G | G | G | T | A | C | C | A | T | G | C | C | A | G | A | G | T | A | G | G | C | T | T | G | |||||||||||||
| 12R | C | C | A | C | A | A | C | C | A | A | C | A | C | C | A | G | T | G | ||||||||||||||||||||
| 13F | G | G | G | G | T | A | C | C | A | A | C | A | C | A | A | T | G | A | A | A | A | G | A | C | A | C | ||||||||||||
| 13R | T | T | G | G | A | G | C | T | T | T | C | C | A | G | G | A | A | C | ||||||||||||||||||||
| 2‘R | A | C | A | G | A | C | A | A | A | A | C | A | T | G | C | G | A | C | ||||||||||||||||||||
| 2"F | G | G | G | G | T | A | C | C | C | C | T | A | C | A | A | C | T | G | C | T | C | A | T | T | A | C | ||||||||||||
| 6‘F | G | G | G | G | T | A | C | C | C | T | T | T | G | T | T | T | A | T | A | C | G | T | T | C | C | |||||||||||||
| 6"F | A | C | T | T | T | A | A | A | T | T | G | C | A | C | T | C | ||||||||||||||||||||||
| 6"R | G | T | G | T | T | T | A | T | T | T | T | A | C | G | T | G | T | T | T | A | G | |||||||||||||||||
| 6‴R | A | A | C | A | A | T | A | T | G | G | A | A | C | G | T | A | T | A | A | A | C | |||||||||||||||||
| 10‘R | A | C | A | A | A | C | A | G | G | A | G | G | C | T | A | T | A | C | ||||||||||||||||||||
| 12‘R | A | C | C | T | G | G | C | A | A | A | T | G | C | G | T | C | A | G | ||||||||||||||||||||
| 12F | G | G | G | G | T | A | C | C | A | A | C | A | C | A | A | T | G | A | A | A | A | G | A | C | A | C | ||||||||||||
In vitro transcription reactions with T3 RNA polymerase were performed using standard protocols. For a 10-μL reaction, 2.5 μL of 800 Ci/ mmole [α-32P]UTP, 1 μL of 50 μM UTP, and 1 μL each of 10 mM ATP, 10 mM CTP, and 10 mM GTP were used. Transcribed RNAs were gel purified on 5% polyacrylamide-urea gels.
Drosophila embryo nuclear extract
Preparation of Drosophila embryo soluble nuclear fraction is a scaled-down version of Kamakaka et al. (1991). Six- to eighteen hour embryos (at 25°C) were harvested for each genotype. The eggs were stored for up to 60 h at 4°C. The two genotypes used were Canton S and elave5wsn; DvORF2, in which ELAV function is supplied by a transgene expressing the D. virilis elav ORF (Yao and White 1991). Two 15 mL Wheaton Scientific Dounce homogenizers were used and all buffers in Kamakaka et al. (1991) additionally contained 2 μg/mL pepstain A and 2 μg/mL leupeptin.
UV cross-linking assays
Cross-linking conditions were adapted from Min et al. (1995). Nuclear extract (2 mg/mL), in 1× binding binding mix (0.4 mM ATP, 20 mM creatine phosphate, 3.2 mM magnesium acetate, 400 μg/mL tRNA) was incubated for 8 min at 30°C. [α-32P]UTP nASI RNA, up to a concentration of 1.5 nM, was added to the nuclear extract and incubated for 15 min at 30°C. Equal CPM of different RNAs were added in a given experiment to reduce bias of RNAs with high U content. The samples were irradiated (254 nm) for 14 min in a Stratalinker (Stratagene). Subsequently, the samples were RNase A digested and analyzed by SDS-PAGE (10%). The gels were dried onto gel blot paper and exposed overnight on a PhosphorImager screen (Molecular Dynamics).
ELAV immunoblot and immunoprecipitation
Affinity-purified rat anti-ELAV polyclonal antibodies were used at a dilution of 1:400 (Robinow and White 1991). Immunoblot analysis was performed using standard techniques and visualized with ECL Western blotting system (Amersham). For ELAV immunoprecipitation, the UV cross-linking assay was followed through to RNAse A digestion, whereupon Mab-7D4E6 (α-ELAV) was added and incubated for 45 min at 30°C. 0.5 volumes of Immunopure Immobilized Protein A (Pierce) in 0.5 M RNA-binding buffer (RBB) (50 mM Tris at pH 7.8, 0.5 M NaCl, and 0.025% NP-40) was then added and incubated for 45 min at 30°C. The beads were washed 5× with 1 mL of 1 M RBB. Subsequent SDS-PAGE gels were transferred onto nitrocellulose filters and exposed to PhosphorImager plates before ELAV immunoblot processing.
Acknowledgments
We thank Drs. L. Iverson and L. Bell for the pCV and P{w+ = hsp–Sxl} plasmids and Dr. M. Hortsch for the nrg cDNA. We thank Dr. J. Belote for the D. virilis genomic library. We thank Dr. I. Kiamos of the Drosophila European Genome project, Crete for cosmids that span the nrg region. We are indebted to Dr. M. Soller for comments on the manuscript and Drs. M. Moore and M. Rosbash for discussions and reviewers for helpful suggestions. We thank J.-R. Neptune and D. Bordne for excellent technical help and E. Dougherty for help with graphics. This work was supported by NIH grants NS36179 and GM33205.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL white@brandeis.edu; FAX (781) 736-3107.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.903101.
References
- Abe R, Sakashita E, Yamamoto K, Sakamoto H. Two different RNA-binding activities for the AU-rich element and the poly(A) sequence of the mouse neuronal protein mHuC. Nucleic Acids Res. 1996;24:4895–4901. doi: 10.1093/nar/24.24.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara SG, Jonas V, Rosenfeld MG, Ong ES, Evans RM. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982;298:240–244. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- Antic D, Keene JD. Embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation, and posttranscriptional gene expression. Am J Hum Genet. 1997;61:273–278. doi: 10.1086/514866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antic D, Lu N, Keene JD. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes & Dev. 1999;13:449–461. doi: 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashaw GJ, Baker BS. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell. 1997;89:789–798. doi: 10.1016/s0092-8674(00)80262-7. [DOI] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL. Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell. 1992;69:795–807. doi: 10.1016/0092-8674(92)90291-j. [DOI] [PubMed] [Google Scholar]
- Buckanovich RJ, Yang YY, Darnell RB. The onconeural antigen Nova-1 is a neuron-specific RNA-binding protein, the activity of which is inhibited by paraneoplastic antibodies. J Neurosci. 1996;16:1114–1122. doi: 10.1523/JNEUROSCI.16-03-01114.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crenshaw EB, Russo AF, Swanson LW, Rosenfeld MG. Neuron-specific alternative RNA processing in transgenic mice expressing a metallothionein-calcitonin fusion gene. Cell. 1987;49:389–398. doi: 10.1016/0092-8674(87)90291-1. [DOI] [PubMed] [Google Scholar]
- Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferns M, Hoch W, Campanelli JT, Rupp F, Hall ZW, Scheller RH. RNA splicing regulates agrin-mediated acetylcholine receptor clustering activity on cultured myotubes. Neuron. 1992;8:1079–1086. doi: 10.1016/0896-6273(92)90129-2. [DOI] [PubMed] [Google Scholar]
- Ford LP, Watson J, Keene JD, Wilusz J. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes & Dev. 1999;13:188–201. doi: 10.1101/gad.13.2.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB, Carson CC, Levine T, Keene JD. Selection of a subset of mRNAs from combinatorial 3′ untranslated region libraries using neuronal RNA-binding protein Hel-N1. Proc Natl Acad Sci. 1994;91:11207–11211. doi: 10.1073/pnas.91.23.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski PJ. Splicing regulation in neurons: Tinkering with cell-specific control. Cell. 1998;92:709–712. doi: 10.1016/s0092-8674(00)81399-9. [DOI] [PubMed] [Google Scholar]
- Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398:579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- Hiromi Y, Kuroiwa A, Gehring WJ. Control elements of the Drosophila segmentation gene fushi tarazu. Cell. 1985;43:603–613. doi: 10.1016/0092-8674(85)90232-6. [DOI] [PubMed] [Google Scholar]
- Horabin JI, Schedl P. Regulated splicing of the Drosophila Sex-lethal male exon involves a blockage mechanism. Mol Cell Biol. 1993;13:1408–1414. doi: 10.1128/mcb.13.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz DS, Krainer AR. Mechanisms for selecting 5′ splice sites in mammalian pre-mRNA splicing. Trends Genet. 1994;10:100–106. doi: 10.1016/0168-9525(94)90233-x. [DOI] [PubMed] [Google Scholar]
- Hortsch M, Bieber AJ, Patel NH, Goodman CS. Differential splicing generates a nervous system-specific form of Drosophila neuroglian. Neuron. 1990;4:697–709. doi: 10.1016/0896-6273(90)90196-m. [DOI] [PubMed] [Google Scholar]
- Inoue K, Hoshijima K, Sakamoto H, Shimura Y. Binding of the Drosophila Sex-lethal gene product to the alternative splice site of transformer primary transcript. Nature. 1990;344:461–463. doi: 10.1038/344461a0. [DOI] [PubMed] [Google Scholar]
- Iverson LE, Rudy B. The role of the divergent amino and carboxyl domains on the inactivation properties of potassium channels derived from the Shaker gene of Drosophila. J Neurosci. 1990;10:2903–2916. doi: 10.1523/JNEUROSCI.10-09-02903.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson LE, Tanouye MA, Lester HA, Davidson N, Rudy B. A-type potassium channels expressed from Shaker locus cDNA. Proc Natl Acad Sci. 1988;85:5723–5727. doi: 10.1073/pnas.85.15.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain RG, Andrews LG, McGowan KM, Gao F, Keene JD, Pekala PP. Hel-N1, an RNA-binding protein, is a ligand for an A + U rich region of the GLUT1 3′ UTR. Nucleic Acids Symp Ser. 1995;33:209–211. [PubMed] [Google Scholar]
- Jain RG, Andrews LG, McGowan KM, Pekala PH, Keene JD. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. Mol Cell Biol. 1997;17:954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, Darnell RB. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Kamakaka RT, Tyree CM, Kadonaga JT. Accurate and efficient RNA polymerase II transcription with a soluble nuclear fraction derived from Drosophila embryos. Proc Natl Acad Sci. 1991;88:1024–1028. doi: 10.1073/pnas.88.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J, Kim J, Kim YJ. Requirement of RBP9, a Drosophila Hu homolog, for regulation of cystocyte differentiation and oocyte determination during oogenesis. Mol Cell Biol. 1999;19:2505–2514. doi: 10.1128/mcb.19.4.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika SP, Lisbin MJ, White K. ELAV, a Drosophila neuron-specific protein, mediates the generation of an alternatively spliced neural protein isoform. Curr Biol. 1996;6:1634–1641. doi: 10.1016/s0960-9822(02)70787-2. [DOI] [PubMed] [Google Scholar]
- Koushika SP, Soller M, White K. The neuron-enriched splicing pattern of Drosophila erect wing is dependent on the presence of ELAV protein. Mol Cell Biol. 2000;20:1836–1845. doi: 10.1128/mcb.20.5.1836-1845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- Levine TD, Gao F, King PH, Andrews LG, Keene JD. Hel-N1: An autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy NS, Chung S, Furneaux H, Levy AP. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- Lisbin MJ. Drosophila ELAV is a neuron-specific splicing factor that regulates the splicing of neuroglian and binds to four sites on the neuroglian alternatively spliced intron. Ph.D. thesis. Waltham, MA: Brandeis University; 1999. [Google Scholar]
- Lisbin MJ, Gordon M, Yannoni YM, White K. Function of RRM domains of Drosophila melanogaster ELAV. RNP1 mutations and RRM domain replacements with ELAV family proteins and sxl. Genetics. 2000;155:1789–1798. doi: 10.1093/genetics/155.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H, Gagel RF, Berget SM. An intron enhancer recognized by splicing factors activates polyadenylation. Genes & Dev. 1996;10:208–219. doi: 10.1101/gad.10.2.208. [DOI] [PubMed] [Google Scholar]
- Madueno E, Papagiannakis G, Rimmington G, Saunders RD, Savakis C, Siden-Kiamos I, Skavdis G, Spanos L, Trenear J, Adam P, et al. A physical map of the X chromosome of Drosophila melanogaster: Cosmid contigs and sequence tagged sites. Genetics. 1995;139:1631–1647. doi: 10.1093/genetics/139.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley JL, Tacke R. SR proteins and splicing control. Genes & Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- McKeown M, Madigan SJ. Sex determination and differentiation in invertebrates: Drosophila and Caenorhabditis elegans. Curr Opin Cell Biol. 1992;4:948–954. doi: 10.1016/0955-0674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Min H, Chan RC, Black DL. The generally expressed hnRNP F is involved in a neural-specific pre- mRNA splicing event. Genes & Dev. 1995;9:2659–2671. doi: 10.1101/gad.9.21.2659. [DOI] [PubMed] [Google Scholar]
- Mottes JR, Iverson LE. Tissue-specific alternative splicing of hybrid Shaker/lacZ genes correlates with kinetic differences in Shaker K+ currents in vivo. Neuron. 1995;14:613–623. doi: 10.1016/0896-6273(95)90318-6. [DOI] [PubMed] [Google Scholar]
- Mount SM, Burks C, Hertz G, Stormo GD, White O, Fields C. Splicing signals in Drosophila: Intron size, information content, and consensus sequences. Nucleic Acids Res. 1992;20:4255–4262. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer VE, Fan XC, Steitz JA. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, Berget SM. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes & Dev. 1991;5:2086–2095. doi: 10.1101/gad.5.11.2086. [DOI] [PubMed] [Google Scholar]
- Niwa M, Rose SD, Berget SM. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes & Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- Okano HJ, Darnell RB. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J Neurosci. 1997;17:3024–3037. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Yang ES, Kim-Ha J, Kim YJ. Down regulation of extramacrochaetae mRNA by a Drosophila neural RNA binding protein Rbp9 which is homologous to human Hu proteins. Nucleic Acids Res. 1998;26:2989–2994. doi: 10.1093/nar/26.12.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Vectors for P-mediated transformation in Drosophila. Biotechnology. 1988;10:437–456. doi: 10.1016/b978-0-409-90042-2.50028-3. [DOI] [PubMed] [Google Scholar]
- Robinow S, White K. The locus elav of Drosophila melanogaster is expressed in neurons at all developmental stages. Dev Biol. 1988;126:294–303. doi: 10.1016/0012-1606(88)90139-x. [DOI] [PubMed] [Google Scholar]
- ————— Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J Neurobiol. 1991;22:443–461. doi: 10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- Robinow S, Campos AR, Yao KM, White K. The elav gene product of Drosophila, required in neurons, has three RNP consensus motifs. Science. 1988;242:1570–1572. doi: 10.1126/science.3144044. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Inoue K, Higuchi I, Ono Y, Shimura Y. Control of Drosophila Sex-lethal pre-mRNA splicing by its own female-specific product. Nucleic Acids Res. 1992;20:5533–5540. doi: 10.1093/nar/20.21.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson ML. Evidence for 3′ untranslated region-dependent autoregulation of the Drosophila gene encoding the neuronal nuclear RNA-binding protein ELAV. Genetics. 1998;150:723–733. doi: 10.1093/genetics/150.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson ML, Lisbin MJ, White K. Two distinct temperature-sensitive alleles at the elav locus of Drosophila are suppressed nonsense mutations of the same tryptophan codon. Genetics. 1995;141:1101–1111. doi: 10.1093/genetics/141.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel CW, Kanaar R, Rio DC. Regulation of tissue-specific P-element pre-mRNA splicing requires the RNA-binding protein PSI. Genes & Dev. 1994;8:1713–1725. doi: 10.1101/gad.8.14.1713. [DOI] [PubMed] [Google Scholar]
- Siebel CW, Admon A, Rio DC. Soma-specific expression and cloning of PSI, a negative regulator of P element pre-mRNA splicing. Genes & Dev. 1995;9:269–283. doi: 10.1101/gad.9.3.269. [DOI] [PubMed] [Google Scholar]
- Sokolowski M, Furneaux H, Schwartz S. The inhibitory activity of the AU-rich RNA element in the human papillomavirus type 1 late 3′ untranslated region correlates with its affinity for the elav-like HuR protein. J Virol. 1999;73:1080–1091. doi: 10.1128/jvi.73.2.1080-1091.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnowski BA, Belote JM, McKeown M. Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell. 1989;58:449–459. doi: 10.1016/0092-8674(89)90426-1. [DOI] [PubMed] [Google Scholar]
- Valcarcel J, Singh R, Zamore PD, Green MR. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature. 1993;362:171–175. doi: 10.1038/362171a0. [DOI] [PubMed] [Google Scholar]
- Wang X, Tanaka Hall TM. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat Struct Biol. 2001;8:141–145. doi: 10.1038/84131. [DOI] [PubMed] [Google Scholar]
- Yannoni YM, White K. Association of the neuron-specific RNA binding domain-containing protein ELAV with the coiled body in Drosophila neurons. Chromosoma. 1997;105:332–341. doi: 10.1007/BF02529748. [DOI] [PubMed] [Google Scholar]
- Yao KM, White K. Organizational analysis of elav gene and functional analysis of ELAV protein of Drosophila melanogaster and Drosophila virilis. Mol Cell Biol. 1991;11:2994–3000. doi: 10.1128/mcb.11.6.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Neural specificity of elav expression: Defining a Drosophila promoter for directing expression to the nervous system. J Neurochem. 1994;63:41–51. doi: 10.1046/j.1471-4159.1994.63010041.x. [DOI] [PubMed] [Google Scholar]
- Yao KM, Samson ML, Reeves R, White K. Gene elav of Drosophila melanogaster: A prototype for neuronal-specific RNA binding protein gene family that is conserved in flies and humans. J Neurobiol. 1993;24:723–739. doi: 10.1002/neu.480240604. [DOI] [PubMed] [Google Scholar]
- Yost HJ, Lindquist S. RNA splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell. 1986;45:185–193. doi: 10.1016/0092-8674(86)90382-x. [DOI] [PubMed] [Google Scholar]
- Zhao G, Hortsch M. The analysis of genomic structures in the L1 family of cell adhesion molecules provides no evidence for exon shuffling events after the separation of arthropod and chordate lineages. Gene. 1998;215:47–55. doi: 10.1016/s0378-1119(98)00273-x. [DOI] [PubMed] [Google Scholar]