Abstract
Pyrroline-carboxy-lysine (Pcl) is a demethylated form of pyrrolysine that is generated by the pyrrolysine biosynthetic enzymes when the growth media is supplemented with D-ornithine. Pcl is readily incorporated by the unmodified pyrrolysyl-tRNA/tRNA synthetase pair into proteins expressed in Escherichia coli and in mammalian cells. Here, we describe a broadly applicable conjugation chemistry that is specific for Pcl and orthogonal to all other reactive groups on proteins. The reaction of Pcl with 2-amino-benzaldehyde or 2-amino-acetophenone reagents proceeds to near completion at neutral pH with high efficiency. We illustrate the versatility of the chemistry by conjugating Pcl proteins with poly(ethylene glycol)s, peptides, oligosaccharides, oligonucleotides, fluorescence, and biotin labels and other small molecules. Because Pcl is genetically encoded by TAG codons, this conjugation chemistry enables enhancements of the pharmacology and functionality of proteins through site-specific conjugation.
Keywords: protein engineering, protein medicinal chemistry, desmethylpyrrolysine, amber suppression
Just as small molecule drug candidates are optimized through medicinal chemistry, protein biotherapeutics need to be optimized in terms of their pharmacological and physiochemical properties, selectivity, and immunogenicity (1–3). Such protein medicinal chemistry efforts may, for example, involve choosing an appropriate expression system to achieve a particular glycosylation state of the protein (2). Changes in the primary protein sequence through recombinant DNA technology can enhance solubility and stability or alter immunogenicity and selectivity of the protein drug. Only a few of the canonical 20 amino acids are commonly used to modify the properties of proteins through chemical derivatization. Conjugation of poly(ethylene glycol)s (PEGs) for serum half-life extension, chemical haptenation to increase the immunogenicity of antigens, and other conjugations to effect the physicochemical and pharmacological properties of proteins drugs and vaccines are primarily performed through lysines, cysteines, and the N terminus (4). Amine reactive reagents are broadly available, but conjugation results in heterogenous mixtures with often substantially reduced activity relative to the unmodified protein. Homogenous products can be produced by optimizing reaction conditions such that only the N terminus is derivatized, but accessibility and effects on activity may limit this site-specific modification approach. Alternatively, a solvent exposed cysteine can be introduced through mutagenesis, but native cysteines may need to be mutated to achieve derivatization at a single desired site. Often, this type of mutagenesis is not a viable option because additional cysteine residues can interfere with protein folding, causing aggregation or dimerization, and in proteins expressed in mammalian cells, often form cysteine or glutathione adducts that need to be disrupted through reduction prior to conjugation with thiol-specific reagents (5). Site-specific bioorthogonal chemical or enzymatic derivatization can also be accomplished through a number of peptide tags (4, 6) but requires adding a stretch of nonendogenous sequence to the protein of interest. Any chemistry utilizing the standard 20 amino acids will face similar hurdles to achieve site-specificity without deleterious effects to the protein of interest.
The most elegant way to generate homogenously, site-specifically modified proteins is the in vivo incorporation of unnatural amino acids (7–9). Over 70 unnatural amino acids featuring a wide array of functionalities can be incorporated at TAG codons using specific tRNA/tRNA synthetase pairs engineered through an in vivo selection process to be orthogonal to the cellular machinery of the host cells. A set of reactive unnatural amino acids carrying keto (10), azido (11), and alkyne (12) moieties are most frequently used for bioconjugation through oxime formation and “click-chemistry.” Although oxime formation requires acidic pH and some click-chemistry reactions require a copper catalyst for efficient conjugation, unnatural amino acid technology provides unprecedented opportunities for protein labeling and conjugation through chemical functionalities that are not available among the common 20 amino acids.
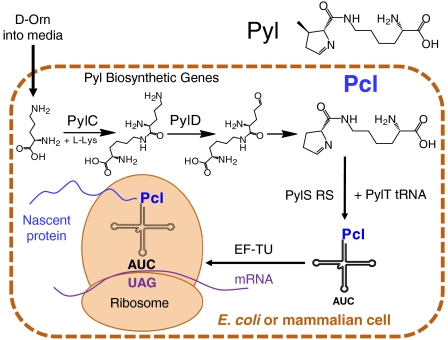
Pyrrolysine (Pyl), the 22nd naturally encoded amino acid, was discovered in several methyltransferases in methanogenic archaea (Fig. 1) (Selenocysteine being the 21st amino acid) (13–17). In Methanosarcinae, the TAG codon, a canonical termination signal, is primarily repurposed to signal Pyl incorporation (18). Two of the three Pyl biosynthetic genes, pylC and pylD, along with the pyrrolysl-tRNA/tRNA synthetase pair can be transferred to Escherichia coli (19) as well as mammalian cells (20), and facilitate the incorporation of pyrroline-carboxy-lysine (Pcl) rather than Pyl when D-ornithine (D-Orn) is added to the growth media (Fig. 1) (20, 21). Here we show that Pcl can be incorporated into a broad range of target proteins at TAG codons in both E. coli and mammalian cells, and we report a specific conjugation chemistry that enables coupling of synthetic compounds to Pcl in recombinant proteins. This combination provides a powerful platform for site-specific modification of proteins.
Fig. 1.
Biosynthetic generation and incorporation of Pcl into proteins produced in E. coli or mammalian cells using the Pyl machinery from Methanosarcinae maize and D-Orn added to the media (20).
Results
Pcl Protein Production in E. coli and Mammalian Cells.
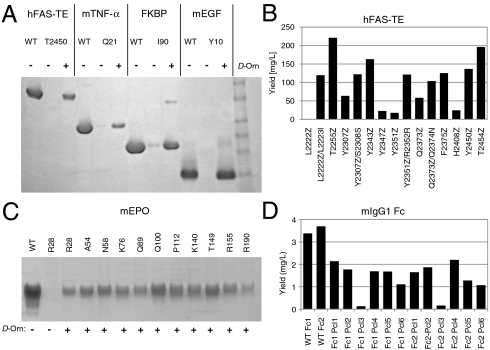
Pyl-containing proteins can only be produced in small amounts from E. coli. Pcl incorporation at a single site was, however, supported at high yields in E. coli and mammalian cells when 1–5 mM D-Orn was added to the growth media (20). So far, more than 350 Pcl mutants of over 30 different proteins have been produced, including enzymes, membrane receptors, growth factors, Fc-fusion proteins, antibodies, and antibody fragments (Fig. 2, SI Appendix). Expression yields in E. coli were typically 25–40% but some reached 80% of wild-type protein using a two-plasmid expression system (Fig. 2 A and B). Transient cotransfection of HEK293 cells with the Pyl machinery regularly produced Pcl protein at 10–20% of the corresponding wild-type proteins with the best yield reaching ∼65% (Fig. 2 C and D). As with any site-directed mutagenesis, yields for a mutant protein may be altered by effects on folding, stability, and solubility. Lower than wild-type yields are expected due to competition between Pcl-charged tRNA and release factor at the TAG codon. In fact, truncated protein products consistent with translation termination were generally observed but were easily separated during purification, for example with the use of a purification tag at the C terminus of the protein of interest. There are likely some contextual effects on the incorporation efficiency in the DNA or mRNA as illustrated by the improved yield of four hFAS-TE mutants with additional nucleotide changes at the +1 codon (Fig. 2B). Incorporation efficiency cannot be predicted at this time but because of the ease of expression, many sites can quickly be scanned for each new application. Our observations for Pcl are consistent with reports (18, 24, 25) that Pyl, unlike selenocysteine, incorporation does not require a conserved mRNA element (22, 23). Expression of Pcl proteins has so far been tested in shake flask culture, small bioreactors and wavebags™ and feasibility of large scale production needs to be evaluated in the future.
Fig. 2.
Incorporation of Pcl into proteins produced in E. coli or mammalian cells (20). (A) Examples of single-site Pcl incorporation into hFAS-TE, mTNF-α, FKBP, and mEGF using E. coli as expression host. Wild-type (WT) and Pcl proteins from equal volume cultures grown with (+) and without (-) D-Orn were compared. (B) hFAS-TE Pcl protein yields as a function of incorporation site. For some sites, yields were increased by changing the first nucleotide after the TAG codon resulting in a silent or conservative mutation. Pcl is abbreviated as Z. Examples of single-site Pcl incorporation into mouse EPO (C) and mIgG1 Fc domain constructs (D) using transient transfection and HEK293 cells. Protein yields varied with incorporation site relative to WT protein. Pcl incorporation was verified by mass spectrometry. For additional information, see SI Appendix.
Pcl-Specific Bioorthogonal Conjugation Chemistry.
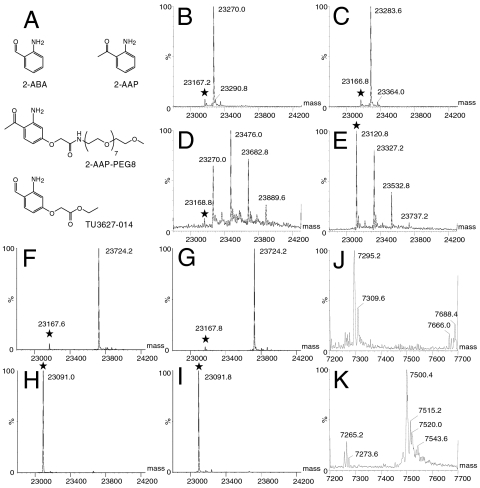
It has been suggested previously that Pyl is chemically reactive (26). Previously, 2-amino-benzaldehyde (2-ABA) was used to derivatize Δ1-pyrroline-5-carboxylate, a molecule analogous to the pyrroline ring of Pyl, in order to study its role in the biosynthesis of proline (27). Here, we show that Pcl (and Pyl) incorporated into proteins can with high efficiency and specificity be derivatized with 2-ABA or 2-amino-acetophenone (2-AAP) moieties (Fig. 3A, SI Appendix). The reactions proceeded to near completion at pH 5 and pH 7.5 at molar ratios of 6∶1 of 2-ABA or 2-AAP using 17 μM Pcl protein (Fig. 3 B and C). Conjugation of 2-ABA to the Pcl residue was confirmed by mass spectrometric (MS) analysis (SI Appendix: Fig. S1). Additional conjugation events were only observed at very high 2-ABA concentration (> 80 mM), both for the Pcl protein (Fig. 3D) and also for a control protein that did not contain Pcl (Fig. 3E). These data suggest that other reactive groups of the protein can only be modified at extreme reactant concentrations. The 2-ABA/2-AAP reaction with Pcl appears to be highly chemoselective as none of the 12 lysines, six cysteines (in three disulfide bonds), or eight histidines of hRBP4 used as test case in Fig. 3 were derivatized.
Fig. 3.
Specific reactions of Pcl- and Pyl-containing proteins with 2-ABA and 2-AAP reagents (A). Shown are intact LC-ESI mass spectra of reaction mixtures. Unmodified proteins are highlighted with stars. 2-ABA (B) and 2-AAP (C) at 6∶1 reactant to protein formed nearly complete adducts [17 μM hRBP4 Phe122Pcl; molecular mass unreacted: 23167.2 Da and 23166.8 Da observed (obs.), 23167.7 Da calculated (cal).; reacted with 0.1 mM 2-ABA (B): 23270.0 Da obs., 23270.8 Da cal., reacted with 0.1 mM 2-AAP (C): 23283.6 Da obs., 23284.8 Da cal., 12 h, 22 °C, pH 5.0)]. At very high concentrations, 2-ABA (or 2-ABA dimers, tetramers and hexamer) formed additional adducts with the Pcl protein (D) (17 μM hRBP4 Phe122Pcl, 80 mM 2-ABA, 12 h, 22 °C, pH 7.5) but also with a control protein (E) that did not contain any Pcl (100 mM 2-ABA, 15,400∶1 reactant to protein; 6.5 μM hRBP4 Phe62OMePhe unreacted: 23120.8 Da obs., 23,122 Da cal., 100 mM 2-ABA, 12 h, 22 °C, pH 7.4). 2-AAP-PEG8 (at 9.1 mM) reacted with hRBP4 Phe122Pcl to ∼94% at pH 7.5 (PBS; 2,100∶1 reactant to protein; 4.3 μM protein; unreacted: 23167.6 Da obs., 23167.7 cal.; reacted: 23724.2 Da obs., 23724.3 Da cal.) (F) and to ∼96% at pH 5.0 (200 mM sodium acetate; 10,500∶1 reactant to protein; 0.86 μM protein; unreacted: 23167.8 Da obs., reacted: 23724.2 Da obs.) (G). Wild-type hRBP4 (23091.2 and 23092.0 Da obs., 23,092 Da cal.) did not react with 9.1 mM 2-AAP-PEG8 at identical conditions (H, pH 7.5 and I, pH 5.0). Expression of mEGF Tyr10TAG in E. coli in the presence of D-Orn and biosynthetic genes pylB, pylC and pylD resulted in a mixture of Pcl and Pyl protein (J). When reacted with 1 mM 2-AAP-derivative TU3627-014 (0.01 mM protein) for 16 h at 22 °C in 10× PBS, pH 7.4, both Pcl and Pyl protein reacted to near completion and to similar extents (K) (mEGF Tyr10Pcl unreacted: 7295.2 Da obs., 7,296 cal.; reacted: 7500.4 Da obs., 7,501 Da cal.; mEGF Tyr10Pyl unreacted: 7309.6 Da obs., 7,310 Da cal; reacted: 7515.2 Da obs., 7,515 Da cal.). See SI Appendix for additional information.
Site-specific conjugation of Pcl proteins also proceeded with high efficiency with derivatized 2-AAP or 2-ABA reagents (Table 1). For example, near complete conjugation to hRBP4 Phe122Pcl protein (4.3 μM or 0.9 μM) occurred for a short PEG (2-AAP-PEG8 at 9.1 mM) at pH 7.5 (Fig. 3F) and pH 5.0 (Fig. 3G) while wild-type protein showed no indication of any reaction (Fig. 3 H and I). Pyl and Pcl had similar reactivity as was demonstrated by the derivatization of a mixture of Pyl- and Pcl-containing mEGF protein with TU3627-014 (Fig. 3 A, J, and K). The reactions of 2-ABA and 2-AAP reagents proceeded with high efficiency at 4 °C, room temperature and 37 °C, at low protein (> 1 μM) and reagent (typically < 1 mM) concentrations and reached near completion after 16 h at neutral pH (Table 1).
Table 1.
Functionalization of Pcl proteins
| Reagent | Structure(s) | Coupling [%] | Fig. |
| PEGs (linear and branched) | 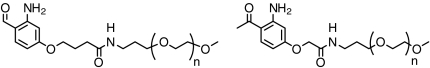 |
50 to > 95 | 3, 5 |
| Disaccharide | 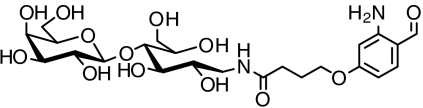 |
> 98 | SI Appendix: Fig. S8 |
| DOPE phospholipid | 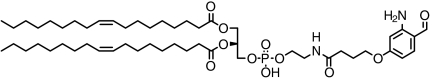 |
> 94 | SI Appendix: Fig. S9 |
| Biotin | 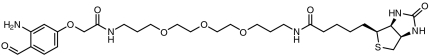 |
> 98 | SI Appendix: Fig. S10 |
| Fluorescein | 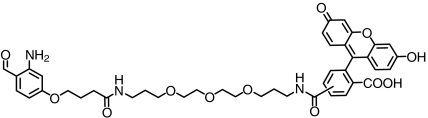 |
> 88 | SI Appendix: Fig. S11 |
| Nitro-phenyl haptens | 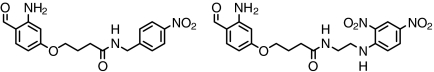 |
> 93 | SI Appendix: Fig. S12 |
| CpG- phosphothioate | 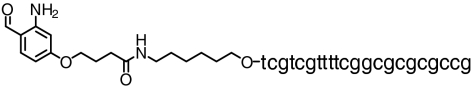 |
∼80 | SI Appendix: Fig. S13 |
| PADRE peptide (T-cell epitope) | 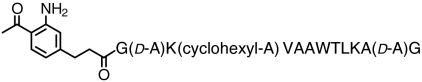 |
> 83 | SI Appendix: Fig. S14 |
Pcl reagents tested and coupling efficiencies at pH 7.4 and 22 °C. For details and results, see Fig. 3, Fig. 5, and SI Appendix: Figs. S8–S14 as indicated
Structure and Stability of Pcl Conjugates.
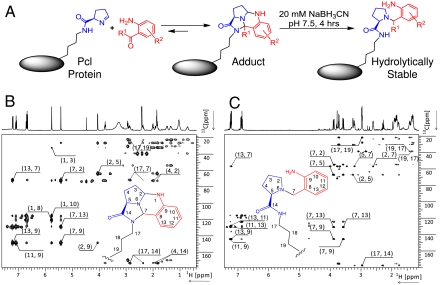
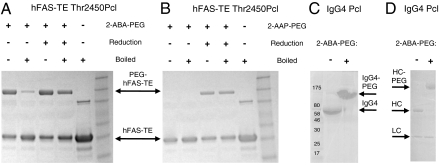
Nuclear magnetic resonance (NMR) spectroscopy studies of the Pcl amino acid reacted with 2-ABA or 2-AAP suggested the structure of the Pcl protein adducts shown in Fig. 4A. The reaction of 2-ABA with Pcl, but not with the alternative imine isomer, rapidly generated a minor and a major product and proceeded to completion (SI Appendix: Fig. S2). A detailed analysis of the purified adduct by 1H-13C heteronuclear multiple bond correlation (HMBC) NMR (Fig. 4B, SI Appendix: Fig. S3) suggested that the formation of a pyrrolo-quinazoline ring was stabilized through cyclization of the 2-ABA/2-AAP carbonyl moiety with the ϵ-amide of Pcl (for NMR data of the 2-AAP adduct, see SI Appendix: Fig. S4 and S5). The formation of the Pcl protein adduct was reversible (SI Appendix: Fig. S6); the stability depended on the chemical structure of the reactant (SI Appendix: Fig. S7). For 2-AAP and its derivatives, 50% of the conjugate reverted to unmodified Pcl protein in 4 to 10 h (at 37 °C, pH 7.4). For derivatized forms of 2-ABA, decay to 50% conjugate occurred over 20 to 30 h. MS analysis and repeated reactions demonstrated regeneration of the Pcl side chain by reversion of the conjugation by simple hydrolysis rather than modification to a new structure. The Pcl-2-ABA (or 2-AAP) linkage was further stabilized by a simple and quantitative reduction step with sodium cyanoborohydride (Fig. 4A and Fig. 5 B and C), a reaction that is commonly used during the N-terminal PEGylation of proteins (28). The structure of this hydrolytically stable form was determined by NMR for 2-ABA and Pcl (Fig. 4C, SI Appendix: Fig. S3). For the respective Pcl protein adduct, no degradation was observed over a period of 96 h at 22 °C and pH 7.4 (SI Appendix: Fig. S7). The ability to fine tune the stability of the coupling by the choice of reactive group and linker provides the opportunity to design releasable conjugates, for example for antibody drug conjugates. Alternatively, reduction results in a hydrolytically stable linkage that cannot be destroyed by boiling in SDS gel buffer (Fig. 5 A and B).
Fig. 4.
Site-specific modification of Pcl proteins through 2-ABA (R1 = H) and 2-AAP (R1 = CH3) derivatives. Examples of derivatives are shown in Table 1. (A) Structures of the protein adduct and the hydrolytically stable form after reduction as inferred from NMR studies with reacted Pcl amino acid. Summary of the key through bond connectivities detected by 1H-13C-HMBC NMR spectroscopy for the 2-ABA/Pcl amino acid adduct before (B) and after reduction (C). The first number of the crosspeak label identifies the 1H resonance while the second number identifies the 13C resonance of the respective atom or group. For numbering and structure, see inserts. Additional information, discussion, and NMR data for the 2-AAP adduct are provided as SI Appendix. Pcl was synthesized as a diastereomeric mixture (20). The stereochemistry of Pcl in the protein is drawn as observed for Pyl in proteins (26).
Fig. 5.
Site-specific PEGylation of Pcl proteins. SDS-PAGE of reaction mixtures of hFAS-TE Pcl protein with 30 kDa-linear 2-ABA-PEG (A) and 2-AAP-PEG (B). hFAS-TE Thr2450Pcl (0.2 mM) was incubated with 1 mM PEG for 60 h at 4 °C in PBS (pH 7.4). Mixtures were either reduced with 50 mM NaBH3CN for 1 h at 37 °C or left untreated. All samples were passed over NAP5 size exclusion columns; some samples were boiled at 95 °C for 5 min prior to loading onto the gel. (C) SDS-PAGE (nonreducing gel) of the reaction mixture of a hIgG4 Pcl mutant (0.01 mM) with a 40 kDa-linear 2-ABA-PEG (0.1 mM) detected near quantitative conjugation after 24 h at 22 °C in 200 mM Na acetate buffer (pH 5.0). (D) SDS-PAGE (reducing gel) of the material after reduction (20 mM NaBH3CN, 4 h, 22 °C) showed PEGylation of the IgG4 heavy chain only. See SI Appendix for additional information.
Versatility of the Pcl Conjugation Chemistry and Applications.
The stability, specificity, and efficiency of the coupling chemistry combined with the ease of Pcl incorporation into proteins expressed in E. coli and mammalian cells, make Pcl a versatile and broadly applicable protein modification platform. To illustrate this versatility, we synthesized and successfully conjugated over 50 different Pcl reactive reagents ranging from low molecular weight molecules for fluorescent labeling to PEG polymers (Table 1, Fig. 5, SI Appendix: Figs. S8–S14).
Site-specific PEGylation is a preferred method to extend the serum half-life of biotherapeutic proteins as it preserves activity and generates a homogeneous product (3, 28). We synthesized 2-AAP and 2-ABA activated PEGs of 0.5, 2.4, 5, 10, 20, 30, and 40 kDa molecular mass and coupled them to Pcl proteins (Fig. 5, SI Appendix). Coupling of 2-ABA-activated PEGs (Fig. 5A) was more efficient and resulted in more stable adducts than coupling with 2-AAP-PEGs. In fact, 2-AAP-PEG/Pcl adducts required NaBH3CN reduction for effective purification (Fig. 5B). The reduced conjugates appeared as single species on SDS-PAGE (Fig. 5). Coupling efficiencies varied with length of the PEG from > 95% for short PEGs (Fig. 3 F and G) to typically 50% for 30 and 40 kDa PEGs (Fig. 5 A and B). In one instance, coupling of a 40 kDa PEG to a hIgG4 Pcl mutant proceeded nearly to completion (Fig. 5C), suggesting that PEGylation efficiency is not limited by the conjugation chemistry but rather by accessibility of the Pcl side chain for reactions with large and flexible PEGs. This example further illustrated the high specificity of the PEG attachment to the Pcl residue in the IgG4 heavy chain as only the heavy chain was shifted on a reducing gel (Fig. 5D), highlighting the homogeneity of the PEGylated protein product.
Another method to modify the pharmacology of protein drugs is glycoslyation. This posttranslational modification occurs naturally in proteins expressed in eukaryotic cells but usually not in E. coli. Glycosylated proteins typically feature a mixture of complex oligosaccharide structures attached at specific serines and threonines (O-linked glycosylation) or asparagines (N-linked glycosylation). To explore the feasibility of chemically conjugating defined sugars to proteins expressed in E. coli, we activated a disaccharide and linked it to a 2-ABA moiety (SI Appendix: Fig. S8). Subsequent coupling of the sugar to a Pcl protein proceeded to completion as monitored by MS.
The conjugation of a phospolipid, to be potentially used as membrane anchor, was also successfully tested (SI Appendix: Fig. S9). In addition, a biotin analog (SI Appendix: Fig. S10) and a fluorescent dye (SI Appendix: Fig. S11) were conjugated nearly to completion using 0.1 to 1 mM 2-ABA reagent and 10 μM protein, illustrating the potential of Pcl conjugation for standard protein labeling and detection strategies.
Chemical haptenation of protein antigens, for example with nitro-phenol reagents, has long been used for the efficient generation of antibodies (29, 30). Similarly, the immunogenicity of vaccine antigens can be enhanced by adjuvants such as aluminum oxide particles and by adding immune stimulators such as toll-like receptor (TLR) agonists to these formulations (31). As colocalization of such reagents with the antigen may boost efficacy (32), we tested whether several immune stimulatory entities can be conjugated to proteins. Indeed, site-specific coupling of 2-ABA nitro-phenyl haptens to model antigens proceeded to > 93% (SI Appendix: Fig. S12) while the conjugation of two CpG oligophosphothioate TLR9 agonists (33) (SI Appendix: Fig. S13) and a T-cell epitope peptide (34) (SI Appendix: Fig. S14) reached approximately 80% completion.
Although we have not fully evaluated the utility of all these modifications, the examples never-the-less established the chemistry to produce site-specific PEG, oligosaccharide, small molecule, oligonucleotide, and peptide-protein conjugates through the Pcl amino acid. In general, the reactions of all reagents with MW < 2,000 Da proceeded to > 95% completion after 16 h (Table 1) in a pH and temperature range where most proteins are stable as long as the solubility of the reagents was not limiting. As with all biopharmaceuticals, the immunogenicity of each protein drug containing unmodified or modified Pcl residues will need to be assessed thoroughly during clinical development. Several applications of the technology are being pursued currently.
Discussion
Strategies for protein modification are indispensible for converting a protein into a research compound or into a pharmaceutical agent. Site-specific protein modification methods are most powerful as they allow the protein designer to choose sites were modifications do not affect the protein’s natural function or do so in a controlled manner. Incorporation of unnatural amino acids is currently the most versatile way to accomplish this task (7–9). The ability to introduce new chemical functionalities through synthetic unnatural amino acids provides tools nearly on par with what traditional medicinal chemistry can do for low molecular weight compounds. Unnatural amino acids can themselves constitute the desired modification (e.g., mimics of posttranslational modifications, spectroscopic probes) or alternatively can be functionalized after protein incorporation through chemical derivatization (8, 9). Here, we show that the discovery of a bioorthogonal conjugation chemistry for Pcl, a biosynthetically generated Pyl analog, provides a competitive method for protein medicinal chemistry and labeling through site-specific protein conjugation.
Pyl was discovered about ten years ago as the 22nd genetically encoded proteinogenic amino acid (13–16). Unexpectedly, we discovered that addition of D-Orn to growth media in the presence of the Pyl biosynthesis genes results in the generation and incorporation of a demethylated Pyl analog, Pcl (20, 21) (Fig. 1) rather than Pyl as had been suggested previously (24). D-Orn appears to feed into the Pyl biosynthetic pathway downstream of PylB, as Pcl incorporation requires only PylC and PylD. Preliminary results from low oxygen fermentations in E. coli suggest that Pyl incorporation is intrinsically more efficient than Pcl (20). However, under standard expression conditions, the activity of PylB, the enzyme responsible for the methylation of the pyrrolysine precursor, or a required cofactor appears to limit the production of Pyl proteins in mammalian and E. coli cells (20).
The pyrrolysyl-tRNA/tRNA synthetase pair has naturally evolved in Methanosarcinae to support amber suppression and to be orthogonal to other endogenous pairs in those species as well as in mammalian cells (35), Saccharomyces cerevisiae (35) and E. coli (19). This history of evolutionary pressure along with the observation that even the unmodified pyrrolysyl-tRNA synthetase PylS is relatively tolerant of nonpyrrolysine substrates [but generally utilizes them with lower efficiency (35–42)], makes it an attractive system for the incorporation of unnatural amino acids either using wild-type (35, 38–42) or mutant pyrrolysyl-tRNA synthetases (35, 39, 43–46). Pcl mutagenesis differs from the unnatural amino acid technology in that a standard metabolite, D-Orn, is processed by the biosynthesis enzymes of a natural amino acid into a close analog of Pyl. The close structural similarity of Pcl to Pyl likely explains the high efficiency of Pcl incorporation using the unmodified pyrrolysyl-tRNA synthetase. In nature, the occurrence of Pyl is limited to a specific position in the active site of methylamine methyltransferases in certain organisms (47). Despite this fact, Pcl can be incorporated with high efficiency and good yields at many different positions in a large number of proteins (Fig. 2).
In addition, Pcl proteins can site-specifically be derivatized with 2-ABA and 2-AAP reagents including PEGs, sugars, peptides, and oligonucleotides (Table 1, Fig. 3, 5). Small molecules especially can be coupled with high efficiency, making site-specific conjugation through Pcl a viable option for the development of antibody drug conjugates and imaging reagents as well as many research reagents. Key advantages of the Pcl protein modification platform are universal and robust protein expression and site-specific incorporation of the reactive amino acid in E. coli and mammalian cells with the same cellular machinery that evolved naturally. In contrast to most unnatural amino acids that need to be chemically synthesized, Pcl is generated biosynthetically in the host cell from a readily available media supplement, D-Orn. Most importantly, these features are combined with an efficient, Pcl-specific conjugation chemistry that works for many applications near neutral pH.
Methods
Reagents.
2-AAP, 2-ABA, and NaBH3CN were purchased from Sigma while D-Orn was purchased for Chem-Impex. Pcl ((2S)-2-amino-6-(3,4-dihydro-2H-pyrrole-2-carboxamido)hexanoic acid) and its isomer (S)-2-amino-6-(3,4-dihydro-2H-pyrrole-5-carboxamido)hexanoic acid were synthesized as described (20). The syntheses of all Pcl reactive reagents are described in SI Appendix.
Production of Pcl Proteins (Fig. 2).
E. coli cells were cotransformed with pAra-pylSTCD and an expression plasmid for the protein of interest as described (20). The thioesterase domain of human fatty acid synthase (hFAS-TE) was expressed using a pMH4 plasmid. Mouse tumor necrosis factor alpha (mTNF-α), human FK506-binding protein 1A (FKBP) and mouse epidermal growth factor (mEGF) were cloned into pET vectors using PIPE (48). TAG codons for single-site Pcl incorporation were introduced as shown in SI Appendix: Table S1. FAS-TE constructs with a purine at the +1 codon position after TAG (CTG2223ATT, TCC2307AGC, CGG2351AGG, and CAG2373AAT) were constructed to reduce termination efficiency (49). hFAS-TE was expressed in HK100 cells (20); mTNF-α, FKBP, and mEGF were expressed in BL21(DE3) cells (Invitrogen) grown in terrific broth media (Sigma) at 37 °C supplemented with 5 mM D-Orn. Proteins were purified using Ni-NTA (Qiagen) chromatography and yields are listed in SI Appendix: Table S2. Pcl incorporation into proteins in HEK293F cells was accomplished by transient transfection (20). TAG codons were introduced into pRS expression vectors for human retinol binding protein (hRBP4), mouse erythropoietin (mEPO), two mIgG1 Fc constructs (Fc1, Fc2), and a human IgG4 antibody (hIgG4) (SI Appendix: Table S3). HEK293F cells were grown in suspension in 293 Freestyle expression media. 5 mM D-Orn was added after transfection. Proteins were purified using Ni-NTA or protein A-affinity chromatography. Pcl incorporation was verified by LC-MS. See SI Appendix for additional details.
Site-Specific Derivatization of Pcl and Pyl Proteins (Fig. 3).
Ten microliter hRBP4 Phe122Pcl protein solutions were mixed with 89 μL 200 mM sodium acetate buffer (pH 5.0) and 1 μL 10 mM 2-ABA (Fig. 3B) or 2-AAP (Fig. 3C) solution, with 80 μL 10× PBS and 10 μL 0.8 M 2-ABA solution (Fig. 3D), with 10 μL 10× PBS and 2 μL 100 mM 2-AAP-PEG8 (TU3205-044) solution (Fig. 3F), or with 90 μL 200 mM sodium acetate buffer and 10 μL 100 mM 2-AAP-PEG8 (Fig. 3G). A 10 μL sample of a hRBP4 O-methyl-phenylalanine (OMePhe) mutant (50) was mixed with 80 μL 10× PBS and 10 μL 1 M 2-ABA (Fig. 3E). The reactions proceeded 14 h at 22 °C followed by 72 h at 4 °C before MS. Reactions with wild-type hRBP4 protein at concentrations of 20 μM (Fig. 3H) and 4 μM (Fig. 3I) were performed in parallel. A mEGF Tyr10Pcl/Pyl protein mixture (Fig. 3J) was reacted similarly with 1 mM of TU3627-014 (Fig. 3K).
Protein and Peptide LC-MS and Verification of 2-ABA/Pcl Adduct Formation.
Intact protein MS was obtained on an automated QTOF II LC-MS system (Waters). Nano-reverse-phase liquid chromatography (LC) MSMS of proteolytic digests was performed using a LTQ Orbitrap hybrid mass spectrometer (ThermoElectron). MS data were processed as described (20). LC-MS analysis of the tryptic digest of the 2-ABA-derivatized hRBP4 Phe122Pcl protein verified conjugation to the Pcl residue (SI Appendix: Fig. S1, Table S4).
NMR Characterization of Pcl Adducts (Fig. 4).
Pcl and 2-ABA solutions in D2O were mixed and reacted to near completion as monitored by NMR (SI Appendix: Fig. S2, Tables S5, S6). The 2-ABA/Pcl adduct was purified by HPLC, redisolved in d6-DMSO, and a 1H-13C HMBC spectrum (Fig. 4B) optimized for the detection of three-bond-correlations using a 50 ms delay (J = 10 Hz) was acquired at 300 K on a 400 MHz Bruker Avance instrument (Bruker Biospin). For the characterization of the reduced 2-ABA/Pcl adduct (Fig. 4C), aliquots of a NaBH3CN solution in D2O were added to a second sample until the adduct resonance at 5.6 ppm (SI Appendix: Fig. S2) completely disappeared. The sample was repeatedly lyophilized from dry D2O and reconstituted in 0.5 mL D2O. The 1H-13C HMBC spectra of the reduced 2-ABA/Pcl adduct (Fig. 4C) was recorded on a 600 MHz Bruker Avance instrument with a delay of 50 ms. Signal assignments and through-bond correlations for nonreduced and reduced 2-ABA/Pcl adducts are summarized in SI Appendix: Fig. S3, Tables S7, S8, S9. Comparison with the 2-AAP adducts of Pcl (SI Appendix: Fig. S4, Table S10) and of 3,4-dihydro-2H-pyrrole-2-carboxylic acid (SI Appendix: Fig. S5, Tables S11, S12) support the adduct structures shown in Fig. 4A. A detailed discussion is provided as SI Appendix.
Functionalization of Pcl Proteins (Table 1).
Reactions were typically carried out in 10× PBS at pH 7.4 and 22 °C using 10 μM Pcl protein and 100 μM 2-ABA/2-AAP reagent. Preparation of reagents, reaction details, and detection of the protein conjugates are given in SI Appendix: Figs. S8–S14.
Supplementary Material
Acknowledgments.
We thank Dr. Sarah Rue and Dr. Marc Nasoff for the clone of the IgG4 antibody modified in this study.
Footnotes
Conflict of interest statement: A patent application regarding the biosynthesis of pyrroline-carboxy-lysine and site-specific protein modifications via chemical derivatization of pyrroline-carboxy-lysine and pyrrolysine residues has been filed.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105197108/-/DCSupplemental.
References
- 1.Walsh G. Second-generation biopharmaceuticals. Eur J Pharm Biopharm. 2004;58:185–196. doi: 10.1016/j.ejpb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Sola RJ, Griebenow K. Glycosylation of therapeutic proteins an effective strategy to optimize efficacy. Biodrugs. 2010;24:9–21. doi: 10.2165/11530550-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beals JM, Shanafelt AB. Enhancing exposure of protein therapeutics. Drug Discov Today . 2006;3:87–94. doi: 10.1016/j.ddtec.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Sletten EM, Bertozzi CR. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew Chem Int Ed Engl. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Junutula JR, et al. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat Biotechnol. 2008;26:925–932. doi: 10.1038/nbt.1480. [DOI] [PubMed] [Google Scholar]
- 6.Hinner MJ, Johnsson K. How to obtain labeled proteins and what to do with them. Curr Opin Biotech. 2010;21:766–776. doi: 10.1016/j.copbio.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 8.Young TS, Schultz PG. Beyond the canonical 20 amino acids: expanding the genetic lexicon. J Biol Chem. 2010;285:11039–11044. doi: 10.1074/jbc.R109.091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 10.Wang L, Zhang Z, Brock A, Schultz PG. Addition of the keto functional group to the genetic code of Escherichia coli. Proc Natl Acad Sci USA. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin JW, et al. Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J Am Chem Soc. 2002;124:9026–9027. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 12.Deiters A, Schultz PG. In vivo incorporation of an alkyne into proteins in Escherichia coli. Bioorg Med Chem Lett. 2005;15:1521–1524. doi: 10.1016/j.bmcl.2004.12.065. [DOI] [PubMed] [Google Scholar]
- 13.Hao B, et al. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 14.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 15.Blight SK, et al. Direct charging of tRNA(CUA) with pyrrolysine in vitro and in vivo. Nature. 2004;431:333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- 16.Ambrogelly A, Palioura S, Soll D. Natural expansion of the genetic code. Nat Chem Biol. 2007;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 17.Bock A, et al. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991;5:515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Baranov PV, Atkins JF, Gladyshev VN. Pyrrolysine and selenocysteine use dissimilar decoding strategies. J Biol Chem. 2005;280:20740–20751. doi: 10.1074/jbc.M501458200. [DOI] [PubMed] [Google Scholar]
- 19.Longstaff DG, et al. A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine. Proc Natl Acad Sci USA. 2007;104:1021–1026. doi: 10.1073/pnas.0610294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cellitti SE, et al. D-ornithine coopts pyrrolysine biosynthesis to make and insert pyrroline-carboxy-lysine. Nat Chem Biol. 2011 doi: 10.1038/nchembio.586. epublished: April 27, www.nature.com/naturechemicalbiology. [DOI] [PubMed] [Google Scholar]
- 21.Gaston MA, Zhang L, Green-Church KB, Krzycki JA. The complete biosynthesis of the genetically encoded amino acid pyrrolysine from lysine. Nature. 2011;471:647–650. doi: 10.1038/nature09918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zinoni F, Heider J, Bock A. Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc Natl Acad Sci USA. 1990;87:4660–4664. doi: 10.1073/pnas.87.12.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allmang C, Wurth L, Krol A. The selenium to selenoprotein pathway in eukaryotes: more molecular partners than anticipated. Biochim Biophys Acta. 2009;1790:1415–1423. doi: 10.1016/j.bbagen.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Namy O, et al. Adding pyrrolysine to the Escherichia coli genetic code. FEBS Lett. 2007;581:5282–5288. doi: 10.1016/j.febslet.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 25.Longstaff DG, Blight SK, Zhang L, Green-Church KB, Krzycki JA. In vivo contextual requirements for UAG translation as pyrrolysine. Mol Microbiol. 2007;63:229–241. doi: 10.1111/j.1365-2958.2006.05500.x. [DOI] [PubMed] [Google Scholar]
- 26.Hao B, et al. Reactivity and chemical synthesis of L-pyrrolysine - the 22(nd) genetically encoded amino acid. Chem Biol. 2004;11:1317–1324. doi: 10.1016/j.chembiol.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 27.Vogel HJ, Davis BD. Glutamic gamma-semialdehyde and delta1-pyrroline-5-carboxylic acid, intermediates in the biosynthesis of proline. J Am Chem Soc. 1952;74:109–112. [Google Scholar]
- 28.Roberts MJ, Bentley MD, Harris JM. Chemistry for peptide and protein PEGylation. Adv Drug Deliv Rev. 2002;54:459–476. doi: 10.1016/s0169-409x(02)00022-4. [DOI] [PubMed] [Google Scholar]
- 29.Landsteiner K. The Specificity of Serological Reactions. 3rd Ed. Cambridge, MA: Harvard University Press; 1962. [Google Scholar]
- 30.Palm NW, Medzhitov R. Immunostimulatory activity of haptenated proteins. Proc Natl Acad Sci USA. 2009;106:4782–4787. doi: 10.1073/pnas.0809403105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Hagan DT, De Gregorio E. The path to a successful vaccine adjuvant—‘The long and winding road’. Drug Discov Today. 2009;14:541–551. doi: 10.1016/j.drudis.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Blander JM, Medzhitov R. Toll-dependent selection of microbial antigens for presentation by dendritic cells. Nature. 2006;440:808–812. doi: 10.1038/nature04596. [DOI] [PubMed] [Google Scholar]
- 33.Vollmer J, et al. Characterization of three CpG oligodeoxynucleotide classes with distinct immunostimulatory activities. Eur J Immunol. 2004;34:251–262. doi: 10.1002/eji.200324032. [DOI] [PubMed] [Google Scholar]
- 34.Alexander J, et al. Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity. 1994;1:751–761. doi: 10.1016/s1074-7613(94)80017-0. [DOI] [PubMed] [Google Scholar]
- 35.Mukai T, et al. Adding l-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem Biophys Res Commun. 2008;371:818–822. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
- 36.Polycarpo CR, et al. Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase. FEBS Lett. 2006;580:6695–6700. doi: 10.1016/j.febslet.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobayashi T, Yanagisawa T, Sakamoto K, Yokoyama S. Recognition of non-alpha-amino substrates by pyrrolysyl-tRNA synthetase. J Mol Biol. 2009;385:1352–1360. doi: 10.1016/j.jmb.2008.11.059. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen DP, et al. Genetic encoding and labeling of aliphatic azides and alkynes in recombinant proteins via a pyrrolysyl-tRNA Synthetase/tRNA(CUA) pair and click chemistry. J Am Chem Soc. 2009;131:8720–8721. doi: 10.1021/ja900553w. [DOI] [PubMed] [Google Scholar]
- 39.Yanagisawa T, et al. Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode N(epsilon)-(o-azidobenzyloxycarbonyl) lysine for site-specific protein modification. Chem Biol. 2008;15:1187–1197. doi: 10.1016/j.chembiol.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Fekner T, Li X, Lee MM, Chan MK. A pyrrolysine analogue for protein click chemistry. Angew Chem Int Ed Engl. 2009;48:1633–1635. doi: 10.1002/anie.200805420. [DOI] [PubMed] [Google Scholar]
- 41.Li X, Fekner T, Chan MK. N6-(2-(R)-propargylglycyl)lysine as a clickable pyrrolysine mimic. Chemistry – An Asian Journal. 2010;5:1765–1769. doi: 10.1002/asia.201000205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Fekner T, Ottesen JJ, Chan MK. A pyrrolysine analogue for site-specific protein ubiquitination. Angew Chem Int Ed Engl. 2009;48:9184–9187. doi: 10.1002/anie.200904472. [DOI] [PubMed] [Google Scholar]
- 43.Chen PR, et al. A facile system for encoding unnatural amino acids in mammalian cells. Angew Chem Int Ed Engl. 2009;48:4052–4055. doi: 10.1002/anie.200900683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding Ne-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen DP, Garcia Alai MM, Kapadnis PB, Neumann H, Chin JW. Genetically encoding N(epsilon)-methyl-L-lysine in recombinant histones. J Am Chem Soc. 2009;131:14194–14195. doi: 10.1021/ja906603s. [DOI] [PubMed] [Google Scholar]
- 46.Neumann H, et al. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rother M, Krzycki JA. Selenocysteine, pyrrolysine, and the unique energy metabolism of methanogenic archaea. Archaea. 2010 doi: 10.1155/2010/453642. doi: 10.1155/2010/453642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klock HE, Koesema EJ, Knuth MW, Lesley SA. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins. 2008;71:982–994. doi: 10.1002/prot.21786. [DOI] [PubMed] [Google Scholar]
- 49.Brown CM, Stockwell PA, Trotman CN, Tate WP. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res. 1990;18:6339–6345. doi: 10.1093/nar/18.21.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu W, Brock A, Chen S, Chen S, Schultz PG. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.