Abstract
ADP-ribosylation factor domain protein 1 (ARD1) is a 64-kDa protein containing a functional ADP-ribosylation factor (GTP hydrolase, GTPase), GTPase-activating protein, and E3 ubiquitin ligase domains. ARD1 activation by the guanine nucleotide-exchange factor cytohesin-1 was known. GTPase and E3 ligase activities of ARD1 suggest roles in protein transport and turnover. To explore this hypothesis, we used mouse embryo fibroblasts (MEFs) from ARD1-/- mice stably transfected with plasmids for inducible expression of wild-type ARD1 protein (KO-WT), or ARD1 protein with inactivating mutations in E3 ligase domain (KO-E3), or containing persistently active GTP-bound (KO-GTP), or inactive GDP-bound (KO-GDP) GTPase domains. Inhibition of proteasomal proteases in mifepristone-induced KO-WT, KO-GDP, or KO-GTP MEFs resulted in accumulation of these ARD1 proteins, whereas KO-E3 accumulated without inhibitors. All data were consistent with the conclusion that ARD1 regulates its own steady-state levels in cells by autoubiquitination. Based on reported growth factor receptor-cytohesin interactions, EGF receptor (EGFR) was investigated in induced MEFs. Amounts of cell-surface and total EGFR were higher in KO-GDP and lower in KO-GTP than in KO-WT MEFs, with levels in both mutants greater (p = 0.001) after proteasomal inhibition. Significant differences among MEF lines in content of TGF-β receptor III were similar to those in EGFR, albeit not as large. Differences in amounts of insulin receptor mirrored those in EGFR, but did not reach statistical significance. Overall, the capacity of ARD1 GTPase to cycle between active and inactive forms and its autoubiquitination both appear to be necessary for the appropriate turnover of EGFR and perhaps additional growth factor receptors.
Keywords: tyrosine receptors, ubiquitination
Vesicular trafficking, which is responsible for intracellular transport of proteins and/or lipids between and among organelles, is of major importance in regulating cell function and fate (1). Extracellular stimuli, either via direct ligand interaction with specific cell-surface receptors (e.g., growth factor) or in response to mechanical or environmental signals (e.g., pH, oxygen), also use vesicular trafficking to produce needed alterations in cellular metabolism, growth, and/or motion (2). For example, after ligand binding, plasma membrane receptors, such as the epidermal growth factor receptor (EGFR) are internalized in clathrin- or caveolin-coated vesicles (3) and can be recycled, after ligand dissociation, to the plasma membrane via multivesicular bodies. Receptor degradation after covalent modification (e.g., phosphorylation and/or ubiquitination) can occur through lysosomal or proteasomal pathways (4).
Vesicle formation is initiated by guanine nucleotide-exchange factor (GEF)-catalyzed activation of ADP-ribosylation factor (ARF) that results in its specific association with membrane sites where ARF–GTP recruits coat proteins and cargo, resulting in membrane deformation and initiation of budding. After movement of a completed vesicle to its target and tethering/docking, each transport step ends with membrane fusion (5). Biogenesis of several types of vesicles uses specific guanine nucleotide-binding GTPases as molecular switches for initiation and termination of diverse pathways that deliver molecules to appropriate intracellular sites specified by targeting sequences, posttranslational modifications, and/or associated molecules (6, 7). Mammalian ARFs (ca. 20 kDa) are grouped as class I (ARFs 1–3), class II (ARFs 4 and 5), and class III (ARF6), based on similarities of gene structure, protein sequences, and phylogenetic relationships (8). The ARFs cycle between active GTP- and inactive GDP-bound forms dependent on the actions of GEFs, which accelerate the replacement of ARF-bound GDP with GTP, and GTPase-activating proteins (GAPs) that enhance intrinsic ARF GTPase hydrolysis of GTP (9). Activated ARF–GTP can interact stably with specific effector molecules, such as vesicle coat protein subunits of COPI coatomer complex (10), and Golgi-associated, γ-adaptin ear-containing, ARF-binding adapter proteins of clathrin-coated vesicles (6).
ADP-ribosylation factor domain protein 1 (ARD1), an atypical member of the ARF family, is a ca. 64-kDa molecule with a C-terminal (amino acids 391–565) approximate 18-kDa ARF domain that is ca. 60% identical to class I ARFs (11). As ARF proteins lack detectable GTPase activity and require a GAP to terminate activation, the intrinsic GTPase activity of ARD1 was soon recognized (12). Direct functional interaction of recombinant 18-kDa ARF domain and ca. 46-kDa remainder (GAP domain) of ARD1 was also shown (12). The 46-kDa N-terminal region of ARD1 (1–390), termed “GAP domain,” although it includes sequences with additional diverse functions (Fig. 1A), can activate ARF-domain GTPase activity to generate ARD1–GDP (12, 13). Three structural motifs (RING, B box, and coiled-coil or RBCC) in this region gave ARD1 the designation TRIM23 in the tripartite motif (TRIM) family (14). RBCC sequences participate in protein–protein interactions, and the ARD1 RING exhibits E3 ubiquitin ligase activity demonstrated as apparent autoubiquitination in assays with recombinant ARD1 or fragments, E1, and an appropriate E2, plus ubiquitin and ATP (15). A recent report that the protein NF-κB essential modulator (NEMO) is a substrate for the E3 activity of ARD1 (16) did not relate that activity to its ARF domain.
Fig. 1.
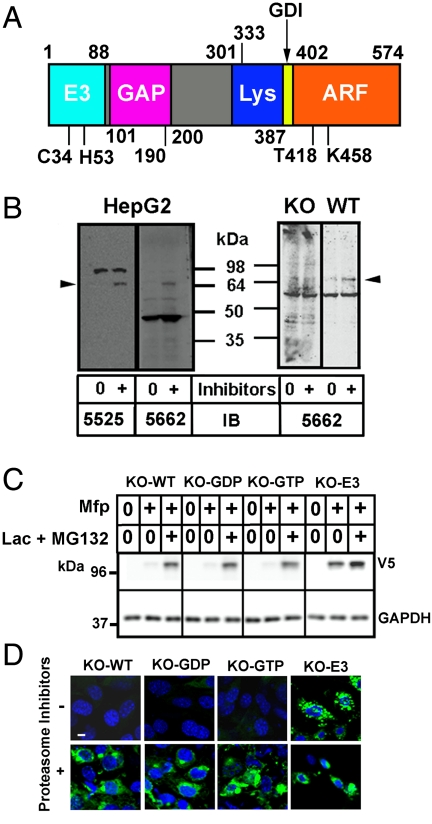
ARD1, a multifunctional protein. (A) Functional domains of 574 amino acid human ARD1 with C-terminal (402–574) ARF sequence. ARD1 contains an E3 ubiquitin ligase motif (positions 31–75) with critical C34 and H53, which were replaced with alanine in E3-inactive mutants. GAP function was assigned to amino acid 101–190, with amino acid 190–333 additionally required for domain interactions that result in GTP hydrolysis. A region required for lysosomal targeting (amino acid 301–402) includes amino acid 387–402 corresponding to the N-terminal amphipathic helix of an ARF molecule, which can act as a GDP-dissociation inhibitior (GDI). Mutations in the ARF region (402–574) that abolish GTP binding (T418N) or GTPase activity (K458I) are shown. (B) Effect of proteasome inhibitors on ARD1 content in HepG2 cells and MEFs. HepG2 cells were incubated 18 h without (0) or with (+) lactacystin and MG132, before preparation of lysates and Western blotting of proteins (40 µg) with ARD1 antibodies 5525 or 5662. MEFs lacking ARD1 (KO) or from wild-type mice (WT) were incubated ca. 18 h without or with proteasome inhibitors before Western blotting using ARD1 antibodies 5662. Arrowhead, endogenous ARD1 protein. (C) ARD1-/- MEF lines stably transfected with indicated GFP–ARD1-V5/His constructs (KO-WT, KO-GDP, KO-GTP, KO-E3) were incubated 18 h without (0) or with (+) Mfp or Mfp plus lactacystin and MG132 before Western blotting (40 µg proteins) with antibodies against V5 or GAPDH. Results were replicated in three experiments. (D) MEFs were incubated with Mfp without or with proteasome inhibitors (as in C) before fixation and preparation for inspection of EGFP–ARD1 by confocal immunofluorescence microscopy.
To explore the biological function(s) of ARD1, we prepared from ARD1-null mice mouse embryo fibroblast (MEF) lines stably transfected with plasmids for mifepristone (Mfp)-inducible expression of ARD1, wild type, or with specific mutations that prevent usual function of the ARF or E3 ligase domains. Here, we report evidence that continuous function of the ARF-domain GTPase cycling is likely required for degradation of endogenous EGF and TGF-β III; receptors in these MEFs. EGFR degradation, although involving proteasomes, was seemingly independent of ARD1 E3 ubiquitin ligase activity that catalyzes its automodification.
Results and Discussion
Endogenous ARD1 Is Unstable in Cells.
For a decade, virtually all of our numerous attempts to detect ARD1 in many cells had failed consistently. To identify an ARD1 function, we had transiently transfected several cell lines with plasmids encoding wild-type ARD1 and mutants (see Fig. 1A) of the ARF domain that were persistently inactive or active. Cells constitutively overexpressing ARD1 protein failed to proliferate, which seems likely, in retrospect, to have been a consequence (at least in part) of elevated levels of its E3 ligase activity. Endogenous ARD1 was finally detected by two different antibodies in HepG2 cells grown overnight with, but not without, proteasome inhibitors (Fig. 1B, arrowhead). This behavior mirrors reports of other RING-type E3 ubiquitin ligases capable of autoubiquitination that causes their instability in cells (17, 18) and detection only after inhibition of proteasomal proteases. Results were similar with fibroblasts grown from WT MEFs, although KO MEFs always, unfortunately, showed extensive nonspecific staining with our antipeptide antibodies (Fig. 1B).
Our ARD1 KO mouse (C57/BL6) was initially generated for alternative exploration of the biological role of ARD1. Mice lacking the ARD1 gene appeared grossly normal and thus far present no obvious pathological phenotype or histopathology of major organs including brain, spleen, heart, kidneys, lung, and axial lymph nodes, or problems with breeding. Fatty liver without identified cause appeared in some older KO mice. ARD1-null MEF lines that inducibly express WT or mutant ARD1 proteins under control of the CMV promoter were, therefore, prepared.
Induced Expression of ARD1 in KO MEFs.
RT-PCR confirmed apparent absence of ARD1 mRNA in KO MEFs (Fig. S1). We used ARD1-/- MEFs for Mfp-inducible expression of murine WT ARD1 or its mutant forms, which do not cycle between GTP- and GDP-bound forms. Mutation T418N interferes with GTP binding, locking ARD1 in its inactive GDP-bound form, whereas K458I abolishes GTPase activity, producing persistently active ARD1–GTP (19). The Mfp-induced double E3 mutant with alanine replacing C34 and H54, amino acids critical for E3 ubiquitin ligase activity, was also generated (15). We report data from eight stably transfected MEF lines referred to, respectively, as KO-WT, KO-GDP, and KO-GTP (two lines of each), and one line each of KO-E3 and KO-GFP (empty vector). No ARD1 protein was detected in any of these MEFs before induction (Fig. 1C), but after 18 h with 10 μM Mfp, Western blot analysis revealed ARD1 in all cells (except KO-GFP) with a KO-E3 level sevenfold that of KO-WT and the ARF-domain mutants (Fig. 1C and Fig. S1). Proteasome inhibition during Mfp induction increased amounts of overexpressed proteins dramatically in all lines except KO-E3, which was increased ≤ 100% (Fig. 1C). Despite the large difference in ARD1 protein levels between KO-E3 and the other MEF lines, ARD1 mRNA levels did not differ among these cell lines (except for KO-GFP, in which no ARD1 mRNA was detected) (Fig. S1).
Intracellular Localization of ARD1.
Consistent with results of Western blotting after induction without proteasome inhibition, ARD1 (GFP-tagged) was barely visible by fluorescence microscopy (Fig. 1D) in any MEFs except KO-E3, where it was clearly more abundant in multiple scattered puncta. With proteasome inhibition, however, GFP–ARD1 was seen in punctate areas throughout cytoplasm of all MEFs with discrete concentrations of KO-WT and KO-GDP proteins in a perinuclear region.
Because ARD1 has E3 ligase activity and had been associated with lysosomes (13, 20), localization of different recombinant ARD1 proteins with that of LysoTracker was assessed. WT and GDP- or GTP-bound ARD1 seemed to colocalize with LysoTracker, but the E3 ligase mutant did not coincide with either LysoTracker (Fig. S2) or lysosomal membrane protein LAMP1 (Fig. S3). E3 ligase mutant ARD1 in cell fractions separated by centrifugation appeared to be largely soluble (Fig. S3), although a significant portion was pelleted after centrifugation for 18 h at 100,000 × g. These data are consistent with the notion that E3 mutant ARD1 protein was associated with intracellular structures resembling exosomes (i.e., sedimented only after prolonged high-speed centrifugation), although a considerable amount of the overexpressed protein in MEFs did remain “soluble.” Analysis of protein composition of the “exosome” fraction is clearly important.
When ARD1 E3 ubiquitin ligase activity was reported (15), its autoubiquitination was suggested, despite failure of the multiubiquitinated protein to react with ARD1 antibodies. Evidence here of ARD1 accumulation in cells treated with proteasome inhibitors (Figs. 1 C and D) is consistent with autoubiquitination of intracellular ARD1 and seems to parallel descriptions of MARCH7 (21) and MARCH5 9 (17) as E3 ligases requiring enzymatic deubiquitination for stabilization or detection. Thus, it appears that ARD1 self-regulates its degradation, presumably subject to additional molecular interactions that also influence ARD1 stability and thereby biological function.
ARD1 Effects on EGFR mRNA and Protein.
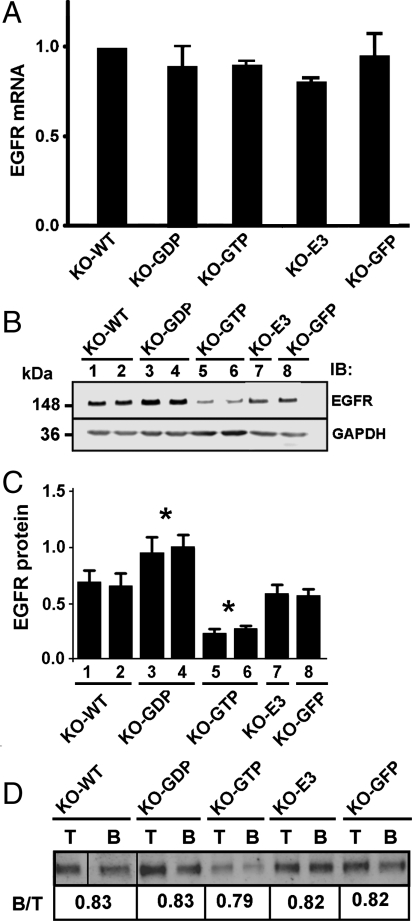
Our studies of ARD1 molecular interactions had revealed its association with the GEF cytohesin-1 (19), which activated its ARF domain. As this cytohesin and others (cytoheins-2, -3) have been implicated in modulating growth factor receptor function (22–24), in addition to activating ARFs, we explored a relationship of ARD1 to kinase growth factor receptors (R), first EGFR. Levels of EGFR mRNA (Fig. 2A) were similar in KO-WT, KO-GDP, KO-GTP, and KO-E3 MEFs. There were, however, clear differences among MEF lines (Fig. 2 B and C) in amounts of EGFR protein. EGFR levels in KO-GDP were > 60% higher, and in KO-GTP ca. 50% lower than those in KO-WT (Fig. 2C). Overall, lack of significant differences in amounts of mRNA perhaps suggests rapid turnover of EGFR protein that enables precise regulation, temporal and spatial, of EGF action. EGFR protein content seemed most related to the activation state of the ARD1 ARF domain, whereas absence of ARD1 E3 ligase activity had no effect. The E3 ligase domain appears responsible for autoubiquitination and, likely, regulation of ARD1 degradation, as well as accumulation of the E3 ligase mutant and other ARD1 proteins after proteasome inhibition. It is tempting to speculate that the ARF domain of ARD1 acts similarly to the class I ARFs in vesicular transport, although we do not know how ARD1 interaction with membranes is regulated.
Fig. 2.
EGFR mRNA and protein in ARD1 KO MEFs overexpressing WT or mutant ARD1 proteins. (A) EGFR mRNAs in stably transfected MEFs induced with Mfp were quantified by real-time PCR. RNA values were normalized to α-tubulin (also used for protein normalization in experiments below). Data were analyzed using Sequence Detector version 1.6.3 program. Data are averages ± 1/2 range for each cell line in two independent experiments. No significant differences were detected. (B) Samples (40 μg) of lysate proteins from eight MEF lines induced with Mfp were analyzed by Western blotting with EGFR and GAPDH antibodies. (C) For EGFR, each densitometric value was expressed relative to that of KO-GFP cells in the same experiment equal to 100. Data are means ± SEM of values for each line in three experiments. *p < 0.001 for difference between each of lanes 3 and 4 (KO-GDP) or 5 and 6 (KO-GTP) from KO-GFP. (D) Biotinylated proteins from MEF lines induced with Mfp, were separated for quantification of immunoreactive EGFR in cell surface (B) and in total lysate (T) samples (2.5% of each). Ca. 80% of EGFR was biotinylated in each cell line regardless of differences in total amounts.
To determine whether amounts of EGFR at the plasma membrane paralleled total cell content, we quantified biotinylated cell-surface EGFR. Even with large differences in total EGFR content of cells overexpressing different ARD1 proteins, amounts at the cell surface were 79–83% of the total in all MEFs (Fig. 2D), consistent with a functional relationship between GTPase cycling of ARD1 and EGFR cycling via cell surface to degradation after endocytosis. Fruitless interaction of endogenous GEF (cytohesin-1) with overexpressed ARD1–GDP that could not be activated would interfere with cytohesin-1 activation of endogenous ARD1 (presumably absent in these KO cells) and additional ARF substrates. Overexpression of ARD1–GTP may have permitted more EGFR degradation than did similar amounts (Fig. 1C) of other ARD1 mutants, although its capacity to support EGFR transport is presumably limited to a single turnover by the inability of this ARD1 to hydrolyze GTP. EGFR activation and subsequent degradation requires ligand binding and probably homodimerization (25–27), although the existence of active dimers even in the absence of ligand has been proposed (28), so that concentrated EGFR, for example, in plasma membranes of KO-GDP MEFs could become activated independent of EGF. A role for ARD1 in regulation of EGFR transport begins to emerge from exploring molecular interactions of the overexpressed proteins, although caution in extrapolation from such observations to the behavior of endogenous molecules is always required.
Effects of Proteasome and Lysosome Inhibitors on Degradation of EGFR.
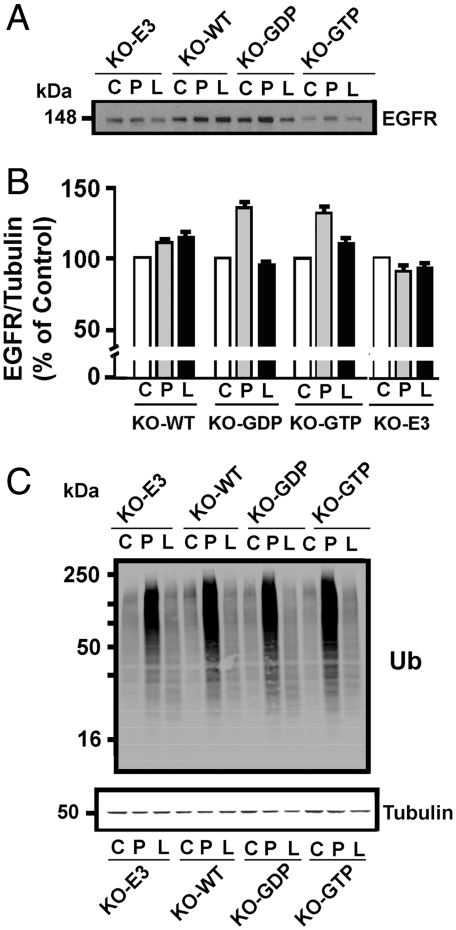
As EGFR is subject to both proteasomal and lysosomal degradation (2), we looked for accumulation of EGFR in MEFs treated with proteasome (lactacystin and MG132) or lysosome (chloroquine) inhibitors (Fig. 3A). EGFR content was clearly greater after incubation of KO-GDP or KO-GTP cells with proteasome inhibitors (Fig. 3B), but levels of EGFR in KO-WT or KO-E3 cells were not appreciably affected and chloroquine lacked significant effects. Multiplicity of ubiquitinated proteins accumulated in all MEF lines (Fig. 3C) with proteasomal degradation inhibited obscures detection of differences in relatively small contributions from EGFR or other specific protein(s). Most of the EGFR appeared to be targeted for proteasomal degradation, possibly with a role for ARD1 GTPase action in its delivery to proteasomes. All observations are consistent with ARD1 participation in EGFR degradation.
Fig. 3.
Effects of proteasome and lysosome inhibitors on EGFR content of MEFs expressing ARD1 proteins. (A) After induction with Mfp without (C) or with proteasome (P) or lysosome (L) protease inhibitors, the indicated ARD1 KO MEFs were analyzed by Western blotting with antibodies against EGFR. (B) Densitometric values for EGFR in P and L samples expressed relative to that of α-tubulin in the same sample were normalized to the value for the same cells (C) without inhibitors equal to 100%. Data are from three experiments. Only after Mfp induction of ARD1–GDP and ARD1–GTP mutants with proteasome inhibitors were ARD1 levels significantly (p < 0.05) different from (higher than) those of the same cells without inhibitors. (C) Immunoblot from a representative experiment with antibodies against ubiquitin (Ub) and α-tubulin.
Effects on Insulin and TGF-β Receptors.
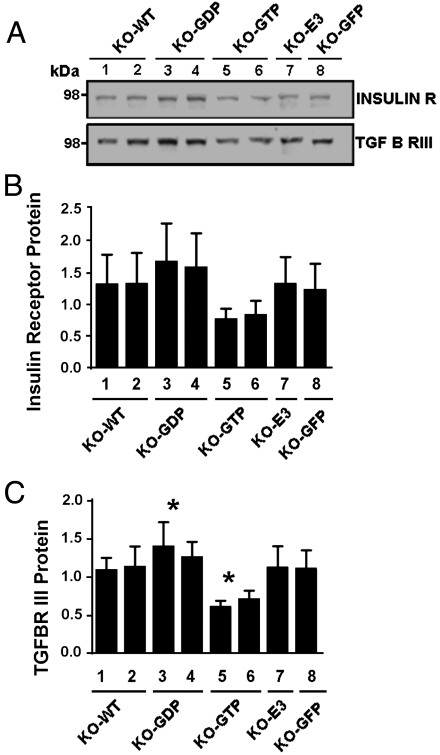
Our EGFR data prompted us to also evaluate effects of ARD1 action on levels of endogenous insulin receptor (IR) and TGF-βR in MEF lines. Findings were similar to those for EGFR, without differences among KO-GFP (empty vector), KO-WT, and KO-E3, but with significantly more TGF-βR in KO-GDP and less in KO-GTP MEFs than in KO-GFP (Fig. 4C). Although the same trend was apparent for the IR (Fig. 4B), the differences were not statistically significant. Each of these kinase receptors is activated and internalized after agonist binding, but pathways of their endocytosis and signaling from endosomes thereafter are not identical (29, 30). Some of the numerous differences in endosomal signaling mechanisms among tyrosine kinase, G-protein-coupled, and toll-like receptors were reviewed by Murphy et al. (29). Differences in usage of signaling pathways also exist among growth factor receptors, which we had thought potentially similar enough to allow identification of shared ARD1 actions. Understanding of such ARD1 function(s), however, may await elucidation of specific molecular interactions of each of those receptors. TGF-βR III, a TGF-β coreceptor also degraded in proteasomes (31), is internalized along with TGF-βR I and II (31–33). Our data are consonant with the notion that ARD1 GTPase could have a role in regulating internalization of and signaling by dimeric or monomeric growth factor receptors. An action of ARD1 in the regulation of growth factor receptors such as those of EGF, TGF-β, and perhaps insulin, seems clearly consistent with our findings, and the effects described here were demonstrably dependent on continued cyclic activation of its ARF domain.
Fig. 4.
Insulin and TGF-β receptor proteins in ARD1–KO MEFs overexpressing WT or mutant ARD1. (A) Samples (40 μg) of proteins from eight MEF lines containing an indicated ARD1 or empty vector (KO-GFP) construct and induced with Mfp were analyzed by Western blotting with antibodies against insulin or TGF-β receptor. Densitometric values in each experiment were expressed relative to that of the same protein in KO-GFP cells equal to 100. Data are means ± SEM of values from three experiments. The amounts of insulin R data (B) did not reach significance. * p < 0.01 and p < 0.05 for differences, respectively, between amounts of TGF-βR (C) in KO-GDP and KO-GTP MEFs.
It may be relevant that cytohesin-1, the only known ARD1-GEF (19, 34), was translocated to plasma membrane of PC-12 cells (derived from rat adrenal pheochromocytoma) in response to EGF and nerve growth factor (22, 24). Perhaps translocation of cytohesin-1 to the plasma membrane in response to specific stimuli is associated with activation of ARD1, which was influenced by specific phospholipids (34). In addition, ARAP1, an ARF GTPase-activating protein, regulates EGFR endocytosis, trafficking, and signaling (35, 36), although the ARD1 molecule does contain its own GAP domain. All data are consistent with the participation of ARFs, including ARD1, in trafficking and function of plasma-membrane-associated receptors. In addition, the finding that ARD1 participates in antiviral defense by ubiquitinating NEMO suggests that ARD1 has more than one function related to innate immunity (16).
Materials and Methods
Cell Culture.
HepG2 cells were grown at 37 °C (5% CO2/95% humidity) in DMEM high glucose (4.5 g/L D-glucose, Invitrogen) with 50 units/mL penicillin, 50 mg/mL streptomycin, and 10% FBS (Hyclone). MEF1 medium contained DMEM high glucose, with 10% mesenchymal growth supplement (MCGS, Lonza) replacing FBS, penicillin, and streptomycin. MEFs stably transfected with ARD1 plasmids were grown in MEF2 [MEF1 plus Zeocin, 0.8 mg/mL (Invitrogen) and hygromycin B, 0.2 mg/mL, (Invitrogen)].
Generation and Growth of MEFs.
The animal studies were performed according to the animal protocol approved by the National Institutes of Health, National Heart, Lung, and Blood Institute animal user and care committee. Day 14 embryos of WT or ARD1-/- (KO) C57BL6 mice were euthanized with CO2 and washed in sterile phosphate-buffered saline. Skin was removed, minced, incubated (30 min, 37 °C) in 2 mL of trypsin/EDTA (Lonza), washed in PBS, and incubated for 2 wk in MEF1 medium (20 mL). Adherent MEFs were split one-fifth at 80% of confluence for continued growth in the same medium and studies at passages 10–20.
Genomic DNA from MEFs (Wizard Genomic DNA Purification Kit, Promega) was template for PCR with primers F1-G and R1-G to generate an ca. 2.4-kb WT ARD1 allele fragment or F1-G and R-NEO for an 2.1-kb fragment from the neomycin-disrupted allele (Table S1). DNA was amplified using the Advantage 2 PCR System (BD Clontech) for 30 cycles of 40 s, 95 °C/45 s, 60 °C/4 min, 68 °C, followed by 10 min, 68 °C. PCR products were separated in 1% agarose gel containing ethidium bromide and viewed by ultraviolet light.
Preparation of Murine ARD1 cDNA, Inducible Plasmids, and Stably Transfected MEFs.
WT mouse ARD1 cDNA, reverse transcribed from total RNA isolated (RNeasy Mini Kit, Qiagen) from a 100-mm dish of 80–90% confluent of WT MEF, was amplified using primers F-RT and R-RT (Table S1). Purified mARD1 cDNA (QIAquick Gel Extraction Kit, Qiagen) was cloned into TOPO vector (Invitrogen) for EcoRI excision of an ca. 1.6-kb fragment that was purified and ligated in frame to pGENE-V5/His-C vector (Invitrogen) already cut with EcoRI yielding pGENE-ARD1-V5/His encoding EGFP in pGENE-ARD1-V5/His by PCR (primers, Table S1). The plasmid pGENE-EGFP–ARD1-V5/His was used to generate ARD1 KO MEFs expressing ARD1–WT protein. Mutations were made in this plasmid using a QuickChange Site-Directed Mutagenesis Kit (Stratagene; primers, Table S1) to generate ARD1–E3 encoding C34A and H53A, replacements that abolish E3 ligase activity. ARD1–GTP with ARF-domain mutation K458I is persistently active and ARD1–GDP with a T418N replacement is inactive. Excision of ARD1 from pGENE-EGFP–ARD1-V5/His produced the control empty vector. Complete coding regions (EGFP through His tag) of all plasmids were sequenced (sequencing primers Table S1) using ABIPIRM377 DNA sequencer (Perkin Elmer).
ARD1 KO MEFs were cotransfected with pSwitch regulatory plasmid and a pGene-GFP-ARD1-V5/His plasmid (37). Each plasmid (8 μg) in 800 μL of Opti-MEM (Invitrogen) was incubated (15 min, room temperature) with 30 μL of Plus reagent before addition of Lipofectamine (32 μL, Invitrogen); 15 min later, the mixture was added to 70–80% confluent ARD1 KO MEFs (100-mm dishes) previously washed with PBS and incubated 1 h with 3–5 mL of Opti-MEM. After 4 h with cells, medium was replaced with fresh MEF1. MEFs were transfected a second time 24 h later and, after 24 h, cells were split 1∶4 for growth in MEF2. Stably transfected MEF lines were screened for expression of expected proteins by incubation of cells for 4–12 h with 10 μM Mfp before analysis of mRNA and protein by RT-PCR and Western blotting, respectively. After 3–4 wk, six MEF lines (from 24 initiated) for each mARD1 construct were selected for further study.
Centrifugal Fractionation of Cells.
Each cell line was plated on 10 150-mm dishes and grown to 70–80% confluency. Mfp (final concentration 10 μM) was added to each dish 24 h before cells were rinsed twice with 7 mL of ice-cold PBS and harvested by scraping in 5 mL of ice-cold PBS. Unless otherwise stated, all subsequent procedures were carried out at 4 °C. Cells were pelleted, dispersed, and homogenized (Dounce tissue grinder) in 10 mL of ice-cold 20 mM Tris•HCl buffer containing 1 mM NaN3, 250 mM sucrose, 0.5 mM EDTA, 1 mM DTT, protease inhibitors (BD Biosciences, catalog no. 554779), and phosphatase inhibitors (Sigma, P5726). Cell lysate was sonicated and fractionated by sequential centrifugation of each supernatant (after the initial 15 min at 1,000 × g to yield 1S) 3,000 × g, 15 min (3S); 20,000 × g, 20 min (20S); 100,000 × g, 60 min (100S); and 100,000 × g, 18 h (100 × S). Samples (0.1% of total lysate, supernatant, and pellet) were analyzed by Western blotting with antibodies against V5 (ARD1), plus markers for nuclei (histone H4), Golgi (GM130), lysosomes (LAMP1), and cytosol (GAPDH).
Western Blotting and Quantification of Proteins.
Proteins (40–60 μg), separated by SDS-PAGE in 4–20% gels, were transferred to nitrocellulose membranes that were incubated (1 h, room temperature) in Tris-buffered saline containing 5% nonfat dry milk (BioRad) and 0.1% Tween 20, then overnight at 4 °C with antibodies against EGFR, IR or TGF-βR III (rabbit, 1∶1,000 dilution, Cell Signalling), nucleoporin and GM130 (mouse monoclonal, 1∶1,000; BD Bioscience), V5 (mouse monoclonal, 1∶1,000; Invitrogen), GAPDH, and α-tubulin (mouse monoclonal, 1∶5,000; Sigma). ARD1 antibodies (21st Century Biochemicals, Inc.), affinity-purified Ab no. 5525 from rabbits immunized with full-length ARD1 protein or Ab no. 5562 against ARD1 (301–319, RQEEMALSVVDAHVREKLC), were diluted 1∶1,000. Horseradish peroxidase-linked secondary antibodies (Promega) were detected using Supersignal West Pico (or Femto) Chemiluminescent Substrate Kits (Pierce). Images captured using a LAS4000 system were quantified with ImageGauge 4.0 (Fujifilm).
Fluorescence Microscopy.
MEFs (3.5 × 104) in 0.5 mL of MEF2 in four-well LabTeK II chamber slides were allowed to adhere overnight (15 h) before incubation (18 h) with 10 μM Mfp in MEF2 without or with MG132 and lactacystin, 50 μM each. After washing with PBS containing CaCl2 and MgCl2 (1 mM each), cells were fixed (20 min) with 4% paraformaldehyde (Electron Microscopy, Inc.), washed with PBS, covered with Vectashield (Vector Laboratories) containing DAPI, and sealed with coverslips or processed for immunostaining as described (19). To visualize lysosomes (acidic interior), cells were incubated with the fluorophore 50 nM LysoTracker for 15 min at 37 °C. Images were collected using a Zeiss LSM 510 laser scanning confocal microscope.
Biotinylation of Cell-Surface Proteins.
A Cell-Surface Protein Isolation Kit (Pierce) was used for biotinylation of proteins according to the manufacturer’s protocol. Briefly, 5.2 × 106 MEFs were grown overnight (ca. 15 h) in MEF2, incubated 18 h in MEF2 with 10 μM Mfp, rinsed three times (20 mL each) in ice-cold PBS, and incubated with EZ-link Sulfo-NHS-SS-Biotin (0.24 mg/mL, 1 h) before adding quenching solution (500 μL/plate). Samples (1/40) of proteins were separated in 4–20% Tris-glycine gels and transferred to nitrocellulose for densitometric quantification.
EGF and EGFR in MEFs.
MEFs (2 × 105/100-mm dish) were incubated overnight (ca. 15 h) in MEF2, then 18 h in serum-free MEF2 (minus MCGS) containing 10 μM Mfp. Medium was replaced with serum-free MEF2 containing purified murine EGF (BD Bioscience), 100 ng/mL, for the indicated time before washing cells with 10 mL of ice-cold PBS and collection by scraping in ice-cold M-PER™ (100 μL, Pierce) containing protease (Roche) and phosphatase (Sigma) inhibitors. After 5 min on ice, samples were sonified (20 s), immediately frozen on dry ice, and stored at -80 °C.
Inhibition of Intracellular Protein Degradation.
MEFs (2 × 105) were grown overnight in 100-mm plates in MEF2 before incubation (18 h, 37 °C) with 10 μM Mfp without or with proteasome inhibitors (lactacystin and MG132, each 50 μM, Sigma) or 100 μM chloroquine (Sigma) to inhibit lysosomal proteases. MEFs harvested by scraping in ice-cold M-PER (Pierce) containing protease and phosphatase inhibitors (100 μL per plate) were sonified (20 s) to break cells. Cell lysates were analyzed by Western blotting and densitometric quantification.
Isolation of RNA and Quantitative PCR.
Total RNA (5 μg), separated from lysates using a total RNeasy isolation kit (Qiagen), was reverse transcribed using a Transcription III Superscript Kit (Invitrogen). For specific PCR amplification, TaqMan Universal PCR master mix and gene-specific TaqMan probes (Applied Biosystems) were used. Data were analyzed using the Sequence Detector version 1.6.3. In most experiments, α-tubulin was an internal control. To evaluate differences among MEF lines, cycle thresholds (Ct) for ARD1and EGFR from each were expressed relative to that for α-tubulin in the same sample. Data for each line were then expressed relative to that for control MEFs (usually “KO-WT”) in the same experiment.
Statistical Analysis.
Densitometer readings (raw data) were normalized to the GAPDH readings (internal control). Each EGFR, IR, or transforming growth factor receptor III reading was divided by the GAPDH value for that gel lane (e.g., EGFR-1/GAPDH-1 or EGFR-2/GAPDH-2, etc.). Experiments were repeated three times. Data are reported as means ± SD of values from the indicated number of experiments. (In later experiments, α-tubulin replaced GAPDH as “sample control” after we recognized GAPDH variations and employed especially conservative interpretation of all receptor values.) ANOVA was used to detect overall differences among group means. If an overall difference was detected (p < 0.05), we proceeded to all pairwise group comparisons using the Tukey-Kramer post hoc test with significance at p ≤ 0.05. SAS 9.2 (SAS Institute, Inc.) and GraphPad Prism version 4.00 for Windows (GraphPad Software) were used for statistical analyses.
Supplementary Material
Acknowledgments.
We thank Drs. Christian Combs and Daniela Malide from the light microscopy core facility National Institutes of Health, National Heart, Lung, and Blood Institute (NHLBI). The research was supported by the Intramural Research Program of the National Institutes of Health, NHLBI.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103867108/-/DCSupplemental.
References
- 1.Rothman JE, Wieland FT. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 2.Eden ER, White IJ, Futter CE. Down-regulation of epidermal growth factor receptor signalling within multivesicular bodies. Biochem Soc Trans. 2009;37:173–177. doi: 10.1042/BST0370173. [DOI] [PubMed] [Google Scholar]
- 3.Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2009;315:683–696. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 4.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spang A. The life cycle of a transport vesicle. Cell Mol Life Sci. 2008;65:2781–2789. doi: 10.1007/s00018-008-8349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 7.Pfeffer SR. Unsolved mysteries in membrane traffic. Annu Rev Biochem. 2007;76:629–645. doi: 10.1146/annurev.biochem.76.061705.130002. [DOI] [PubMed] [Google Scholar]
- 8.Tsuchiya M, Price SR, Tsai SC, Moss J, Vaughan M. Molecular identification of ADP-ribosylation factor mRNAs and their expression in mammalian cells. J Biol Chem. 1991;266:2772–2777. [PubMed] [Google Scholar]
- 9.Moss J, Vaughan M. Molecules in the ARF orbit. J Biol Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- 10.Kirchhausen T. Three ways to make a vesicle. Nat Rev Mol Cell Biol. 2000;1:187–198. doi: 10.1038/35043117. [DOI] [PubMed] [Google Scholar]
- 11.Mishima K, Tsuchiya M, Nightingale MS, Moss J, Vaughan M. ARD 1, a 64-kDa guanine nucleotide-binding protein with a carboxyl-terminal ADP-ribosylation factor domain. J Biol Chem. 1993;268:8801–8807. [PubMed] [Google Scholar]
- 12.Vitale N, Moss J, Vaughan M. ARD1, a 64-kDa bifunctional protein containing an 18-kDa GTP-binding ADP-ribosylation factor domain and a 46-kDa GTPase-activating domain. Proc Natl Acad Sci USA. 1996;93:1941–1944. doi: 10.1073/pnas.93.5.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vitale N, Ferrans VJ, Moss J, Vaughan M. Identification of lysosomal and Golgi localization signals in GAP and ARF domains of ARF domain protein 1. Mol Cell Biol. 2000;20:7342–7352. doi: 10.1128/mcb.20.19.7342-7352.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reymond A, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vichi A, Payne DM, Pacheco-Rodriguez G, Moss J, Vaughan M. E3 ubiquitin ligase activity of the trifunctional ARD1 (ADP-ribosylation factor domain protein 1) Proc Natl Acad Sci USA. 2005;102:1945–1950. doi: 10.1073/pnas.0409800102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arimoto K, et al. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proc Natl Acad Sci USA. 2010;107:15856–15861. doi: 10.1073/pnas.1004621107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lerner M, et al. The RBCC gene RFP2 (Leu5) encodes a novel transmembrane E3 ubiquitin ligase involved in ERAD. Mol Biol Cell. 2007;18:1670–1682. doi: 10.1091/mbc.E06-03-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vitale N, et al. Specific functional interaction of human cytohesin-1 and ADP-ribosylation factor domain protein (ARD1) J Biol Chem. 2000;275:21331–21339. doi: 10.1074/jbc.M909642199. [DOI] [PubMed] [Google Scholar]
- 20.Vitale N, Horiba K, Ferrans VJ, Moss J, Vaughan M. Localization of ADP-ribosylation factor domain protein 1 (ARD1) in lysosomes and Golgi apparatus. Proc Natl Acad Sci USA. 1998;95:8613–8618. doi: 10.1073/pnas.95.15.8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nathan JA, et al. The ubiquitin E3 ligase MARCH7 is differentially regulated by the deubiquitylating enzymes USP7 and USP9X. Traffic. 2008;9:1130–1145. doi: 10.1111/j.1600-0854.2008.00747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bill A, et al. Cytohesins are cytoplasmic ErbB receptor activators. Cell. 2010;143:201–211. doi: 10.1016/j.cell.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 23.Hafner M, et al. Inhibition of cytohesins by SecinH3 leads to hepatic insulin resistance. Nature. 2006;444:941–944. doi: 10.1038/nature05415. [DOI] [PubMed] [Google Scholar]
- 24.Venkateswarlu K, Gunn-Moore F, Tavare JM, Cullen PJ. EGF-and NGF-stimulated translocation of cytohesin-1 to the plasma membrane of PC12 cells requires PI 3-kinase activation and a functional cytohesin-1 PH domain. J Cell Sci. 1999;112:1957–1965. doi: 10.1242/jcs.112.12.1957. [DOI] [PubMed] [Google Scholar]
- 25.Chung I, et al. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464:783–787. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- 26.Alvarado D, Klein DE, Lemmon MA. Structural basis for negative cooperativity in growth factor binding to an EGF receptor. Cell. 2010;142:568–579. doi: 10.1016/j.cell.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leahy DJ. The ins and outs of EGFR asymmetry. Cell. 2010;142:513–515. doi: 10.1016/j.cell.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 29.Murphy JE, Padilla BE, Hasdemir B, Cottrell GS, Bunnett NW. Endosomes: A legitimate platform for the signaling train. Proc Natl Acad Sci USA. 2009;106:17615–17622. doi: 10.1073/pnas.0906541106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 31.Blobe GC, Liu X, Fang SJ, How T, Lodish HF. A novel mechanism for regulating transforming growth factor beta (TGF-beta) signaling. Functional modulation of type III TGF-beta receptor expression through interaction with the PDZ domain protein, GIPC. J Biol Chem. 2001;276:39608–39617. doi: 10.1074/jbc.M106831200. [DOI] [PubMed] [Google Scholar]
- 32.Blobe GC, et al. Functional roles for the cytoplasmic domain of the type III transforming growth factor beta receptor in regulating transforming growth factor beta signaling. J Biol Chem. 2001;276:24627–24637. doi: 10.1074/jbc.M100188200. [DOI] [PubMed] [Google Scholar]
- 33.Chen W, et al. Beta-arrestin 2 mediates endocytosis of type III TGF-beta receptor and down-regulation of its signaling. Science. 2003;301:1394–1397. doi: 10.1126/science.1083195. [DOI] [PubMed] [Google Scholar]
- 34.Pacheco-Rodriguez G, Meacci E, Vitale N, Moss J, Vaughan M. Guanine nucleotide exchange on ADP-ribosylation factors catalyzed by cytohesin-1 and its Sec7 domain. J Biol Chem. 1998;273:26543–26548. doi: 10.1074/jbc.273.41.26543. [DOI] [PubMed] [Google Scholar]
- 35.Daniele T, Di Tullio G, Santoro M, Turacchio G, De Matteis MA. ARAP1 regulates EGF receptor trafficking and signalling. Traffic. 2008;9:2221–2235. doi: 10.1111/j.1600-0854.2008.00823.x. [DOI] [PubMed] [Google Scholar]
- 36.Yoon HY, Lee JS, Randazzo PA. ARAP1 regulates endocytosis of EGFR. Traffic. 2008;9:2236–2252. doi: 10.1111/j.1600-0854.2008.00839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kato J, Zhu J, Liu C, Moss J. Enhanced sensitivity to cholera toxin in ADP-ribosylarginine hydrolase-deficient mice. Mol Cell Biol. 2007;27:5534–5543. doi: 10.1128/MCB.00302-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.