Abstract
MicroRNAs (miRNAs) play a critical role in determining the differentiation fate of pluripotent stem cells and germ cells in mammals. However, the mechanism(s) of miRNA-mediated posttranscriptional regulation with regard to lineage specification and differentiation in chick development require further investigation. Therefore, we conducted miRNA expression profiling to explore specific miRNA signatures in undifferentiated blastoderm and primordial germ cells (PGCs). We identified seven miRNAs that are highly expressed in blastoderm and 10 that are highly expressed in PGCs. In this study, miR-302a and miR-456 for blastoderm and miR-181a* for PGCs were analyzed further for their target transcripts and regulatory pathways. Both miR-302a and miR-456 bound directly to the sex-determining region Y box 11 transcript and could act as posttranscriptional coregulators to maintain the undifferentiated state of the chicken blastoderm through the suppression of somatic gene expression and differentiation. Moreover, miR-181a* showed a bifunctional role in PGCs by binding to two different transcripts. miR-181a* inhibited the somatic differentiation of PGCs by silencing homeobox A1 expression. Additionally, miR-181a* prevented PGCs from entering meiosis through the repression of the nuclear receptor subfamily 6, group A, member 1 transcript. Collectively, our data demonstrate that in chickens miRNAs intrinsically regulate the differentiation fate of blastoderms and PGCs and that the specific timing of germ cell meiosis is controlled through miRNA expression.
Keywords: aves, germline, sex differentiation
At stage X, the chicken blastoderm consists of 40,000–60,000 undifferentiated embryonic cells and is able to develop pluripotent stem cells through in vitro culture (1). During chicken germline development, primordial germ cells (PGCs) first appear from the epiblast in the blastoderm and translocate to the hypoblast area of the pellucida (2, 3). During gastrulation, PGCs circulate through the vascular system and settle down in the gonadal anlagen. Such a differentiation pathway, including germ cell lineage during chicken embryo development, is a systematic process, governed by the concerted action of multiple unknown regulatory mechanisms (4–6).
MicroRNAs (miRNAs) are small, noncoding RNAs ranging from 18 to 23 nucleotides that posttranscriptionally regulate gene expression in various tissues and cell types. Typically, miRNAs act as specific regulators of gene expression and are capable of controlling the fate of cells in a time- and tissue-specific manner (7, 8) through regulation of cellular differentiation, in addition to developmental patterning and morphogenesis (9–11). To date, several miRNA profiles have been classified as ESC-specific miRNAs, including miR-290–295 and miR-302–367 clusters (12, 13). However, both the miRNA expression profiling and posttranscriptional gene regulation for lineage specification, commitment, and differentiation during chicken embryo development remain largely uninvestigated. It has been shown recently that miRNA biogenesis and specific expression are required for PGC and germline development of mouse PGCs (14). Such miRNAs regulate the gain of lineage-specific differentiation in germ cells, in addition to the loss of pluripotent potential in stem cells. However, the intricate posttranscriptional network of miRNA expression for lineage specification and differentiation during chicken embryo development has yet to be investigated in detail.
Understanding the cellular and molecular mechanism(s) that underlie the developmental fate of early embryos is critical for the practical use of genetic modifications. In the current study, to identify miRNA signatures for the maintenance of the undifferentiated state of blastoderms and the germline lineage of chicken PGCs, global miRNA expression profiles using miRNA expression microarrays were analyzed. The miRNAs that were significantly expressed in the undifferentiated blastoderm and chicken PGCs were examined further to investigate the regulatory pathways of their expression. We demonstrate that posttranscriptional regulation through miRNA expression is important for the control of differentiation in both undifferentiated blastoderm and chicken PGCs.
Results
Preparation of PGCs from Embryonic Gonads.
To collect purified chicken PGCs, FACS was performed using a chicken PGC-specific marker, anti–stage-specific embryonic antigen 1 (anti–SSEA-1) antibody, and confirmed by staining with anti–SSEA-1 antibody and the periodic acid-Schiff (PAS) reaction, which are specific to chicken PGCs (15, 16). The percentage of SSEA-1+ and PAS+ cells after FACS analysis were 93 ± 1.4% and 96 ± 0.8, respectively, and the viability of the sorted PGCs was 95.0 ± 0.8% (Fig. S1 and Table S1).
Identification of miRNAs Highly Expressed in Stage X Blastoderm and Chicken PGCs Based on Customized Chicken miRNA Expression Microarray.
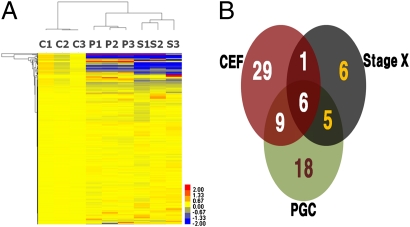
To identify certain miRNAs and miRNA clusters that specifically appear in undifferentiated blastoderm and chicken PGCs, we compared the miRNA expression profiles of stage X blastoderms, SSEA-1+ PGCs, and chicken embryonic fibroblasts (CEFs). The hierarchical clustering showed that the clustered miRNA expression pattern of the undifferentiated blastoderm was more closely related to PGCs than to CEFs (Fig. 1A). Based on the comparison with CEF expression profiles, we identified six miRNAs that were specifically up-regulated in undifferentiated blastoderm and 18 miRNAs that were specifically up-regulated in chicken PGCs (Fig. 1B and Tables S2 and S3; Welch's t test: P < 0.05). Additionally, five miRNAs that were significantly expressed in both stage X blastodermal cells and PGCs were identified and selected (Table S4).
Fig. 1.
Data analysis of the customized chicken miRNA expression microarray. (A) Hierarchical clustering of the chicken miRNA expression microarray. Clustering was performed from the normalized expression data of all probes. Each row represents the relative gene expression of a single miRNA. C, CEF; P, PGCs; S, Stage X. High expression levels of miRNAs compared with the mean are indicated in red, low expression levels in blue, and mean expression levels in yellow. Clustering results are displayed using the TreeView software. (B) Venn diagram illustrations of miRNA increases in SSEA+ chicken PGCs, stage X blastoderm cells, and CEFs from the customized chicken miRNA expression microarray. The common genes from the miRNA microarray experiments were up-regulated at least twofold (P < 0.05) in each cell type.
Validation of Selected miRNAs by Quantitative Real-Time PCR Analyses.
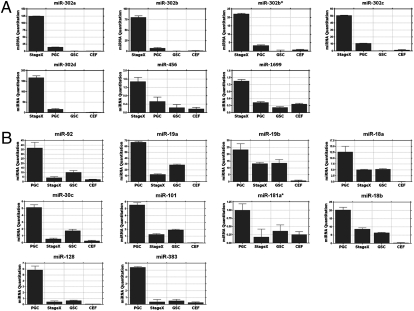
To verify the expression of miRNAs identified by the microarray, we performed real-time PCR using undifferentiated blastoderm, PGCs, gonadal stroma cells (GSCs), and CEFs. In candidate stage X-specific miRNAs, the expression levels of miR-456 and miR-1699 were relatively higher in the blastoderm than in other tissues (Fig. 2A). In chicken PGCs, 10 of the 18 miRNAs were confirmed to be predominantly expressed (Fig. 2B). Finally, when the expression levels of five miRNAs specifically expressed in both undifferentiated blastoderm and PGCs were examined by real-time PCR, all were highly expressed in the stage X blastoderm and PGCs compared with somatic cells such as CEFs and GSCs. Interestingly, the expression levels of five miRNAs were higher in the undifferentiated blastoderm than in the chicken PGCs (Fig. 2A). We therefore classified these five miRNAs as miRNAs that were highly expressed in the undifferentiated blastoderm. Based on the miRNA microarray and real-time PCR analysis, we identified seven miRNAs that were highly and specifically expressed in the chicken stage X blastoderm and 10 miRNAs that were highly and specifically expressed in PGCs.
Fig. 2.
Quantitative expression analysis of miRNAs highly expressed in undifferentiated blastoderm cells (A) and PGCs (B). Real-time PCR analysis was conducted in triplicate and normalized to the expression levels of small nucleolar RNA (snoRNA). miRNA expression was compared among stage X blastodermal cells, PGCs, GSCs, and CEFs. PGCs, GSCs, and CEFs were retrieved from 6-d-old embryos (stage 29). Error bars indicate SE of triplicate analyses.
Knockdown of miR-302a and miR-456 in Undifferentiated Chicken Blastodermal Cells.
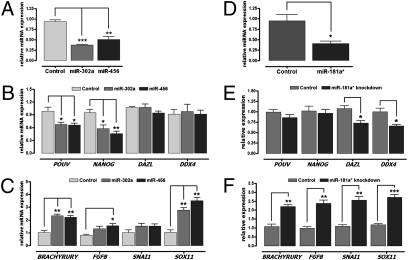
Among stage X miRNAs, the miR-302 cluster acts as a critical regulator for pluripotency through regulation of the cell cycle and methylation of related genes in mammals. miR-456 has been characterized in chickens but not in mammals. Also, target binding sites of miR-302a and miR-456 were localized to the sex-determining region Y box 11 (SOX11) 3′ UTR based on computational prediction of target miRNA. To examine the function of the miRNAs in the maintenance of the undifferentiated state in early development, miR-302a and miR-456 were selected, and their knockdown probes were introduced into undifferentiated blastodermal cells. Both knockdown probes were able to silence target miRNA expression significantly. The knockdown of miR-302a and miR-456 resulted in a significant decrease, 61.0 ± 0.3% (P < 0.001) and 52.0 ± 1.8% (P < 0.01), respectively, compared with knockdown of control miRNA (Fig. 3A). Second, we validated changes in the expression levels of pluripotency-related genes [POU domain class 5 transcription factor 1 (POUV) and nanog homeobox (NANOG)] and germ cell-associated genes [deleted azoospermia-like (DAZL) and DEAD box polypeptide 4 (DDX4)]. The knockdown of both miR-302a and miR-456 significantly decreased the expression of pluripotency-related genes POUV (miR-302a, P < 0.05, and miR-456, P < 0.05) and NANOG (miR-302a, P < 0.05, and miR-456, P < 0.01) but not the germ cell genes DAZL or DDX4 (Fig. 3B). Next, the gene expression patterns of somatic genes including brachyury homolog (BRACHYURY), fibroblast growth factor 8 (FGF8), snail homolog 1 (SNAI1), and SOX11 were analyzed in the knockdown blastodermal cells. The expression levels of all somatic genes except SNAI1 were significantly higher (P < 0.05) in miR-302a and miR-456 knockdown cells than in the control miRNA (Fig. 3C).
Fig. 3.
Quantitative expression analysis after knockdown of miR-302a and miR-456 in undifferentiated blastoderm cells and after knockdown of miR-181a* in chicken PGCs. (A) Relative miRNA expression in stage X blastoderm cells following silencing with the knockdown probes for miR-302a or miR-456. (B) Relative expression of pluripotent genes (POUV and NANOG) and germ cell-related genes (DAZL and DDX4) in stage X blastoderm cells after transfection of the knockdown probe for miR-302a or miR-456. (C) Relative expression of somatic genes (BRACHYRURY, FGF8, SNAI1, and SOX11) in stage X blastoderm cells following transfection of the miR-302a or miR-456 knockdown probes. (D) Relative miRNA expression in PGCs after miR-181a* silencing. (E) Relative expression of pluripotent genes and germ cell-related genes in PGCs after miR-181a* knockdown. (F) Relative expression of somatic genes in PGCs after miR-181a* silencing. Noncomplementary sequences in the chicken genome were used as a control. real-time PCR was conducted in triplicate and normalized to control expression of snoRNA (A and D) or GAPDH (B, C, E, and F). Significant differences between control and treatment groups are indicated as ***P < 0.001, **P < 0.01, and *P < 0.05. Error bars indicate the SE of triplicate analyses.
We also examined the functional activity of miR-302a and miR-456 in chicken PGCs using the same knockdown probes. The efficacy of both knockdown probes in silencing target miRNA was similar to that of blastodermal cells. Interestingly, like blastodermal cells, effects of the knockdown of miR-302a and miR-456 in chickens were similar to those of blastodermal cells (Fig. S2). These results indicate that silencing both miR-302a and miR-456 not only induces a decrease in the expression of pluripotency-related genes but also increases expression of somatic genes in both undifferentiated blastodermal cells and PGCs.
Knockdown of miR-181a* in Chicken PGCs.
Germ cell specification involves molecular events that are programmed by somatic cells. For example, the homeobox (HOX) and mesoderm genes are repressed by transcriptional regulators such as B-lymphocyte-induced maturation protein 1 (Blimp1). We hypothesized that miR-181a* was a candidate posttranscriptional regulator because the binding site of miR-181a* was localized to the homeobox A1 (HOXA1) 3′ UTR, based on target prediction. To investigate the function of miR-181a* in chicken PGCs, the miR-181a* transcript was silenced, and the expression levels of germ cell-related genes were analyzed (Fig. 3D). In contrast to the knockdown of miR-302a and miR-456, miR-181a* silencing specifically decreased the expression levels of DAZL and DDX4; however, no change was detected in the expression of POUV or NANOG (Fig. 3E). Additionally, miR-181a* silencing dramatically up-regulated the expression levels of all the somatic genes (BRACHYRURY, FGF8, SNAI1, and SOX11) in chicken PGCs (Fig. 3F). These results indicate that miRNA181a* is involved in the regulation of germ cells but not pluripotent cells.
Specific Analysis of the Target Gene Transcript for the Regulatory Roles of miR-302a and miR-456.
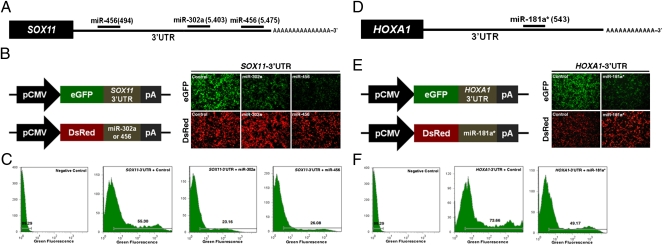
To examine the posttranscriptional gene regulation of miR-302a and miR-456, we validated the binding activity and down-regulation of the miRNA target site using the 3′ UTRs of the SOX11 transcript, which was predicted to be a miR-302a and miR-456 target transcript using the computational miRNA target prediction database TargetScan (Fig. 4A; http://www.targetscan.org). We also subcloned the 3′ UTR of the SOX11 transcript to generate a reporter–target construct in which the expression of a GFP transcript was coupled with the miRNA target 3′ UTR (Fig. 4B). To perform a dual fluorescence reporter assay, coexpression vectors containing Discosoma sp. red fluorescent protein (DsRed) and miR-302a or miR-456 were constructed (Fig. 4B). After cotransfection of SOX11 eGFP-3′-UTR and DsRed-miRNA for miR-302a or miR-456, GFP and DsRed expression were analyzed by fluorescence microscopy and FACS (Fig. 4C). The percentage of GFP+ cells (55.3% in control vs. 23.16% in miR-302a and 26.08% and miR-456) and the density of GFP fluorescence decreased in miR-302a– or miR-456–transfected cells compared with control samples. These results indicate that both miR-302a and miR-456 bind directly to the 3′ UTR of the SOX11 transcript and regulate SOX11 function during somatic cell differentiation.
Fig. 4.
In vitro target assay of miR-302a, miR-456, and miR-181a*. (A) Diagram of miR-302a and miR-456 binding sites in the SOX11 3′ UTR. (B) Expression vector maps for eGFP with SOX11 3′ UTR and DsRed with each miRNA. The 3′ UTR of the SOX11 transcript was subcloned between the eGFP gene and the polyA tail to generate the fusion construct of the GFP transcript following the miRNA target 3′ UTR (pcDNAeGFP3′UTR) (Upper), and the miRNA expression vector was designed to coexpress DsRed and each miRNA (pcDNADsRedmiRNA) (Lower). After cotransfection of pcDNA-eGFP-3′-UTR for the SOX11 transcript and pcDNA-Ds-Red miRNA for miR-302a or miR-456, the fluorescence signals of GFP and DsRed were detected using fluorescence microscopy (Right) and FACS (C). (D) Diagram of miR-181a* binding site in the HOXA1 3′ UTR. (E) The expression vector map for eGFP with the HOXA1 3′ UTR, DsRed with miR-181a*, and the fluorescence signals of GFP and DsRed using fluorescence microscopy. (F) After cotransfection of pcDNA-eGFP-3′-UTR for the HOXA1 transcript and pcDNA-DsRed-miRNA for miR-181a*, the fluorescence signals of GFP and DsRed were detected using FACS.
Specific Analysis of the Target Gene Transcript for miR181a*.
To analyze the miR-181a* target transcript, we predicted the target gene(s) of miR-181a* from a computational target prediction database (TargetScan; Fig. 4D; Gene Expression Omnibus: GSE 15830) (Table S5). From this analysis, both HOXA1 (a somatic gene involved in somatic cell differentiation) and nuclear receptor subfamily 6, group A, member 1 (NR6A1 (a gene involved in germ cell differentiation) were selected and analyzed further.
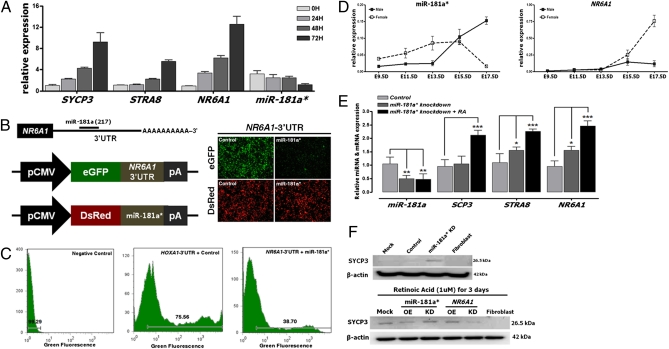
After the cotransfection of HOXA1-eGFP-3′UTR and DsRed-miRNA-181a* (Fig. 4E), the percentage of GFP+ cells (73.66% in control vs. 49.17% in miR-181a*) decreased, compared with the controls (Fig. 4 E and F). To assess NR6A1, which is involved in retinoic acid (RA) signaling, a critical regulator for meiosis in early germ cells, we analyzed the changes in meiotic gene expression in chicken PGCs after RA treatment. The mRNA expression of meiotic genes including synaptonemal complex protein 3 (SYCP3), stimulated by retinoic acid gene 8 homolog (STRA8), and NR6A1 increased gradually as the incubation time with RA increased; however, miR-181a* levels were down-regulated (Fig. 5A). This result suggested that miR-181a* could be closely related to the regulatory pathway of meiotic differentiation in chicken PGCs, through down-regulation of NR6A1. Interestingly, a binding site for miR-181a* was located in the 3′ UTR of the NR6A1 transcript (Fig. 5B). Based on these results, we generated a reporter–target construct in which the eGFP transcript accompanied the miR-181a* target 3′ UTR and a coexpression vector containing the DsRed gene and miR-181a* for dual fluorescence reporter assays (Fig. 5B). We observed that the percentage of GFP-expressing cells (75.56% in control vs. 38.70% in miR-181a*) was reduced by the cotransfection of miR-181a*, compared with controls (Fig. 5 B and C). Collectively, these results indicate that miR-181a* bound directly to the 3′ UTR where it regulated NR6A1 function during cell differentiation or meiosis.
Fig. 5.
In vitro target assay of miR-181a* on the NR6A1 transcript and a quantitative expression analysis of meiotic genes following the knockdown of miR-181a* in chicken PGCs. (A) Expression levels of the meiotic genes, including SYCP3, STRA8, and NR6A1, were analyzed in chicken PGCs 0–72 h after RA treatment. Expression of endogenous miR-181a* in PGCs was down-regulated after RA treatment. (B) Diagram of the miR-181a* binding site in the NR6A1 3′ UTR and the expression vector maps for eGFP with NR6A1 3′ UTR and DsRed with miR-181a*. After cotransfection of pcDNA-eGFP-3′-UTR for the NR6A1 transcript and pcDNA-DsRed-miRNA for miR-181a*, the fluorescence signals of GFP and DsRed were detected using fluorescence microscopy and FACS (C). (D) Quantitative expression pattern of miR181a* and NR6A1 in male and female chicken embryonic gonads from E9.5 to E17.5. Error bars indicate the standard error (SE) of triplicate analysis. (E) Relative expression of SYCP3, STRA8, and NR6A1 after silencing with the knockdown probes for miR-181a* at 72 h, followed by RA treatment in chicken PGCs. *P < 0.05, **P < 0.01, and ***P < 0.001: significant difference compared with control. (F) Expression of SYCP3 protein after silencing with the knockdown probes for miR-181a* at 72 h, followed by RA treatment in chicken PGCs examined by Western blotting. OE, overexpression, KD, knockdown.
Real-Time PCR Analysis of miR181a* and NR6A1 During Chicken Embryonic Gonad Development.
To confirm the functional relationship between miR181a* and NR6A1 in vivo, the expression patterns of miR181a* and NR6A1 transcripts were analyzed by real-time PCR during chicken embryonic gonad development. In the female embryo, miR181a* expression was gradually up-regulated by embryonic day (E) 15.5 during development, and the NR6A1 transcript levels markedly increased from E13.5 when chicken PGCs entered meiosis (Fig. 5D). In the male embryonic gonads, the miR181a* transcript increased between E13.5 and E17.5, and NR6A1 expression was unchanged at these time points. To confirm the functional relationship between miR181a* and NR6A1 in vitro, expression patterns were analyzed by NR6A1 silencing and overexpression. In contrast to knockdown of miR-181a*, NR6A1 silencing specifically decreased the expression levels of mir-181a* and meiotic genes (SYCP3 and STRA8), although no change was detected in the expression of DAZL or DDX4 (Fig. S2 D–G and Table S6). NR6A1 overexpression specifically decreased the expression levels of pluripotency-related, germ cell-associated, and somatic genes, although increased expression of meiotic genes was detected (Fig. S2 H–K). These data indicate that miR181a* and NR6A1 regulate each other negatively during germ cell development.
Functions of miR-181a* in Meiotic Differentiation of Germ Cells.
To confirm the function of miR-181a* during meiotic events, the expression patterns of meiotic transcripts were analyzed by knocking down miR-181a* during RA-induced meiotic differentiation of germ cells. Without RA, miR-181* knockdown increased expression levels of meiotic transcripts (STRA8, NR6A1) except for SYCP3, but miR-181a* silencing dramatically induced all meiotic transcripts including SYCP3 during RA-induced meiotic differentiation of germ cells (Fig. 5E). To investigate further the synergistic interaction between miR-181* and other pathways regulating gene expression, SYCP3 protein expression was analyzed. SYCP3 protein was increased by miR-181a* knockdown in chicken PGCs. In RA-induced meiotic differentiation of germ cells, SYCP3 protein was increased by miR-181a* silencing and NR6A1 overexpression. However, SYCP3 protein decreased with miR-181a* overexpression and in NR6A1 knockdown cells (Fig. 5F).
Discussion
miRNAs function as major regulators and pivotal determinants in numerous critical cellular processes (7, 17, 18). Several studies have suggested that embryonic development correlates with diverse and dynamic expression patterns of miRNAs in many organisms and multiple tissues (19–21). In this study, a chicken miRNA expression microarray was developed that contained 479 miRNA sequence probes based on the miRBase Sequence database, version 14 (Sanger Institute). The array data obtained were consistent and highly reproducible and may be useful for global miRNA expression analyses in a variety of avian species.
Based on the chicken miRNA microarray and qRT-PCR analyses, we identified seven miRNAs and undifferentiated chicken stage X blastoderm and 10 miRNAs that were highly expressed in PGCs. Interestingly, five of these miRNAs were selected as candidate miRNA signatures specifically expressed in both stage X blastoderm and chicken PGCs. qRT-PCR analyses also confirmed that the five miRNAs were highly expressed in the stage X blastoderm and PGCs compared with somatic cells, CEFs, and GSCs. However, the expression levels of the five miRNAs in undifferentiated blastoderm cells were increased compared with their levels in chicken PGCs (Fig. 2A). Thus, these five miRNAs (miR-302a, miR-302b, miR-302b*, miR-302c, and miR-302d) were classified as miRNAs that were highly expressed in the undifferentiated blastoderm (Table S4). The miR-302 cluster is known to contain critical stem cell-specific miRNAs that maintain an undifferentiated state in ESCs (13). To evaluate the putative functions of these miRNAs in the undifferentiated state of chicken embryonic cells, miR-302a and miR-456 were silenced. This silencing decreased the expression of the pluripotency-related genes but not of the germ cell-associated genes (Fig. 3B). Additionally, somatic gene expression increased significantly in miR-302a and miR-456 knockdown cells (Fig. 3C), and the knockdown of miR302a in chicken PGCs showed similar expression patterns (Fig. S2). These results indicate that the expression of the miR-302 cluster and miR-456 are stem cell markers in chicken as well as in mammals and may prevent stage X blastodermal cells and PGCs from differentiating into the somatic cell lineages.
In contrast to other somatic cell lineages, PGCs maintain functional characteristics similar to those of the undifferentiated ESCs. Furthermore, an additional pluripotent cell type, embryonic germ cells, has been derived and established from PGCs in mammals and chickens (22–24). Thus, the miR-302 cluster that is highly expressed in undifferentiated blastodermal cells may be up-regulated in chicken PGCs, even though it is expressed at lower levels than in the stage X blastoderm. In this study, the miRNA-17–92 cluster was expressed predominantly in chicken PGCs (Fig. 2B) but not in the stage X blastoderm. It has been reported that the miR-17–92 cluster is highly expressed in mouse and human ESCs and many cancerous tissues and has a seed sequence similar to ESC-specific cell-cycle miRNAs (25–29). However, the chicken miRNA-92 cluster was specifically expressed in PGCs but not in undifferentiated blastoderm cells (Fig. 2 A and B). Recently, it has been reported that the expression of miR-17–92 affects the balance between ESC self-renewal and differentiation (30–32). Thus, specific miRNAs and potential miRNA targets of the miRNA-92 cluster could be involved in the regulation of cell-cycle progression in chicken PGCs but not in undifferentiated blastodermal cells. In the current study, we also observed that miR-181a* was expressed specifically in chicken PGCs. In knockdown experiments, we hypothesized that silence of miR-181a* altered the expression networks that could cause cellular differentiation, apoptosis, and proliferation. Interestingly, miR-181a* silencing specifically decreased the expression levels of the germ cell-related genes. However, no change in the expression of the pluripotency-related genes was observed (Fig. 3E), although miR-181a* knockdown markedly increased the expression of all somatic genes in chicken PGCs (Fig. 3F). These results indicate that miR181a* in chicken PGCs not only represses somatic differentiation through the silencing of somatic gene transcripts but also promotes germ cell differentiation. miR181a* appears not to be involved in the regulation of pluripotency-related gene expression in chicken PGCs. However, further investigation is necessary for a comprehensive understanding of regulation networks and gene–miRNA interactions for maintaining the properties of germ cells and pluripotent cells.
The knockdown of miR-302a, miR-456, and miR-181a* induced the up-regulation of somatic gene expression in undifferentiated blastoderm and PGCs. Thus, we investigated further the biological pathways governed by these chicken miRNAs. For candidate target somatic genes, SOX11 3′ UTR for miR-302a and miR-456 and HOXA1 3′ UTR for miR-181a* were predicted using a comprehensive database for predicting target miRNAs. In cotransfection experiments using the eGFP-3′ UTR and miRNA, the number of GFP+ cells and the density of green fluorescence decreased, suggesting that each miRNA bound directly to the target transcript 3′ UTR and silenced its expression. These results indicate that the miRNAs prevent stage X blastodermal cells and PGCs from differentiating into somatic cell lineages with somatic gene silencing. Collectively, to maintain an undifferentiated state, ESCs, stage X blastodermal cells, and germ cells not only activate pluripotency-related genes but also repress somatic genes (Fig. 6).
Fig. 6.
Schematic illustrating the current working hypothesis regarding the regulation of miR-302a, miR-456, and miR-181a*. miR181a* prevents chicken PGCs from entering meiosis by downregulating the NR6A1 transcript that triggers the meiosis process in chicken PGCs through RA signaling from gonadal stroma cells. miR181a* also suppresses the somatic differentiation fate in chicken PGCs by silencing the somatic gene expression, including HOXA1. Both miR-302a and miR-456 regulate the pluripotency in chicken blastodermal cells and PGCs by silencing the somatic gene expression, including SOX11.
In mice, the germline is directly specified from the epiblast, and germ cell specification requires bone morphogenic protein signaling from the extraembryonic ectoderm and visceral endoderm (33–36). As a critical determinant of the germ cell lineage in mice, Blimp1 interacts with protein arginine methyltransferase 5 (Prmt5), an arginine-specific histone methyltransferase, and the Blimp1–Prmt5 complex represses the somatic gene expression of HOX genes and downstream targets during the formation of the initial PGC population (37). Similar to these reports, we found that the somatic HOXA1 transcript was a target of miR-181a*, which down-regulated its expression in chicken PGCs (Fig. 4). Taken together with knockdown experiments and functional analyses of the miR181a* target transcript, we propose that these miRNAs play important roles in posttranscriptional gene regulation associated with the suppression of somatic differentiation during early germ cell development in chickens.
In the current study, we confirmed the function of another miR-181a* target transcript, NR6A1, which is related to germ cell development. NR6A1 is involved in RA signaling and is a critical regulator of meiosis in developing germ cells (38). Interestingly, after the treatment of chicken PGCs with RA, we found that the expression levels of meiotic genes as well as NR6A1 were increased dramatically in accordance with the incubation time with RA (Fig. 5A). Additionally, the in vivo expression profiles of miR-181a* and NR6A1 transcripts showed an inverse relationship during chicken embryonic gonad development (Fig. 5D). In the female embryo, miR181a* expression was down-regulated gradually as female embryonic gonads developed; however, the NR6A1 transcript was increased dramatically, particularly around day 13.5. Smith et al. (39) reported that germ cells in the chicken female gonad initiate meiosis at around E12.5, at which time STRA8 is up-regulated in the female left gonad. In contrast to the female embryo, miR181a* was highly expressed in male embryonic gonads; however, NR6A1 expression was suppressed after E13.5 (39). Thus, we propose that miR-181a* and NR6A1 could be closely related to the regulatory pathways of meiotic differentiation in chicken PGCs. The meiotic pathway was suppressed by silencing the NR6A1 transcript (Fig. 6). In addition to the repression of the NR6A1 meiotic gene and HOXA1 somatic gene transcript, miR-181* may modulate the different regulatory pathway(s) for meiosis in chicken early germ cells. The mechanisms by which miR-181a* and NR6A1 regulate germ cell differentiation in chicken thus require further investigation (Fig. 6).
Here we report miRNA expression profiling and miRNA functional analysis in the stage X blastoderm and PGCs in chickens. Furthermore, we demonstrated that the germ cell differentiation pathway of meiosis initiation is controlled in a time-dependent manner by these miRNAs. Collectively, our data indicate that miRNAs are key posttranscriptional regulators for the control of differentiation of both undifferentiated blastodermal cells and germ cells in chickens.
Materials and Methods
The preparation of PGCs and additional methods are described in SI Material and Methods.
Design of Chicken miRNA Expression Microarray and Expression Profile Analysis.
We developed chicken miRNA expression chips (AMDID 027206; Agilent) that contained 479 miRNA sequence probes, based on the miRBase database, version 14 (Sanger Institute). For miRNA microarray analysis, total RNA was labeled with cyanine 3-pGp (Cy3) using the Agilent miRNA Complete Labeling and Hyb Kit (Agilent Technologies) and then was placed on the chicken miRNA v.14 chips (AMDID 027206; Agilent). Slides were hybridized for 16 h at 42 °C in the Agilent hybridization system and washed with 0.0005% Triton X-102 for 5 min followed by a second washing for 5 min. The slides then were centrifuged and dried at room temperature.
miRNA expression profiles were analyzed using the GeneSpring GX v.11 (Agilent Technologies). All data were analyzed with the standard normalization method for one-channel microarrays, namely, percentile median normalization. Fold-change values were calculated for unpaired comparisons between normal and treated groups. The fold-change filters included the requirement that miRNAs should be increased at least 150% compared with the controls for the up-regulated miRNAs and decreased by 66.67% compared with the controls for the down-regulated miRNAs. Welch's t test was performed to identify significant (P < 0.05) changes in expression.
Supplementary Material
Acknowledgments
This work was supported by a grant from the The Next Generation BioGreen 21 Program (No. PJ008142), Rural Development Administration, and by Grant R31-10056 from the World Class University Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology, Republic of Korea.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106141108/-/DCSupplemental.
References
- 1.Lavial F, et al. Ectopic expression of Cvh (Chicken Vasa homologue) mediates the reprogramming of chicken embryonic stem cells to a germ cell fate. Dev Biol. 2009;330:73–82. doi: 10.1016/j.ydbio.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. 1951. Dev Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- 3.Ginsburg M, Eyal-Giladi H. Temporal and spatial aspects of the gradual migration of primordial germ cells from the epiblast into the germinal crescent in the avian embryo. J Embryol Exp Morphol. 1986;95:53–71. [PubMed] [Google Scholar]
- 4.Lee BR, et al. A set of stage-specific gene transcripts identified in EK stage X and HH stage 3 chick embryos. BMC Dev Biol. 2007;7:60–70. doi: 10.1186/1471-213X-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim H, et al. MPSS profiling of embryonic gonad and primordial germ cells in chicken. Physiol Genomics. 2007;29:253–259. doi: 10.1152/physiolgenomics.00067.2006. [DOI] [PubMed] [Google Scholar]
- 6.Han JY, et al. Gene expression profiling of chicken primordial germ cell ESTs. BMC Genomics. 2006;7:220–225. doi: 10.1186/1471-2164-7-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 10.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zovoilis A, Smorag L, Pantazi A, Engel W. Members of the miR-290 cluster modulate in vitro differentiation of mouse embryonic stem cells. Differentiation. 2009;78:69–78. doi: 10.1016/j.diff.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Card DA, et al. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi K, et al. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One. 2008;3(3):e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park TS, et al. Improved germline transmission in chicken chimeras produced by transplantation of gonadal primordial germ cells into recipient embryos. Biol Reprod. 2003;68:1657–1662. doi: 10.1095/biolreprod.102.006825. [DOI] [PubMed] [Google Scholar]
- 16.Kim JN, et al. Enriched gonadal migration of donor-derived gonadal primordial germ cells by immunomagnetic cell sorting in birds. Mol Reprod Dev. 2004;68:81–87. doi: 10.1002/mrd.20051. [DOI] [PubMed] [Google Scholar]
- 17.Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 18.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 19.Wienholds E, et al. MicroRNA expression in zebrafish embryonic development. Mech Dev. 2005;122:S149–S150. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 20.Mineno J, et al. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joglekar MV, Parekh VS, Mehta S, Bhonde RR, Hardikar AA. MicroRNA profiling of developing and regenerating pancreas reveal post-transcriptional regulation of neurogenin3. Dev Biol. 2007;311:603–612. doi: 10.1016/j.ydbio.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Resnick JL, Bixler LS, Cheng L, Donovan PJ. Long-term proliferation of mouse primordial germ cells in culture. Nature. 1992;359:550–551. doi: 10.1038/359550a0. [DOI] [PubMed] [Google Scholar]
- 23.Shamblott MJ, et al. Derivation of pluripotent stem cells from cultured human primordial germ cells. Proc Natl Acad Sci USA. 1998;95:13726–13731. doi: 10.1073/pnas.95.23.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park TS, Han JY. Derivation and characterization of pluripotent embryonic germ cells in chicken. Mol Reprod Dev. 2000;56:475–482. doi: 10.1002/1098-2795(200008)56:4<475::AID-MRD5>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 25.Laurent LC, et al. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells. 2008;26:1506–1516. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- 26.Marson A, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, et al. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 32.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 33.Ying Y, Zhao GQ. Cooperation of endoderm-derived BMP2 and extraembryonic ectoderm-derived BMP4 in primordial germ cell generation in the mouse. Dev Biol. 2001;232:484–492. doi: 10.1006/dbio.2001.0173. [DOI] [PubMed] [Google Scholar]
- 34.Ying Y, Liu XM, Marble A, Lawson KA, Zhao GQ. Requirement of Bmp8b for the generation of primordial germ cells in the mouse. Mol Endocrinol. 2000;14:1053–1063. doi: 10.1210/mend.14.7.0479. [DOI] [PubMed] [Google Scholar]
- 35.Saitou M, Barton SC, Surani MA. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- 36.Lawson KA, et al. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohinata Y, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 38.Barreto G, Borgmeyer U, Dreyer C. The germ cell nuclear factor is required for retinoic acid signaling during Xenopus development. Mech Dev. 2003;120:415–428. doi: 10.1016/s0925-4773(03)00018-2. [DOI] [PubMed] [Google Scholar]
- 39.Smith CA, Roeszler KN, Bowles J, Koopman P, Sinclair AH. Onset of meiosis in the chicken embryo; evidence of a role for retinoic acid. BMC Dev Biol. 2008;8:85–103. doi: 10.1186/1471-213X-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.