Abstract
The photoreceptor phytochrome (phy) A has a well-defined role in regulating gene expression in response to specific light signals. Here, we describe a new Arabidopsis mutant, laf1 (long after far-red light 1) that has an elongated hypocotyl specifically under far-red light. Gene expression studies showed that laf1 has reduced responsiveness to continuous far-red light but retains wild-type responses to other light wavelengths. As far-red light is only perceived by phyA, our results suggest that LAF1 is specifically involved in phyA signal transduction. Further analyses revealed that laf1 is affected in a subset of phyA-dependent responses and the phenotype is more severe at low far-red fluence rates. LAF1 encodes a nuclear protein with strong homology with the R2R3–MYB family of DNA-binding proteins. Experiments using yeast cells identified a transactivation domain in the C-terminal portion of the protein. LAF1 is constitutively targeted to the nucleus by signals in its N-terminal portion, and the full-length protein accumulates in distinct nuclear speckles. This accumulation in speckles is abolished by a point mutation in a lysine residue (K258R), which might serve as a modification site by a small ubiquitin-like protein (SUMO).
Keywords: Signal transduction, phytochrome A, Arabidopsis, MYB, transcription factor, nuclear speckles
Plants are sessile organisms that depend on light not only as their source of energy but also for the timing of important developmental processes such as seed germination, stem elongation, and the transition to reproductive growth. To monitor variations in the wavelength, intensity, direction, and period of light plants have evolved different photoreceptors, among which the phytochromes are probably the best studied photoreceptors (Fankhauser 2001). Phytochromes have the capacity to reversibly convert from a red-light-absorbing form, Pr, to a far-red-light-absorbing form, Pfr, through the absorption of either red (R) or far-red (FR) light. This mechanism allows phytochromes to sense different R:FR light ratios and to act as a light-responsive developmental switch (Furuya 1993; Quail et al. 1995; Smith 2000). Among the five members of the Arabidopsis phytochrome protein family (phyA–phyE; Clack et al. 1994), phyA is the only one that can perceive FR light, and it is involved mainly in the regulation of de-etiolation (Whitelam et al. 1993; Reed et al. 1994). Recent findings have shown that phyA can be transported to the nucleus in a FR light–dependent manner (Kircher et al. 1999; Hisada et al. 2000; Kim et al. 2000). After import into the nucleus on FR light illumination, the phyA–GFP forms numerous speckles that dissolve after transfer to darkness (Nagy et al. 2001).
Various approaches have been taken to understand how the FR light signal is transduced by phyA to activate gene expression. Two-hybrid screens using phytochrome as a bait have led to the identification of three proteins, PIF3, PKS1, and nucleoside diphosphate kinase 2 (NDPK2) that interact with both phyA and phyB (Ni et al. 1998; Choi et al. 1999; Fankhauser et al. 1999). Genetic analyses have yielded several Arabidopsis mutants that are specifically disrupted in phyA signal transduction. The mutants fhy1, fhy3, fin2, fin219, far1, pat1, rsf1/hfr1/rep1, and laf6 (Whitelam et al. 1993; Soh et al. 1998, 2000; Hudson et al. 1999; Bolle et al. 2000; Fairchild et al. 2000; Fankhauser and Chory 2000; Hsieh et al. 2000; Møller et al. 2001) show reduced responses under FR light conditions, whereas spa1 and eid1 (Hoecker et al. 1998; Büche et al. 2000) show exaggerated responses.
The identified genetic components of phyA signaling fall into three classes according to the cellular locations of their encoded products: PKS1, NDPK2, PAT1, and FIN219 localize preferentially in the cytoplasm, LAF6 is imported into plastids, and FAR1, SPA1, PIF3, RSF1/HFR1/REP1, and EID1 are nuclear (Ni et al. 1998; Choi et al. 1999; Fankhauser et al. 1999; Hoecker et al. 1999; Hudson et al. 1999; Bolle et al. 2000; Fairchild et al. 2000; Hsieh et al. 2000; Soh et al. 2000; Dieterle et al. 2001; Møller et al. 2001). The functions of most of these proteins are not obvious from their sequences and remain to be determined. FAR1 and PSK1 are novel proteins with no sequence similarities to other proteins in the database, SPA1 is a WD-40 repeat protein, and PAT1 is a member of the plant-specific GRAS (GAI, RGA, and Scarecrow) protein family, some members of which regulate gibberellin response or root development. LAF6 shows homology with ABC transporters, FIN219 is a member of the GH3 protein family, which is involved in auxin signaling, and EID1 is a F-box containing protein. PIF3 and RSF1/HFR1/REP1 show homology with the basic helix–loop–helix (bHLH) class of transcription factors. This array of proteins and their diverse intracellular localization reflect the complexity of phyA responses and suggest that FR light-activated phyA initiates a signaling cascade with several branches.
Many physiological modifications observed after activation of phyA signal transduction are caused by changes in gene expression (Kuno and Furuya 2000). Although some sequences necessary for light responsiveness have been defined, no single consensus motif nor a single transcription factor has been identified for such regulation and no cis-element has yet been linked to a single photoreceptor. Among the transcription factors implicated in light-regulated transcription in plants, most have been isolated by their ability to bind to specific promoter sequences. These include the MYB protein CCA1, several leucine zipper proteins (bZIPs such as ATHB-2/4, CPRF and GBF1) and GT-1 (Weisshaar et al. 1991; Gilmartin et al. 1992; Schindler et al. 1992; Carabelli et al. 1993; Wang et al. 1997). Nevertheless, genetic evidence for the functional involvement of most of the DNA binding proteins in light-regulated gene expression and photomorphogenesis remains to be shown.
Another approach to investigating the function of specific transcription factors is to analyze mutants disrupted in genes encoding these proteins and to characterize their light responses. Examples are the bZIP HY5, whose deficiency in plants resulted in elongated hypocotyls under all light conditions, the MYB protein LHY1, which is involved in circadian rhythm, and the bHLH proteins PIF3 and RSF1/HFR1/REP1 (Oyama et al. 1997; Schaffer et al. 1998; Halliday et al. 1999; Fairchild et al. 2000; Soh et al. 2000; Spiegelman et al. 2000).
Here, we report the isolation of the laf1 mutant, which is specifically impaired in phytochrome A signal transduction. Our results show that LAF1 belongs to the R2R3–MYB transcription family of proteins and likely functions as a positive component of phyA signaling. This transcription activator may be responsible for regulating the expression of a specific subset of phyA target genes. Moreover, we show that a LAF1–GFP fusion can localize to subnuclear speckles, and this localization is abolished by the K258R point mutation in LAF1.
Results
Mutant screening and isolation of laf mutants
To identify genes involved in the phyA signal transduction pathway, we screened a collection of independent Arabidopsis gene trap lines, which were generated using the Ds-based system of Sundaresan et al. (1995). Mutants with elongated hypocotyls under FR light and/or resistance to FR light-induced killing were selected (Barnes et al. 1996a; Bolle et al. 2000; Møller et al. 2001). The progeny of putative mutants was tested under continuous FR and red (R) light conditions, and only mutants with long hypocotyls under FR light were considered to be specifically impaired in phyA signaling. Here, we describe the isolation and characterization of one such mutant, called laf1 (long after far-red light 1).
Physiological characterization of the laf1 mutation
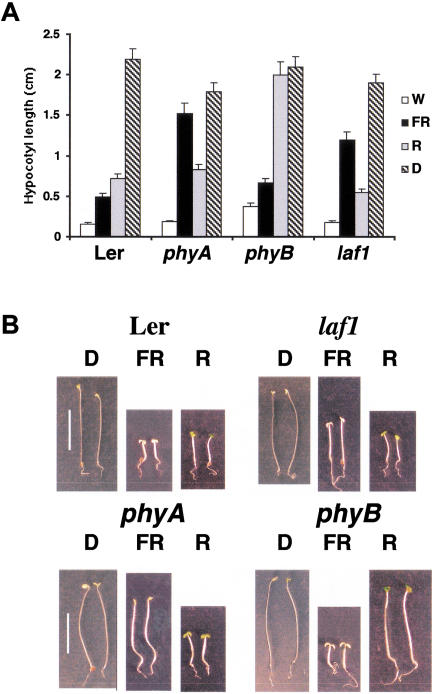
The mutant laf1 is defective in several seedling responses when grown under continuous FR light. FR suppresses hypocotyl elongation in both the wild-type (WT) and the laf1 mutant, but this response was reduced significantly in the latter (Fig. 1). In contrast, a phyA photoreceptor mutant (phyA) is completely blind to FR, resulting in long hypocotyls under these conditions (Fig. 1; Whitelam et al. 1993; Shinomura et al. 2000). The suppression of hypocotyl elongation in R (Fig. 1A,B), white (W; Fig. 1A), and blue (B) light (data not shown) was not altered in laf1. Mutant seedlings also showed a normal etiolated phenotype when grown in the dark (D; Fig. 1A,B).
Figure 1.
laf1 seedlings are specifically impaired in FR-induced de-etiolation. (A) Hypocotyl lengths of laf1 compared with WT (Ler), phyA, and phyB under white (W; 15 μmole/m2 sec), far-red light (FR; 3 μmole/m2 sec), red light (R; 35 μmole/m2 sec) conditions and darkness (D). Error bars show standard deviations. (B) Phenotypes of laf1. Seedlings of WT (Ler), laf1, phyA, and phyB were grown for 4 d either in complete darkness under FR (3 μmole/m2 sec) or under R light (35 μmole/m2 sec). Bar, 10 mm.
The ability to green in white light after a prolonged FR light treatment is a specific feature of mutants defective in the phyA photoreceptor or blocked in phyA signaling (Barnes et al. 1996a). We tested laf1 for its ability to green after exposure to different fluencies of FR light. At FR fluencies lower than 2 μmole/m2 sec, laf1 seedlings were resistant to the FR-induced killing, whereas under fluencies higher than 2 μmole/m2 sec they were sensitive and died. This is in contrast with WT seedlings, which were sensitive to FR fluencies even lower than 2 μmole/m2 sec (data not shown). Under FR light, WT plants synthesize anthocyanin, whereas phyA mutants are blocked in this process (Barnes et al. 1996b). We tested for anthocyanin accumulation in laf1 seedlings grown under three different FR fluencies (1.5, 3, and 6 μmole/m2 sec). Although laf1 was able to synthesize anthocyanin under all three light conditions, the levels were reduced by 40%–50% compared with WT (data not shown).
We also examined other seedling responses triggered specifically by phyA, such as FR-dependent apical hook opening, cotyledon unfolding and expansion, and gravitropism (for review, see Neff et al. 2000; Smith 2000). These responses were not altered in laf1 in comparison with WT over a range of fluencies. Furthermore, there was no obvious adult phenotype in laf1 plants, other than the fact that mutant plants appear to have a slightly shorter inflorescence than WT. This did not affect the flowering time of laf1, which was very similar to that of the WT when measured under long day (16 h day/8 h night) conditions.
The loss of responsiveness to FR observed in laf1 could, in principle, result from a reduction in phyA levels or spectral activity. No difference in phyA protein levels between WT and laf1 was observed in etiolated seedlings (data not shown), and the mutation is not linked to the phyA locus. These results, together with the fact that laf1 is impaired only in some FR light/phyA specific responses, indicate that the mutant is not compromised in the perception of FR light but rather in the transduction of its signal.
Genetic characterization of laf1
Genetic analysis showed that the mutant phenotype (i.e., elongated hypocotyls in FR light and partial resistance to FR-induced killing) cosegregated with the kanamycin resistance marker present on the single Ds insertion (Sundaresan et al. 1995). Backcrosses established that the laf1 mutant phenotype was recessive. The mutation was located on chromosome IV and does not correspond to any other mutation that produces a FR-specific long-hypocotyl phenotype (fhy1, fhy3, fin2, far1, pat1, rsf1/hfr1/rep1, fin219, laf6).
Cloning of the LAF1 gene
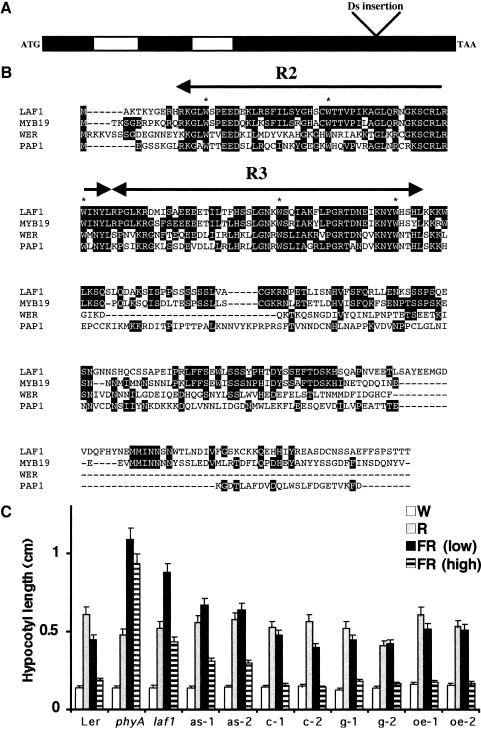
We used the Ds tag in laf1 to clone the LAF1 locus. The nucleotide sequence flanking the insertion was identical to that of a region on chromosome IV containing an ORF that corresponds to a MYB gene previously identified as AtMYB18 (GenBank accession no. Z95744; Kranz et al. 1998). After isolation of the cDNA by RT–PCR and its comparison to the genomic sequence deposited in the database by the Arabidopsis Genome Initiative, we determined that the LAF1 gene contained three exons and two introns (Fig. 2A). The Ds element was inserted in the third exon at nucleotide position 506 from the translation start point.
Figure 2.
LAF1 encodes a protein with homology with R2R3–MYB proteins. (A) A schematic representation of the genomic organization of laf1. The exon (black)/ intron (white) structure and the insertion site of the Ds element (506 bp from the ATG start codon) are shown. (B) Sequence comparison of LAF1 with other members of the R2R3–MYB protein family in Arabidopsis: AtMYB 19 (MYB19; GenBank accession no. Z95745), WEREWOLF (WER; Lee and Schiefelbein 1999), and Production of Anthocyanin Pigment 1 (PAP1; Borevitz et al. 2000). Conserved amino acid residues are highlighted in black. (Arrows) MYB domains (R2 and R3) ; (asterisks) conserved tryptophan residues within these domains. (C) Hypocotyl lengths of laf1 complemented with the 35S–LAF1 cDNA (c-1, c-2) or the 35S–LAF1 genomic transgene (g-1, g-2) and transgenic WT lines with the LAF1 antisense construct (as-1, as-2) or the 35S–LAF1 cDNA overexpressed (oe-1, oe-2). In comparison, the hypocotyl lengths of WT (Ler), phyA, and laf1 are shown. Light conditions are W (15 μmole/m2 sec), R (5 μmole/m2 sec), FR light at low fluence (1.5 μmole/m2 sec), and FR light at high fluence (6 μmole/m2 sec). Error bars show standard deviations.
The LAF1 gene encodes a protein of 283 amino acids with two MYB domains located at the N-terminal region. Each MYB domain consists of an ∼50-amino-acid helix–turn–helix motif, and each contains tryptophan residues in characteristic positions (Fig. 2B). In these respects, LAF1 is similar to the two-repeat (R2R3-type) MYB proteins, more than 130 members of which so far have been identified in the Arabidopsis genome (Martin and Paz-Ares 1997; Riechmann et al. 2000). Overall, LAF1 is most similar to AtMYB19 from Arabidopsis whose function is unknown (Fig. 2B; Kranz et al. 1998).
Expression pattern of LAF1
The steady state level of the LAF1 transcript is very low. Reverse Northern analysis described by Kranz and coworkers (1998) indicated no strong induction of AtMYB18/LAF1 expression after treatment with hormones or elicitors or exposure to abiotic stresses. The only tissues in which weak expression could be detected were cauline leaves. Using Northern blot hybridization with poly(A) RNA from cauline leaves of WT plants, we could detect a single band of ∼850 nucleotides, which corresponds to the appropriate size of the LAF1 cDNA. No LAF1 transcript could be detected in laf1 mutant plants even with RT–PCR suggesting that laf1 is a true null mutant (data not shown).
Genetic complementation of laf1 and analyses of transgenic antisense plants
To test for complementation, we transformed laf1 with either a LAF1 genomic DNA (1018 bp long) or a LAF1 cDNA (852 bp long) under the control of a 35S promoter. Seven independent transgenic lines from each transformation experiment were characterized. Figure 2C shows representative transgenic lines, which display WT hypocotyl lengths under low and high FR fluencies, showing complementation of the mutant phenotype. Interestingly, plants overexpressing the LAF1 gene do not show any clear phenotype under any analyzed light conditions, although the transcript levels are elevated (Fig. 2C).
The third exon of the LAF1 gene, which shows no high sequence homology with any other gene in the databank, was cloned in the antisense orientation under the control of a 35S promoter, and this expression cassette was transferred into WT plants. Four independent LAF1 antisense transgenic lines showed hypocotyl lengths 1.5-fold longer than WT under FR light, but WT hypocotyl lengths under white, R and B light, and in darkness (Fig. 2C). Together, these results confirm that indeed the disruption of the LAF1 gene is responsible for the laf1 mutant phenotype.
Altered expression of several phyA-regulated genes
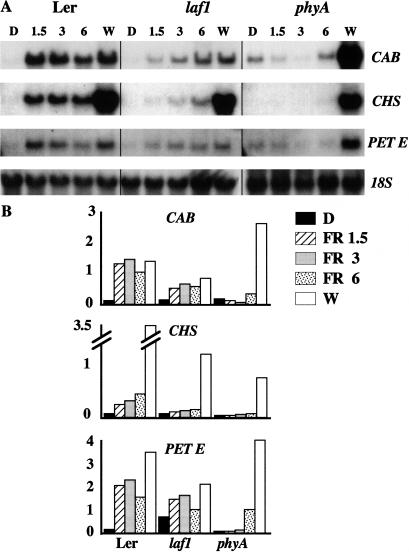
Regulation of developmental processes requires fine tuning of gene expression regulating cell elongation and cell differentiation. Previous pharmacological experiments in tomato and soybean cells have indicated that phyA regulates gene expression by at least three pathways: a cGMP-dependent pathway mediating chalcon synthase (CHS) gene expression, a Ca2+/calmodulin-dependent pathway that is necessary for chlorophyll a/b-binding protein (CAB) and ribulosebisphosphate carboxylase small subunit (RBCS) gene expression, and a third pathway, which requires both cGMP and Ca2+/calmodulin to induce plastocyanin (PET E) and ferredoxin:NADP(+) oxidoreductase (PET H) gene expression (Neuhaus et al. 1993; Bowler et al. 1994). To locate the site of action of LAF1, we performed Northern blot hybridizations with CHS, CAB, and PET E probes by using laf1 seedlings grown in the dark followed by exposure to different FR fluencies for 18 h. Figure 3 shows that the expression levels of all three target genes were reduced in the laf1 mutant compared with WT. However, the reduction was not as severe as that of the phyA mutant, which was used as a control. These results confirm the morphological phenotype under FR light, showing that laf1 is deficient in phyA signaling over a wide range of fluencies.
Figure 3.
Expression of CAB, CHS , and PET E in laf1. Four-day-old seedlings were grown either in the dark under different FR light fluencies (1.5, 3, and 6 μmole/m2 sec) or in W light for 18 h. Each lane contained 10 μg of total RNA. CAB, CHS, and PET E were used as probes. (A) Representative Northern blots. The hybridization with the 18S rDNA probe was used as a loading control. (B) Quantitative expression levels of CAB, CHS, and PET E in WT (Ler), laf1, and phyA. Northern blots were quantified with a PhosphoImager, and the expression levels were normalized with respect to the 18S rRNA level.
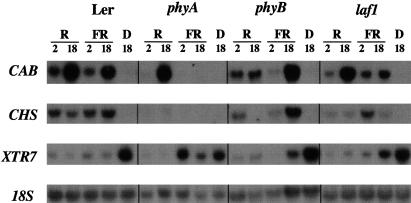
We also analyzed the expression of CAB, CHS, and XTR7 induced by either R or FR light (Fig. 4). CAB gene expression is reduced under FR light conditions in the phyA mutant and under R light conditions in the phyB mutant. In laf1, a reduction in CAB levels can be observed only under FR light. CHS gene expression is dependent on a functional phyA signaling pathway under R and FR light conditions (Barnes et al. 1996b), and accordingly it is reduced under R and FR light conditions in laf1. XTR7, which is involved in cell elongation as it encodes a xyloglucan endotransglucosylase-related protein (Xu et al. 1996), is negatively regulated by phyA and phyB in FR and R light, respectively (Kuno et al. 2000). Expression of XTR7 in the laf1 mutant was reduced in R light similar to WT, but less reduced in FR light. These data confirm the specificity of LAF1 for phyA-specific pathways. Note that the effects on FR-dependent gene regulation of CAB and XTR7 in the laf1 mutant are not strong after 2 h but very pronounced after 18 h.
Figure 4.
The laf1 mutant is specifically disrupted in phyA-dependent gene regulation. Four-day-old seedlings (Ler, phyA, phyB, and laf1) were grown in darkness and then, where indicated, transferred to R (35 μmole/m2 sec) or FR light (3 μmole/m2 sec) for 2 or 18 h. Each lane contained 10 μg of total RNA. CAB, CHS, and XTR7 were used as probes. The 18S rRNA band was used as a loading control.
LAF1 acts as a transcriptional activator
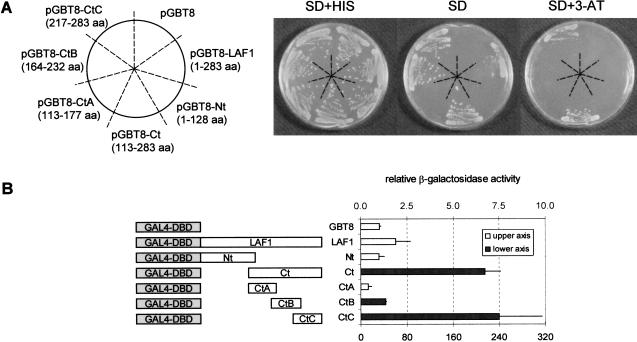
All MYB transcription factors contain one to three conserved MYB domains at the N terminus where the DNA binding domain resides (Rosinski and Atchley 1998; Rabinowicz et al. 1999). The C terminus, on the other hand, shows a high degree of variability among the various MYB proteins and is suggested to be important for protein–protein interaction. To determine whether LAF1 is a transcriptional activator and to identify the region of this protein responsible for this function, we fused the full-length LAF1 cDNA and LAF1 deletion mutants to sequences encoding the GAL4 DNA binding domain. These DNA fusion constructs were tested in yeast for their ability to promote expression of the lacZ and HIS3 reporter genes by binding to upstream activation regions. This allows assessment of growth on medium without histidine and measurement of β-galactosidase activity. Figure 5A shows that the full-length GAL4–LAF1 protein has low levels of transcriptional activity, but the C-terminal fragment GAL4–LAF1/113–283 (Ct) showed high levels of activation as measured by growth without histidine and in presence of 30 mM 3-AT. The LAF1 N-terminal fragment (Nt; GAL4–LAF1/1–128) and a C-terminal fragment (CtA; GAL4–LAF1/113–177) alone were unable to transactivate the reporter gene and prevented growth in the absence of histidine. However, GAL4–LAF1/164–232 (CtB) and GAL4–LAF1/217–283 (CtC) promoted better growth without histidine than the full-length LAF1. Only CtC, however, could promote growth in the presence of 30 mM 3-AT, which increased the stringency of the experiment. These results were confirmed by β-galactosidase assays (Fig. 5B). These data show that the C-terminal domain (amino acids 164–283) of LAF1 contains sequences with transactivation function. Furthermore, the results suggest the existence of two possibly distinct transactivation regions, one between amino acid residues 164 and 232 and the other, which shows six times higher β-galactosidase activity, between amino acid residue 217 and the C terminus. As the full-length GAL4–LAF1 fusion shows less activity than the C-terminal fragment alone (∼100-fold reduction in β-galactosidase activity), the results suggest a negative regulatory function associated with the N-terminal part of the protein, which contains the DNA-binding domain.
Figure 5.
LAF1 acts as a transcriptional activator. (A) Transactivation analysis of LAF1 in yeast. Different LAF1 cDNA fragments were fused to sequences encoding the Gal4 DNA-binding domain in the yeast vector pGBT8 and transformed into yeast strain HF7c. Transformants were plated onto SD plates with histidine and incubated for 3 d. Three different colonies were transferred to SD plates with histidine (SD + HIS), without histidine (SD) and without histidine plus 30 mM 3-AT (SD + 3-AT), and cell growth ability was analyzed after incubation for 4 d. (aa) Amino acid. (B) β-Galactosidase liquid culture assay from yeast carrying a pGBT8 vector either empty or with different LAF1 cDNA fragments by using ONPG as a substrate. The obtained β-galactosidase activity obtained from each construct was depicted relative to the basal levels obtained from the pGBT8 vector (=1). The data represent average values from three independent experiments in which several colonies were used to initiate the cultures.
Nuclear localization of LAF1
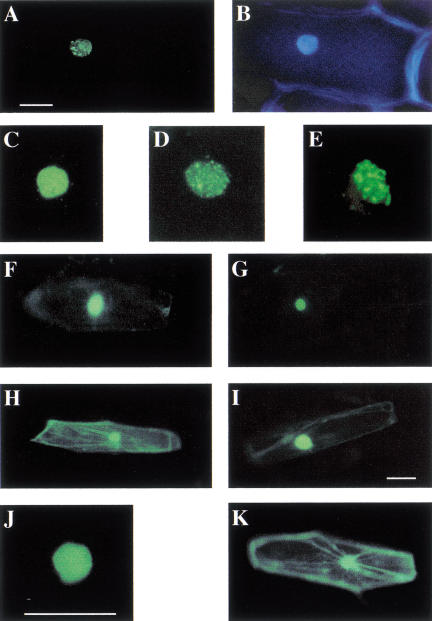
The predicted LAF1 protein contains three potential monopartite nuclear localization signals (Fig. 6). The presence of these signals and the fact that LAF1 is a MYB-type protein with transcriptional activity suggest that LAF1 might function in the nucleus. We used a LAF1–GFP fusion protein to assay for the subcellular localization of the protein. A 35S–LAF1–GFP construct was transfected into onion epidermis cells by particle bombardment, and the treated onion peels were incubated in the dark or in W light. In contrast with the cytoplasmic/nuclear distribution of the GFP protein, which was used as a control, we found that the LAF1–GFP protein is localized in the nucleus irrespective of the light conditions (Figs. 6,7). Moreover, we observed that LAF1–GFP localizes to subnuclear foci (speckles), which are distributed throughout the nucleus.
Figure 6.
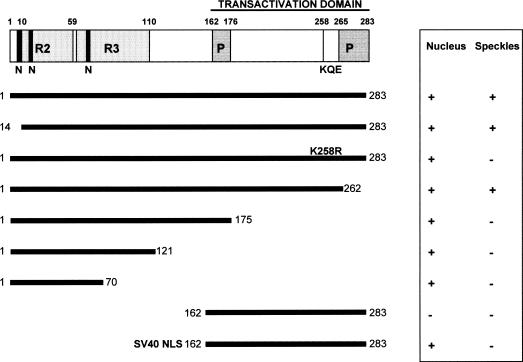
Structure of LAF1–GFP fusion proteins and their respective localization to the nucleus and to nuclear speckles. A schematic diagram of the structure of the LAF1 protein. The MYB domain (R2, R3), the PEST domains (P), the nuclear localization signals (N), the putative sumoylation site (KQE), and the transactivation domain are indicated at the top (numbers indicate amino acid residues). The LAF1 fragments that were fused to the N terminus of GFP are illustrated by bars. SV40-NLS indicates a fusion with the SV40 nuclear localization signal. For analysis of the localization of fusion proteins at least 50 transformed cells were examined in at least three independent transient expression assays by using onion epidermal cells. The table on the right shows the localization of the various GFP fusion proteins (+) positive; (−) negative.
Figure 7.
Representative images of subcellular location of LAF1–GFP fusions in transiently transformed onion epidermal cells. All experiments, except (D) were assayed 6–12 h after bombardment. (A) LAF1/1–283–GFP. (B) The same cell as in (A) stained with DAPI to show the location of the nucleus. (C) LAF1/1–283 3–6 h after bombardment. (D,E) LAF1/1–283 6–12 h after bombardment. (F) LAF1/1–70, (G) LAF1/1–175, (H) LAF1/162–283, (I) SV40-LAF1/162–283, (J) LAF1/1–283 K258R, (K) A control cell expressing GFP alone. Bars in A, I, and J, 50 μm.
This formation of speckles is time dependent. Four to six hours after bombardment of the onion cells with the full-length LAF1–GFP construct, GFP staining was evenly distributed throughout the nucleus (Fig. 7C). Nuclear speckle formation was observed only after ∼8–10 h (Fig. 7D,E). At this time point, in ∼90% of the nuclei examined, the GFP signal was detected exclusively in these foci with no significant background signal elsewhere in the nucleus. Four to eight h later, usually no GFP staining was visible. These results suggest that the formation of speckles might precede degradation of the protein.
To test the functions of the putative NLS and the signals that direct the LAF1–GFP fusion protein to nuclear speckles, we made deletion constructs of LAF1 and fused them to the GFP coding sequence. An N-terminal fragment of LAF1 (LAF1/1–70) containing only the R2 domain was sufficient to direct nuclear localization, but the fusion protein did not accumulate in nuclear speckles (Figs. 6,7F). The same result was obtained for LAF1/1–113, LAF1/1–121, LAF1/1–161, and LAF1/1–175 (Fig. 7G). Only the deletion construct LAF1/1–262 again was able to localize to nuclear speckles (Fig. 6).
The C terminus of LAF1 (amino acids 162–283) fused to GFP was no longer able to localize to the nucleus (Figs. 6,7H), and its distribution throughout the cell resembled the localization of the GFP protein alone (Fig. 7K). The most conserved putative nuclear localization signal (NLS) occurs between amino acids 9 and 12 (RHRK). However, an N-terminal deletion of LAF1 (LAF1/14–283) still was able to localize to the nucleus and to speckles (Fig. 6). Furthermore, this putative NLS (ERHRKG; amino acids 8–13) alone could not direct GFP to the nucleus efficiently (data not shown). Therefore, nuclear localization signals other than the one between amino acids 9–12 are needed to direct LAF1 to the nucleus, and these are located between amino acids 14–70, within the MYB domain R2.
Results from our deletion experiments suggest that sequences between amino acids 176 and 260 are responsible for localization to nuclear speckles. Nonetheless, certain sequences at the N terminus of LAF1 are, in addition, necessary for subnuclear localization, as fusion of an NLS from SV40 (APKKKRKVG; van der Krol and Chua 1991) to LAF1/162–283 caused accumulation in the nucleus but not in speckles (Figs. 6,7I).
Studies in mammalian cells have identified several proteins that localize to nuclear speckles (for review, see Lamond and Earnshaw 1998). In a few cases, for example, PML, it has been shown that these proteins accumulate in speckles only when they are conjugated with a ubiquitin-like protein called SUMO (small ubiquitin-like modifier; Müller et al. 1998). SUMO has been implicated in directing proteins to specific nuclear structures and stabilizing them rather than being involved in protein degradation as is ubiquitin (Melchior 2000; Müller et al. 2001).
We observed a consensus motif for sumoylation (ΨKXE; Melchior 2000; Rodriguez et al. 2001) at amino acid residues 257–260 (KKQE), without the hydrophobic amino acid (Ψ) preceding the target lysine (K). A glycine residue, which is usually in the vicinity of a sumoylation signal, can be found five amino acids upstream of this site. As SUMO usually is conjugated to a lysine residue, we mutated K258 to an arginine in the context of the full-length LAF1 protein fused to GFP. The LAF1 (K258R) mutant localized to the nucleus but not to speckles, suggesting that sumoylation plays a role in the localization of LAF1 to nuclear speckles (Figs. 6,7J).
Discussion
Although phyA is one of the best characterized photoreceptors in higher plants, the protein components involved in the signal transduction pathway are just beginning to emerge (Deng and Quail 1999; Smith 2000; Neff et al. 2000; Fankhauser 2001; Nagy et al. 2001). Isolation and characterization of mutants, especially from Arabidopsis, has proven to be one of the most powerful tools in dissecting the phyA signaling pathway.
laf1 is a novel mutant specific for the phytochrome A signal transduction pathway
Here, we report the genetic identification of a LAF1, which is involved in phyA signal transduction. The most obvious phenotype of laf1 is the reduced inhibition of hypocotyl elongation when germinated under FR light. LAF1 is a specific phyA signaling intermediate as the hypocotyl length is not affected under any other light condition. This is underscored by the molecular analysis, which showed that expression of CHS, CAB, PET E, and XTR7 is only affected in a phyA-dependent way.
The genetic analysis performed here indicates that LAF1 is not allelic to photomorphogenic mutants reported previously. The genetic evidence also indicates that the laf1 mutation is recessive, and that the insertion of a Ds element into the third exon of the gene caused a complete loss-of-function mutation, as no LAF1 transcript can be detected in the mutant.
Nevertheless, laf1 is not completely blocked in phyA signaling, because the hypocotyl elongation is still responsive to FR light and even under low FR light the hypocotyl is not as long as that of a phyA mutant. Under higher FR light fluencies laf1 seedlings are also sensitive to the FR-killing effect, which they are not under low fluencies (<2 μmole/m2 sec). This fluence dependency is confirmed by hypocotyl length analysis (Fig. 2C) and the CAB gene expression pattern (Fig. 3). Furthermore, the laf1 mutant is affected only in a subset of phyA-mediated responses, as under FR light hook opening is not impaired, cotyledons are completely unfolded and expanded (Fig. 1B), and the seedlings show no loss of FR-dependent gravitropism.
As the loss of LAF1 in this null mutant leads to a reduction in phyA signaling, LAF1 must be either an integral component of the transduction pathway or a positive regulator of it. The results suggest that LAF1 could be an element of a pathway that contributes only to some aspects of de-etiolation and needs to act in concert with other pathways for full effect. On the other hand, the partial block of signaling can be explained by the fact that other proteins might have overlapping functions with LAF1; therefore, the loss of LAF1 has only a mild effect on physiological responses. LAF1 appears to be more important at lower FR fluencies compared with high fluencies. LAF1 could be rate limiting at low FR light intensities, but at higher light intensities other factors might be able to substitute. This also might explain why the phenotype is not as strong as in the phyA mutant.
LAF1, a R2R3–MYB protein, acts as a transcriptional activator
The LAF1 gene encodes a protein with sequence homology with the large R2R3–MYB protein family (Romero et al. 1998), corresponding to AtMYB18, previously named by Kranz et al. (1998). The constitutive nuclear localization of transiently expressed LAF1 in onion epidermal cells and the presence of the putative DNA binding domain suggest that LAF1 might act as a transcription factor. We tested whether LAF1 can transactivate a reporter gene when fused to the DNA binding domain of GAL4. The results showed that the domain necessary for transactivation is located in the C terminus of the protein (amino acids 164–283) and especially between amino acid 217 and the C terminus. In other MYB transcription factors, this region also has been shown to be important for transactivation, although no sequence homology is apparent (Lee and Schiefelbein 1999). These results suggest that LAF1 might be involved in the fine tuning of a certain subset of genes as a transcription factor.
Although we understand well how MYB factors bind to target DNA (Rosisnsky and Atchley 1998), little is known about their functions. In vertebrates, for example, many MYB proteins have been found to play an essential regulatory role in cell proliferation and differentiation (Thompson and Ramsay 1995). The functions of MYB genes in plants appear to be far more diverse. Several members have been implicated in the regulation of secondary metabolism, cellular morphogenesis or the control of cell differentiation and cell cycle, signal transduction in plant growth, and responses to hormones, stress, and defense (Jin and Martin 1999).
To our knowledge, this report describes the first R2R3–MYB transcription factor shown to be involved in light signal transduction. So far, few transcription factors have been shown to function in light signaling in Arabidopsis. PIF3 and RSF1/HFR1/REP1 each contain a bHLH motif (Ni et al. 1998; Fairchild et al. 2000; Soh et al. 2000; Spiegelman et al. 2000), CCA1 and LHY1 each contain a single MYB domain (Wang et al. 1997; Schaffer et al. 1998), and HY5 is a bZIP (Oyama et al. 1997). PIF3 was identified as a phytochrome-interacting factor in a yeast two-hybrid screen. Bound to a DNA target site, PIF3 can interact directly with phytochrome in the Pfr form in vitro, suggesting that one mode of phytochrome signal transduction is the direct transcriptional regulation of target genes (Martinez-Garcia et al. 2000). Although PIF3 originally was identified as a protein interacting with both phyA and phyB, more recent biochemical and physiological data suggest that PIF3 plays a more prominent role in phyB signaling than in phyA signaling (Halliday et al. 1999; Zhu et al. 2000). In contrast, based on their respective loss-of-function phenotypes, RSF1/HFR1/REP1 and LAF1 are implicated in phyA signaling, but not phyB signaling.
The effect of the laf1 mutation on CAB, PET E, CHS, and XTR7 gene expression does not allow the distinction whether LAF1 has a direct or indirect impact on the transcription of these genes. The difference in the gene expression pattern of laf1 as compared with WT is more pronounced after 18 h than after 2 h incubation in FR. This suggests that LAF1 might control the sustained transcription of these genes under FR light. A similar observation has been made in the rep1 mutant, where at the 3-h time point the induction of CAB gene expression is similar to WT, but after 6 h the induction is strongly reduced (Soh et al. 2000).
LAF1 localizes to nuclear speckles
Recent studies have shown that besides transcriptional regulation, post-translational modifications such as phosphorylation or protein degradation also play critical roles in the regulation of plant transcription factors (Callis and Vierstra 2000; Hardtke and Deng 2000). One indication that LAF1 accumulation or activity might be regulated is that transgenic plants overexpressing a 35S–LAF1 transgene do not show any obvious phenotype nor any hypersensitivity to FR light, although the LAF1 transcript levels are elevated (Fig. 2C).
One interesting observation is that the LAF1 protein not only localizes to the nucleus, but is directed to distinct nuclear speckles in a time-dependent manner. Nuclear speckles are formed for different reasons. One class of speckles is localized within the interchromatin space and enriched in splicing factors (Lewis and Tollervey 2000). Nuclear speckles also have been implicated in protein modifications caused by SUMO, a small ubiquitin-like modifier, which is conjugated to target proteins by an isopeptide bond (Melchior 2000; Müller et al. 2001). In contrast with ubiquitination, however, the covalent attachment of SUMO does not lead to protein degradation. Only a few SUMO target proteins have been identified, and so far, to our knowledge, none in plants. The exact function of SUMO modification, or sumoylation, is not known. In some cases (e.g., IκBα and p53), conjugation of SUMO could lead to protein stabilization and protection from degradation, whereas in other cases (PML, SP100, RanGAP1, and HIPK2), SUMO conjugation could lead to a different subcellular localization of the modified protein, especially to nuclear speckles (summarized in Melchior 2000; Müller et al. 2001). Surprisingly, many SUMO targets, such as RanGAP1, PML, and HIPK2, contain PEST sequences, which are stretches of at least 12 amino acids rich in P, E, D, S, or T but lacking positively charged amino acids (Rechsteiner and Rogers 1996). In LAF1, two putative PEST sequences can be identified (Fig. 5).
The covalent modification of a target protein by SUMO has been shown to occur at a lysine residue within a minimal consensus sequence, ΨKX(E,D) (Melchior 2000; Rodriguez et al. 2001). LAF1 contains the sequence KKQE (257–260), which is a good match with the consensus sequence, although lacking the hydrophobic amino acid. On changing K258 to R258, thereby disrupting the putative SUMO conjugation site, we observed diffuse nuclear staining and rare speckle formation. These results suggest that recruitment of LAF1 to nuclear speckles requires K258, and this lysine might act as a modification site for SUMO. The domain that is important for nuclear speckle localization is distinct from the nuclear localization signals, which are in the N-terminal portion. However, the C-terminal sequence, containing the domain with the putative sumoylation signal is not sufficient for targeting to nuclear speckles. An SV40 NLS fused to LAF1/171–283 directed the protein to the nucleus but not to speckles. It appears that a functional DNA-binding domain is needed for localization of LAF1 to nuclear speckles.
So far, the only plant proteins that have been identified to localize to speckles are COP1, all phytochromes, CRY2, a blue-light photoreceptor, and RPN6, a component of the proteasome (von Arnim et al. 1998; Mas et al. 2000; Nagy et al. 2001; Peng et al. 2001). COP1 is a RING-finger protein with WD-40 repeats acting as a negative regulator of photomorphogenic development. The COP1 protein has been compared with PML because of their conserved domain structure and similar localization to speckles (Reyes 2001). Analysis of COP1 deletion mutants identified a 50-amino-acid long domain (SNLS) that is necessary for the localization in speckles (Stacey and von Arnim 1999). Although this domain shows no obvious homology with LAF1, it contains a putative sumoylation signal, RKME.
In summary, we present molecular and genetic evidence that LAF1, a nuclear protein containing two MYB motifs, is necessary for a branch of phyA signaling that regulates various photoresponses, including inhibition of hypocotyl elongation as well as CAB, PET E, XTR7, and CHS gene expression. The localization of LAF1 suggests an evolving theme that transcription factors are regulated on the level of protein stability and/or partitioning. Further analysis of the genes that are regulated by LAF1, and the factors that interact with LAF1 should provide important clues for identifying molecular intermediates, which lead from phytochrome photoconversion to alterations in gene expression.
Materials and methods
Plant material, growth conditions, and light sources
A collection of ∼4000 Ds insertion mutants of Arabidopsis thaliana (L.) Heynh. var. Landsberg erecta (generated as described by Sundaresan et al. 1995), was used for genetic screens. The laf1 mutant corresponds to line GT1968. Null mutants of phyA (phyA-201) and phyB (phyB-1) in Ler were used as controls (provided by the Arabidopsis Biological Resource Center). Growth conditions and light sources were described in Bolle et al. (2000). Unless otherwise indicated fluence rates were as follows: FR, 3 μmole/m2 sec; R, 35 μmole/m2 sec; W, 15 μmole/m2 sec. Individual lines were screened as described in Møller et al. (2001). For hypocotyl length measurements, experiments were repeated at least three times, each time measuring more than 20 seedlings per genotype. For Northern blot analysis, 5-day-old etiolated seedlings were either kept in continuous D or transferred into R or FR light for 2 or 18 h.
Genetic analysis
The laf1 mutant was crossed with WT (Ler), and the F2 progeny was analyzed for kanamycin resistance and the laf (long after FR light) phenotype. Cosegregation of the two traits was observed among 120 F2 seedlings, indicating a close linkage of a single Ds element insertion and the laf1 mutation.
Extraction of DNA and RNA
Plant genomic DNA was isolated using the Genomic-tip-100 Kit according to the manufacturer's protocol (QIAGEN). Total RNA was extracted using the RNeasy Plant minikit (QIAGEN), and 10 μg of total RNA was used to isolate poly(A) RNA with the Oligotex kit (QIAGEN).
Isolation of LAF1 cDNA and sequence analysis
DNA sequences flanking the left border of the Ds element were obtained by inverse PCR of genomic DNA from laf1 mutant plants, using the primers and restriction enzymes described in Sundaresan et al. (1995). PCR was performed with Takara LA Taq (Panvera) as recommended by the supplier. DNA samples were amplified using 35 cycles (94 °C for 20 sec, 60 °C for 30 sec, 68 °C for 8 min) followed by elongation at 68 °C for 10 min. A resulting 400-bp long fragment was cloned into pGEM-Teasy vector (Promega) and sequenced. Database searches were performed at the U.S. National Center for Biotechnology Information or Arabidopsis Information Resource (TAIR) with the BLAST program (Altschul et al. 1990) and showed that the Ds element was inserted into the third exon of a MYB transcription factor on chromosome IV (GenBank accession no. Z95744).
A 1018-bp genomic clone and a 852-bp cDNA were amplified from a genomic DNA or a cDNA library, respectively, using primers flanking the ORF predicted in the genome database. The cDNA library was made from Arabidopsis seedlings grown under either FR or W light by using the protocol described by the manufacturer for the Marathon cDNA Amplification Kit (Clontech). RT–PCR using this cDNA library confirmed the correct annotation of the predicted ORF in the genebank. Amino acid alignments were performed using the ClustalW program (DNAstar).
Northern blot analysis
Northern blot analyses were performed as described in Bolle et al. (2000). The bands were quantified by a PhosphorImager (Molecular Dynamics) using 18S RNA as internal standard. As a probe for LAF1, the sequence of the third exon was used as a probe. CHS, CAB, and PET E probes were described in Barnes et al. (1996b). A 300-bp fragment from the 3′ end of the XTR7 cDNA was used to ensure specificity (Xu et al. 1996).
Constructs for complementation and antisense gene
A 35S–LAF1–NOS gene cassette in a binary vector containing a basta-resistance gene (Kost et al. 1998) was used for complementation. Both full-length LAF1 genomic DNA and cDNA were used. Both fragments were generated by using primers to amplify the coding region of LAF1 from the ATG start codon, adding a XhoI site, to the end of the coding region, adding a SpeI site. For the antisense construct, the third exon of the LAF1 gene was expressed in the reverse orientation by using the cauliflower mosaic virus (CaMV) 35S promoter in a binary vector containing a kanamycin-resistance gene (van der Krol and Chua 1991). The constructs were verified by sequencing. The binary vectors were used to transform WT Ler and laf1 plants by vacuum infiltration (after Clough and Bent 1998). T1 transformants were selected on either basta or kanamycin-containing medium, grown to maturity, and selfed. Ten independent transgenic lines were generated with the construct containing the genomic DNA, seven with the cDNA and 10 with the antisense sequence. The presence of the transgene transcript was verified by Northern blot hybridizations. Homozygous T3 seedlings were analyzed for physiological responses.
Transactivation experiments
Full-length LAF1 cDNA and deletion mutants were amplified by PCR, and appropriate restriction sites (5′ BamHI, 3′ NheI) were introduced. PCR was performed using Pfu DNA polymerase (Stratagene) and appropriate primers under the conditions described by the manufacturer. The oligonucleotides used for LAF1/1–283 were (5.1) 5′-CTCTGGATCCATGGCGAAGAC GAAATATGG-3′ and (3.1) 5′-GACCGCTAGCTTACGTCGT TGTTGATGGAG-3′, for Nt (LAF1/1–128) (5.1) and 5′-GACC GCTAGCGGGAAATAGATTTTGCATC-3′, for Ct (LAF1/113–283) (5.2) 5′-CTCTGGATCCAGAAATGGCTCAAGTC TC-3′ and (3.1), for CtA (LAF1/113–177) (5.2) and 5′-GACCGC TAGCATTGATGGCTGTTATTTCCG-3′, for CtB (LAF1/164–232) 5′-CTCTGGATCCCTTCATCTCCCTCACAAGAAAG 3′ and 5′-GACCGCTAGCCTGATCAACATCACCCATTTC 3′, and for CtC (LAF1/217–283) 5′-CTCTGGATCCTGTC GAAGAGACTCTCTCAG-3′ and (3.1). Inserts were fused in-frame to sequences encoding the Gal4 DNA binding domain by cloning them into pGBT8 (Clontech).
We used the yeast strain HF7c, which contains the LacZ and His3 reporter genes under the control of GAL4 17 mers (×3) or GAL1 UAS, respectively (Feilotter et al. 1994). Yeast LiAc-mediated transformation and β-galactosidase liquid culture assays using o-nitrophenyl β-D-galactopyranoside (ONPG) were performed as described in the Clontech Yeast Protocols Handbook (Palo Alto, CA). The synthetic dropout (SD) medium was used either alone or with addition of 40 mg/L histidine (HIS) or 30 mM 3-AT.
Transient expression of GFP fusions in onion cells
The GFP coding sequence (Kost et al. 1998) was fused in-frame to the 3′ end of the LAF1 cDNA. LAF1 cDNA was generated by using primers to amplify the coding region from the ATG start codon (LAF1–ATG Xho 5′-CTCGAGATGGCGAAGACGAAA TATGG-3′) with an additional XhoI site, to the TAA stop codon (LAF1–TGA Kpn 5′-GGTACCCGTCGTTGTTGATGGAG-3′) adding a KpnI site whereas deleting the stop codon. C-terminal deletions of LAF1 were generated by the same method using the LAF1–ATG Xho primer and variable reverse primers: LAF1/1–262 (5′-GGTACCATGATGCTCCTGCTTCTTACATTTGG-3′), LAF1/1–175 (5′-GGTACCGCTGTTATTTCCGTTGCTTTC-3′), and LAF1/1–161 (5′-GGTACCGTTCTCTAGAAGTCTCTGG-3′). To amplify the constructs LAF1/1–70 and LAF1/1–121, we used a 5′ primer that contained an XbaI site instead of an XhoI site, and the 3′ primers were 5′-CTCGAGTGCACTAATCAT ATCCCTCTTTAACC-3′ and 5′-CTCGAGTTGTAAGCTCT GAGACTTGAGC-3′, respectively, introducing an XhoI site. For the N-terminal deletions, variable 5′ primers were used together with LAF1–TGA Kpn: LAF1/14–283 (5′-CTCGAGAT GTTATGGTCACCTGAAGAAGACG-3′) and LAF1/162–283 (5′-CTCGAGATGAAATCTTCATCTCCCTCACAAGAAAGC3′). To generate the LAF1/8–13 fragment, we annealed two oligonucleotides (5′-CTAGATGGAGAGACATAGGAAAGGGC 3′ and 5′-TCGAGCCCTTTCCTATGTCTCTCCAT-3′), encoding the amino acid residues MERHRKG. To generate the SV40-NLS, we annealed the following oligonucleotides: 5′-CTAGAA CAATGGCTCCCAAGAAGAAGAGAAAGGTAC-3′ and 5′-TCGAGTACCTTTCTCTTCTTCTTGGGAGCCATTGTT-3′, encoding the amino acid residues MAPKKKRKVG (van der Krol and Chua 1991). The two NLSs, LAF1/1–70 and LAF1/1–120, were cloned either into a vector containing only the GFP gene (Kost et al. 1998) or upstream of the LAF1/162–283–GFP construct utilizing the XbaI and XhoI restriction sites. The K258R mutation in the context of LAF1/1–283 was generated by using a primer (5′-ATGATGCTCCTGCCTCTTACATTTGGAACC-3′) that introduced the point mutation in the appropriate position and the GeneEditor system (Promega).
Onion epidermal cells were transfected with DNA constructs containing the different LAF1–GFP fusions by using a helium biolistic gun (Kost et al. 1998). Unless stated otherwise, treated epidermal cells were kept in the dark for 6–16 h until the GFP localization was evaluated using an Axioskop microscope (Carl Zeiss).
Acknowledgments
We thank Li-Fang Huang (Rockefeller University) and Wendy B. Gagliano (Cold Spring Harbor Laboratory) for excellent technical assistance and Peter Hare for helpful comments and critical reading of the manuscript. M.L.B. and L.M.L. were supported by postdoctoral fellowships from the Spanish Ministerio de Educación y Ciencia. These studies were supported by an NIH Grant GM-44640 to N.-H.C. and support of the Cold Spring Harbor President's Council, the European Molecular Biology Organization, and the Human Frontiers of Science Program to U.G.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL chua@rockvax.rockefeller.edu; FAX (212) 327-8327.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.915001.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barnes SA, Nishizawa NK, Quaggio RB, Whitelam GC, Chua NH. Far-red light blocks greening of Arabidopsis seedlings via a phytochrome A-mediated change in plastid development. Plant Cell. 1996a;8:601–615. doi: 10.1105/tpc.8.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes SA, Quaggio RB, Whitelam GC, Chua NH. fhy1 defines a branch point in phytochrome A signal transduction pathways for gene expression. Plant J. 1996b;10:1155–1161. doi: 10.1046/j.1365-313x.1996.10061155.x. [DOI] [PubMed] [Google Scholar]
- Bolle C, Koncz C, Chua NH. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes & Dev. 2000;14:1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Neuhaus G, Yamagata H, Chua NH. Cyclic GMP and calcium mediate phytochrome phototransduction. Cell. 1994;77:73–81. doi: 10.1016/0092-8674(94)90236-4. [DOI] [PubMed] [Google Scholar]
- Büche C, Poppe C, Schäfer E, Kretsch T. eid1: A new Arabidopsis mutant hypersensitive in phytochrome A-dependent high-irradiance responses. Plant Cell. 2000;12:547–558. [PMC free article] [PubMed] [Google Scholar]
- Callis J, Vierstra RD. Protein degradation in signaling. Curr Opin Plant Biol. 2000;3:381–386. doi: 10.1016/s1369-5266(00)00100-x. [DOI] [PubMed] [Google Scholar]
- Carabelli M, Sessa G, Baima S, Morelli G, Ruberti I. The Arabidopsis Athb-2 and -4 genes are strongly induced by far-red-rich light. Plant J. 1993;4:469–479. doi: 10.1046/j.1365-313x.1993.04030469.x. [DOI] [PubMed] [Google Scholar]
- Choi G, Yi H, Lee J, Kwon YK, Soh MS, Shin B, Luka Z, Hahn TR, Song PS. Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature. 1999;401:610–613. doi: 10.1038/44176. [DOI] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: The sequences and expression of PHYD and PHYE. Plant Mol Biol. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Deng XW, Quail PH. Signalling in light-controlled development. Semin Cell Dev Biol. 1999;10:121–129. doi: 10.1006/scdb.1999.0287. [DOI] [PubMed] [Google Scholar]
- Dieterle M, Zhou YC, Schäfer E, Funk M, Kretsch T. EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes & Dev. 2001;15:939–944. doi: 10.1101/gad.197201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes & Dev. 2000;14:2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C. The phytochromes, a family of red/far-red absorbing photoreceptors. J Biol Chem. 2001;276:11453–11456. doi: 10.1074/jbc.R100006200. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. RSF1, an Arabidopsis locus implicated in phytochrome A signaling. Plant Physiol. 2000;124:39–45. doi: 10.1104/pp.124.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Yeh KC, Lagarias JC, Zhang H, Elich TD, Chory J. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science. 1999;284:1539–1541. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- Feilotter HE, Hannon GJ, Ruddell CJ, Beach D. Construction of an improved host strain for two hybrid screening. Nucleic Acids Res. 1994;22:1502–1503. doi: 10.1093/nar/22.8.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya M. Phytochromes: Their molecular species, gene families, and functions. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:617–645. [Google Scholar]
- Gilmartin PM, Memelink J, Hiratsuka K, Kay SA, Chua NH. Characterization of a gene encoding a DNA binding protein with specificity for a light-responsive element. Plant Cell. 1992;4:839–849. doi: 10.1105/tpc.4.7.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday KJ, Hudson M, Ni M, Qin M, Quail PH. poc1: An Arabidopsis mutant perturbed in phytochrome signaling because of a T DNA insertion in the promoter of PIF3, a gene encoding a phytochrome-interacting bHLH protein. Proc Natl Acad Sci. 1999;96:5832–5837. doi: 10.1073/pnas.96.10.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Deng XW. The cell biology of the COP/DET/FUS proteins. Regulating proteolysis in photomorphogenesis and beyond? Plant Physiol. 2000;124:1548–1557. doi: 10.1104/pp.124.4.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisada A, Hanzawa H, Weller JL, Nagatani A, Reid JB, Furuya M. Light-induced nuclear translocation of endogenous pea phytochrome A visualized by immunocytochemical procedures. Plant Cell. 2000;12:1063–1078. doi: 10.1105/tpc.12.7.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Xu Y, Quail PH. SPA1: A new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell. 1998;10:19–33. doi: 10.1105/tpc.10.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH. SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science. 1999;284:496–499. doi: 10.1126/science.284.5413.496. [DOI] [PubMed] [Google Scholar]
- Hsieh HL, Okamoto H, Wang M, Ang LH, Matsui M, Goodman H, Deng XW. FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes & Dev. 2000;14:1958–1970. [PMC free article] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH. The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes & Dev. 1999;13:2017–2027. doi: 10.1101/gad.13.15.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol. 1999;41:577–585. doi: 10.1023/a:1006319732410. [DOI] [PubMed] [Google Scholar]
- Kim L, Kircher S, Toth R, Adam E, Schäfer E, Nagy F. Light-induced nuclear import of phytochrome-A:GFP fusion proteins is differentially regulated in transgenic tobacco and Arabidopsis. Plant J. 2000;22:125–133. doi: 10.1046/j.1365-313x.2000.00729.x. [DOI] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schäfer E, Nagy F. Light quality and quantity dependent nuclear import of phytochrome-A and B. Plant Cell. 1999;11:1445–1456. doi: 10.1105/tpc.11.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH. A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 1998;16:393–401. doi: 10.1046/j.1365-313x.1998.00304.x. [DOI] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, et al. Towards functional characterisation of the members of the R2R3–MYB gene family from Arabidopsis thaliana. Plant J. 1998;16:263–276. doi: 10.1046/j.1365-313x.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- Kuno N, Furuya M. Phytochrome regulation of nuclear gene expression in plants. Semin Cell Dev Biol. 2000;11:485–493. doi: 10.1006/scdb.2000.0205. [DOI] [PubMed] [Google Scholar]
- Kuno N, Muramatsu T, Hamazato F, Furuya M. Identification by large-scale screening of phytochrome-regulated genes in etiolated seedlings of Arabidopsis using a fluorescent differential display technique. Plant Physiol. 2000;122:15–24. doi: 10.1104/pp.122.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;99:473–483. doi: 10.1016/s0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Tollervey D. Like attracts like: Getting RNA processing together in the nucleus. Science. 2000;288:1385–1389. doi: 10.1126/science.288.5470.1385. [DOI] [PubMed] [Google Scholar]
- Martin C, Paz-Ares J. MYB transcription factors in plants. Trends Genet. 1997;13:67–73. doi: 10.1016/s0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- Mas P, Devlin PF, Panda S, Kay SA. Functional interaction of phytochrome B and cryptochrome 2. Nature. 2000;408:207–211. doi: 10.1038/35041583. [DOI] [PubMed] [Google Scholar]
- Melchior F. SUMO—Nonclassical ubiquitin. Annu Rev Cell Dev Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- Møller SG, Kunkel T, Chua NH. A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes & Dev. 2001;15:90–103. doi: 10.1101/gad.850101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Matunis MJ, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Hoege C, Pyrowolakis G, Jentsch S. SUMO, ubiquitin's mysterious cousin. Nat Rev Mol Cell Biol. 2001;2:202–210. doi: 10.1038/35056591. [DOI] [PubMed] [Google Scholar]
- Nagy F, Kircher S, Schäfer E. Intracellular trafficking of photoreceptors during light-induced signal transduction in plants. J Cell Sci. 2001;114:475–480. doi: 10.1242/jcs.114.3.475. [DOI] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J. Light: An indicator of time and place. Genes & Dev. 2000;14:257–271. [PubMed] [Google Scholar]
- Neuhaus G, Bowler C, Kern R, Chua NH. Calcium/calmodulin-dependent and -independent phytochrome signal transduction pathways. Cell. 1993;73:937–952. doi: 10.1016/0092-8674(93)90272-r. [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes & Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Staub JM, Serino G, Kwok SF, Kurepa J, Bruce BD, Vierstra RD, Wei N, Deng XW. The cellular level of pr500, a protein complex related to the 19s regulatory particle of the proteasome, is regulated in response to stresses in plants. Mol Biol Cell. 2001;12:383–392. doi: 10.1091/mbc.12.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH, Boylan MT, Parks BM, Short TM, Xu Y, Wagner D. Phytochromes: Photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- Rabinowicz PD, Braun EL, Wolfe AD, Bowen B, Grotewold E. Maize R2R3 Myb genes: Sequence analysis reveals amplification in the higher plants. Genetics. 1999;153:427–444. doi: 10.1093/genetics/153.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1994;104:1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JC. PML and COP1—Two proteins with much in common Trends Biochem. Sci. 2001;26:18–20. doi: 10.1016/s0968-0004(00)01732-1. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- Romero I, Fuertes A, Benito MJ, Malpica JM, Leyva A, Paz-Ares J. More than 80R2R3–MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 1998;14:273–284. doi: 10.1046/j.1365-313x.1998.00113.x. [DOI] [PubMed] [Google Scholar]
- Rosinski JA, Atchley WR. Molecular evolution of the Myb family of transcription factors: Evidence for polyphyletic origin. J Mol Evol. 1998;46:74–83. doi: 10.1007/pl00006285. [DOI] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- Schindler U, Terzaghi W, Beckmann H, Kadesch T, Cashmore AR. DNA binding site preferences and transcriptional activation properties of the Arabidopsis transcription factor GBF1. EMBO J. 1992;11:1275–1289. doi: 10.1002/j.1460-2075.1992.tb05171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, Uchida K, Furuya M. Elementary processes of photoperception by phytochrome A for high-irradiance response of hypocotyl elongation in Arabidopsis. Plant Physiol. 2000;122:147–156. doi: 10.1104/pp.122.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. Phytochromes and light signal perception by plants—An emerging synthesis. Nature. 2000;407:585–591. doi: 10.1038/35036500. [DOI] [PubMed] [Google Scholar]
- Soh MS, Hong SH, Hanzawa H, Furuya M, Nam HG. Genetic identification of FIN2, a far red light-specific signaling component of Arabidopsis thaliana. Plant J. 1998;16:411–419. doi: 10.1046/j.1365-313x.1998.00307.x. [DOI] [PubMed] [Google Scholar]
- Soh MS, Kim YM, Han SJ, Song PS. REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell. 2000;12:2061–2073. doi: 10.1105/tpc.12.11.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman JI, Mindrinos MN, Fankhauser C, Richards D, Lutes J, Chory J, Oefner PJ. Cloning of the Arabidopsis RSF1 gene by using a mapping strategy based on high-density DNA arrays and denaturing high-performance liquid chromatography. Plant Cell. 2000;12:2485–2498. doi: 10.1105/tpc.12.12.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MG, von Arnim AG. A novel motif mediates the targeting of the Arabidopsis COP1 protein to subnuclear foci. J Biol Chem. 1999;274:27231–27236. doi: 10.1074/jbc.274.38.27231. [DOI] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, Ma H, Martienssen R. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes & Dev. 1995;9:1797–1810. doi: 10.1101/gad.9.14.1797. [DOI] [PubMed] [Google Scholar]
- Thompson MA, Ramsay RG. Myb: An old oncoprotein with new roles. Bioessays. 1995;17:341–350. doi: 10.1002/bies.950170410. [DOI] [PubMed] [Google Scholar]
- van der Krol AR, Chua NH. The basic domain of plant B-ZIP proteins facilitates import of a reporter protein into plant nuclei. Plant Cell. 1991;3:667–675. doi: 10.1105/tpc.3.7.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW, Stacey MG. Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene. 1998;221:35–43. doi: 10.1016/s0378-1119(98)00433-8. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell. 1997;9:491–507. doi: 10.1105/tpc.9.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshaar B, Armstrong GA, Block A, da Costa e Silva O, Hahlbrock K. Light-inducible and constitutively expressed DNA-binding proteins recognizing a plant promoter element with functional relevance in light responsiveness. EMBO J. 1991;10:1777–1786. doi: 10.1002/j.1460-2075.1991.tb07702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Campbell P, Vargheese AK, Braam J. The Arabidopsis XET-related gene family: Environmental and hormonal regulation of expression. Plant J. 1996;9:879–889. doi: 10.1046/j.1365-313x.1996.9060879.x. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Tepperman JM, Fairchild CD, Quail PH. Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix-loop-helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc Natl Acad Sci. 2000;97:13419–13424. doi: 10.1073/pnas.230433797. [DOI] [PMC free article] [PubMed] [Google Scholar]