Abstract
To investigate the basis for enhancer–promoter specificity, we compared the ability of enhancers to activate transcription in vivo from core promoters containing either downstream promoter element (DPE) or TATA box motifs. To eliminate position effects, we generated and analyzed pairs of sister Drosophila lines that contain a DPE- or TATA-dependent reporter gene at precisely the same genomic position relative to each enhancer. These studies revealed transcriptional enhancers that are specific for promoters that contain either DPE or TATA box elements. Thus, the core promoter not only mediates the initiation of transcription, but also functions as a regulatory element.
Keywords: Enhancer, transcription, RNA polymerase II, DPE, TATA, core promoter
The regulation of gene expression at the level of transcription is a key control point for many cellular processes. In eukaryotes, there are tens of thousands of protein-coding genes, each of which has its unique program of transcription. The cis-acting DNA sequences that encode these transcriptional programs include transcriptional enhancers, proximal promoters, and core promoters. Enhancers and proximal promoters are recognized by sequence-specific DNA-binding proteins that regulate transcription (see, e.g., Blackwood and Kadonaga 1998; Lee and Young 2000; Lemon and Tjian 2000; Malik and Roeder 2000). Enhancers are often located many kilobase pairs (kbp) upstream or downstream of the transcription start site, whereas proximal promoters are typically within a couple hundred base pairs (bp) of the start site. Core promoters encompass the transcription start site and specify the site of transcription initiation by the basal transcriptional machinery (see, e.g., Orphanides et al. 1996; Smale 1997; White 2001).
The core promoter is at a unique and important position in the transcription process, as it is the eventual target of the action of the many sequence-specific factors and coregulators that control the transcriptional activity of each gene. There are several core promoter elements, including the TATA box, TFIIB recognition element (BRE), Initiator (Inr), and downstream promoter element (DPE). The TATA box is an A/T-rich region that is located about 30 nucleotides (nt) upstream of the transcription start site and is recognized by the TATA-binding protein (TBP) subunit of the TFIID complex. The BRE is located immediately upstream of the TATA box of some TATA-containing promoters, and it increases the affinity of TFIIB for the core promoter (Lagrange et al. 1998). The Inr is a conserved sequence, encompassing the transcription start site, that functions to direct accurate transcription initiation either by itself or in conjunction with a TATA or DPE motif (Smale and Baltimore 1989). The DPE is located about 30 nt downstream of the start site and is recognized by TBP-associated factor (TAF) subunits of TFIID (Burke and Kadonaga 1996, 1997).
There are some intriguing similarities and differences between TATA and DPE elements. Both the TATA and DPE are recognition sites for the binding of TFIID. Core promoters that contain either a TATA or a DPE motif are typically dependent on these elements for transcriptional activity and are hence termed TATA- or DPE-dependent promoters. For basal transcription activity, the addition of a DPE at its normal downstream position to a TATA-dependent core promoter can compensate for the mutation of the TATA box. An analysis of the occurrence of TATA and DPE motifs in Drosophila core promoters revealed that the DPE is about as common as the TATA box, with the majority of core promoters containing either a TATA box or a DPE (Kutach and Kadonaga 2000). Moreover, NC2 (negative cofactor 2; also known as Dr1-Drap1), which had been identified initially as a repressor of TATA-dependent transcription, was found to activate DPE-dependent transcription (Willy et al. 2000).
In this study, we compared the transcriptional properties of TATA and DPE elements in vivo in Drosophila. Specifically, we investigated the ability of transcriptional enhancers to discriminate between core promoters that are dependent on either a TATA box or a DPE motif.
Results and Discussion
Why might a promoter have a DPE or a TATA box? It is possible, for instance, that some transcriptional enhancers might function specifically with either DPE- or TATA-dependent core promoters, as depicted in Figure 1. To test this hypothesis, we generated and analyzed transgenic Drosophila by using a specialized P-element transformation vector, termed the waffle vector (Siegal and Hartl 1996, 1998), in conjunction with enhancer trapping techniques (O'Kane and Gehring 1987; Bellen et al. 1989; Bier et al. 1989). The waffle vector contains two recombination systems (FLP/FRT and Cre/loxP) and was designed for the generation of pairs of sister Drosophila lines by the use of FLP or Cre recombinase in vivo (Fig. 2A). Importantly, each pair of sister lines contains the allelic transgenes at precisely the same genomic position.
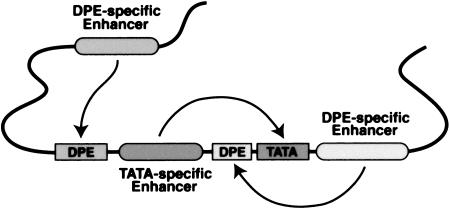
Figure 1.
A model for enhancer-core promoter specificity. This diagram depicts transcriptional enhancers that function specifically with DPE- or TATA-dependent core promoters.
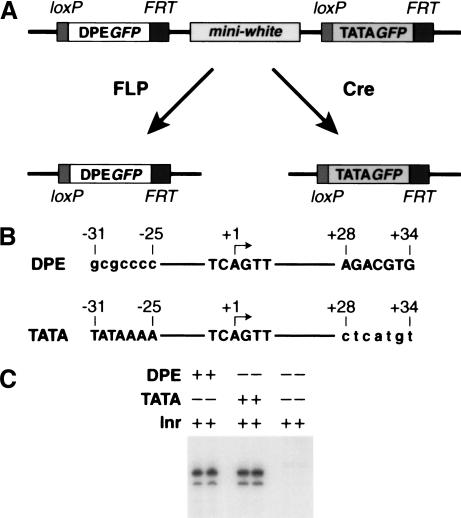
Figure 2.
The use of the waffle vector to generate allelic transgenes that contain DPE- or TATA-dependent reporter genes. (A) Selective excision of the TATA–GFP or DPE–GFP reporter genes from the P-TATA/DPE transposon. The P-TATA/DPE transposon (top) contains both TATA–GFP and DPE–GFP reporter genes. FLP recombinase recognizes the FRT sites and excises the mini-white and TATA–GFP genes. Cre recombinase recognizes the loxP sites and excises the DPE–GFP and mini-white genes. The two resulting sister lines thus contain either DPE–GFP or TATA–GFP at precisely the same genomic position. (B) Sequences of analogous DPE- or TATA-containing core promoters. The segment from the Inr through the DPE is from the Drosophila Antennapedia P2 core promoter, whereas the upstream TATA box is from the adenovirus major late promoter. The two promoters are identical except for 7 bp at the TATA region (−31 to −25) and 7 bp at the DPE region (+28 to +34). The lower case letters denote mutations in the TATA or DPE motifs. (C) In vitro transcription analysis of core promoters containing a DPE, a TATA box, or neither a DPE nor a TATA box.
To use the waffle vector for the comparative analysis of DPE- versus TATA-dependent transcription, we generated analogous TATA- and DPE-containing promoters that were identical except for the presence or absence of TATA and DPE motifs (Fig. 2B). By in vitro transcription analysis, we determined that the DPE- and TATA-containing promoters possess comparable transcriptional activity and that transcription from these promoters is dependent on their respective DPE and TATA motifs (Fig. 2C). Then, the DPE- and TATA-dependent core promoters were used to create DPE–GFP and TATA–GFP reporter genes that were subcloned subsequently into the waffle vector to yield the P-TATA/DPE transposon (Fig. 2A, upper line).
The P-TATA/DPE transposon, which contains the DPE–GFP and TATA–GFP reporter genes with flanking FRT and loxP sites, was introduced into the Drosophila genome by P element-mediated germ-line transformation. Initially, we obtained 29 independent transgenic lines with the P-TATA/DPE transposon and used one of these transgenic fly lines (jb25) as a starting point for an enhancer trap that yielded an additional 114 P-TATA/DPE transposon fly lines. Next, we determined that 76 out of the 143 (29 + 114) lines had the P-TATA/DPE transposon on the third chromosome. We proceeded with these 76 transgenic lines because the subsequent FLP and Cre excision steps would be straightforward with the transposon on the third chromosome. We then identified 54 lines (out of 73 lines tested) with trapped enhancers, as revealed by the presence of GFP transcripts in embryos.
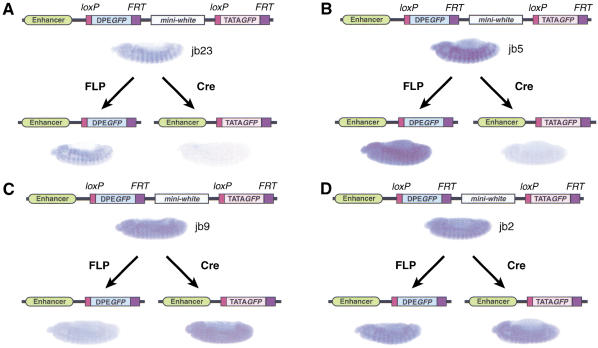
To determine the effects of the trapped enhancers on DPE–GFP relative to TATA–GFP, we performed FLP and Cre excisions, as depicted in Figure 2A, to yield sister lines that contained either DPE–GFP or TATA–GFP at the same genomic position. These experiments revealed transcriptional enhancers that can function specifically with either DPE-dependent or TATA-dependent core promoters. The analysis of 18 pairs of sister lines yielded three DPE-specific enhancers (Fig. 3A,B), one TATA-specific enhancer (Fig. 3C), and 14 apparently nonspecific enhancers (Fig. 3D; data not shown). In each of the experiments shown in Figure 3A–C, the DPE- or TATA-specific enhancer appeared to activate transcription only from its cognate core promoter, as the staining that was seen in the non-cognate line (e.g., DPE-specific enhancer with TATA-dependent promoter, or TATA-specific enhancer with DPE-dependent promoter) was identical to the amount of background staining of wild-type embryos (lacking a GFP gene) that was performed in parallel. It should also be noted that some apparently nonspecific enhancers could be a composite of DPE- and TATA-specific enhancers.
Figure 3.
Identification of DPE- and TATA-specific enhancers. (A,B) The jb23 and jb5 fly lines have DPE-specific trapped enhancers. (C) The jb9 line has a TATA-specific trapped enhancer. (D) The jb2 line has a trapped enhancer that has no apparent specificity for DPE or TATA motifs.
To assess the validity of the enhancer-core promoter specificity, we further analyzed two DPE-specific fly lines (jb23 and jb5), the TATA-specific fly line (jb9), and two of the nonspecific fly lines (jb2 and jb4). First, to determine whether the FLP and Cre recombination events had occurred properly, we performed Southern blot experiments with each set of parental and sister lines and the results were consistent with the expected recombination events as well as with the integration of a single transposon into the genome (data not shown). Second, to test the remote possibility that the core promoter regions of the reporter genes had mutated spontaneously, we amplified the promoter regions of each of the sister lines by PCR and then determined their DNA sequences. These experiments indicated that the core promoter sequences of the DPE–GFP and TATA–GFP reporter genes were not altered. Lastly, by using a PCR technique termed RAGE (rapid amplification of genomic DNA ends; Mizobuchi and Frohman 1993), we determined that the trapped DPE-specific enhancers in the jb23 and jb5 lines were located at different genomic positions (C.Y. Lim, J. Butler, and J. Kadonaga, unpubl.). Hence, we did not trap coincidentally the same enhancer in these two lines.
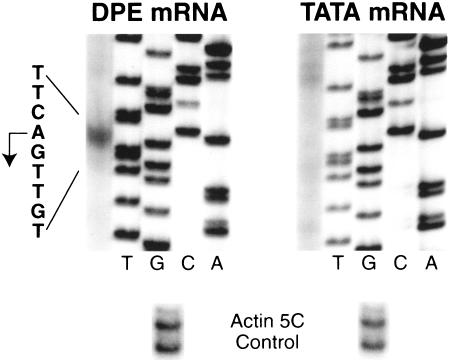
To determine whether the enhancer-specific transcription initiated from the correct start site, we analyzed the locations of the transcription start sites from the DPE–GFP transgenes in jb23 and jb5 by primer extension analysis of polyA+ RNA obtained from embryos. As shown in Figure 4, transcription from the DPE–GFP transgene in the jb23 line does indeed initiate at the correct DPE-dependent start site. In contrast, there was no detectable specific transcription from the TATA–GFP transgene in the sister line. The polyA+ RNA that was used in these primer extension experiments was derived from several thousand embryos; consequently, these data reflect the average amounts of RNA in a population of embryos. Thus, these data also provide a quantitative assessment of the ability of the DPE-specific enhancer to activate transcription from the DPE-dependent core promoter but not from the TATA-dependent core promoter. Identical results were obtained with the jb5 line (data not shown).
Figure 4.
The DPE-specific enhancer in the jb23 fly line initiates transcription from the correct +1 start site in vivo. Poly A+ mRNA was purified from embryos of the DPE and TATA sister lines and then subjected to primer extension analysis. There is no detectable specific transcription from the TATA–GFP reporter gene. As a control for the integrity of the poly A+ RNA, transcripts derived from the Drosophila actin 5C gene were analyzed in parallel, as shown at the bottom.
Therefore, we have identified transcriptional enhancers that act specifically with DPE- or TATA-dependent core promoters. Although specificity between enhancers and promoter regions has been observed (see, e.g., Li and Noll 1994; Merli et al. 1996), it was not determined whether the effects were caused by the proximal promoters (which are bound by sequence-specific factors) or to the core promoters. In other studies, promoter competition experiments revealed that the AE1 and IAB5 enhancers activate transcription preferentially from a TATA-containing promoter relative to a TATA-less promoter (Ohtsuki et al. 1998), but in the absence of competition, both enhancers were able to activate transcription from the TATA-less promoter. Hence, the core-promoter-specific enhancers that have been identified in this study show unique properties that have not been shown previously.
Why might an enhancer need to be specific for a promoter? For example, if there is a cluster of genes (such as those depicted in Fig. 1) that are regulated differentially, then their enhancers would need to function specifically with their cognate promoters. Alternatively, if an enhancer is located tens of kbp away from its cognate promoter, then an additional level of specificity would assist in the establishing the desired interaction between the enhancer and the promoter. It seems likely, however, that many enhancers will not need to activate transcription in a promoter-specific manner. Consistent with this notion, we found that 14 out of the 18 enhancers that we tested did not show specificity for DPE or TATA motifs. Thus, core-promoter-specificity appears to be a special property of a select group of transcriptional enhancers.
This concept of enhancer-core promoter specificity extends beyond the DPE and TATA core promoter motifs. For example, when compared with Sp1, Gal4–VP16 shows a preference for activation of TATA/Inr promoters relative to Inr promoters (Emami et al. 1995). In addition, transcriptional activation by the upstream D‘ element of the terminal deoxynucleotidyltransferase gene (which has a TATA-less, Inr-containing core promoter) was found to depend on the Inr element in the core promoter, even if a TATA box is added to the promoter at the −30 region (Garraway et al. 1996). Hence, it will be important to consider the TATA, BRE, Inr, DPE, and other core promoter motifs in the regulation of enhancer function.
In conclusion, these studies support the notion that the core promoter not only mediates the initiation of transcription by the basal transcriptional machinery, but also functions as a regulatory element. Therefore, in the analysis of gene expression, the core promoter should be considered an active component in the proper regulation of gene activity, rather than a segment of DNA that functions only to direct the accurate initiation of transcription. In the future, it will be interesting to determine the mechanistic basis for enhancer-core promoter specificity. For instance, transcription factors that bind to DPE-specific enhancers might recruit NC2 (Dr1-Drap1), a factor that activates DPE transcription and represses TATA transcription (Willy et al. 2000). From a more global perspective, the functional diversity of the core promoter increases the range of mechanisms by which eukaryotes can control the activity of each of their tens of thousands of genes.
Materials and methods
Construction of the P-TATA/DPE transposon
To construct the P-TATA/DPE transposon, DPE–GFP and TATA–GFP alleles were prepared in pUC119 with DPE- and TATA-dependent minimal core promoters and then the DPE–GFP and TATA–GFP alleles were inserted into the waffle vector (Siegal and Hartl 1996). A detailed description of the construction of the plasmids used in this work is available on request.
Generation and analysis of the sister transgenic fly lines
Germ-line transformants were obtained by co-injection of P-TATA/DPE transposon DNA and P-transposase helper plasmid Δ2,3 into white− embryos (yw67c23/yw67c23). A total of 3758 embryos were injected and 29 independent transformants were obtained. Homozygous stocks were established by repeatedly self-crossing flies with the reddest eyes. Homozygous males were crossed to female yw67c23/yw67c23 to determine if the transposon was integrated into the X chromosome. If the transposon had integrated into the X chromosome, then all the resulting male progeny would have white eyes and all the females would have orange eyes. Three such lines were identified and one line (jb25) was used as the starting point for the enhancer trapping experiments, which were performed by following a modified version of the procedure described by Bier et al. (1989). In addition, 16 of the fly lines from the initial set of transformants were found to have the P-TATA/DPE transposon integrated into the third chromosome and were used in the subsequent FLP and Cre excision steps. Fly lines with the P-TATA/DPE transposon integrated into the third chromosome and a trapped enhancer were subjected to a series of genetic crosses, according to previously described methods (Siegal and Hartl 1996, 1998), to excise either the TATA–GFP allele (with FLP recombinase) or the DPE–GFP allele (with Cre recombinase). GFP transcripts were detected by in situ hybridization analysis with a digoxigenin-UTP-labeled antisense RNA probe (Tautz and Pfeifle 1989; Bergson and McGinnis 1990). Generally, we were not able to detect fluorescence of GFP in the embryos.
Acknowledgments
We thank Bill McGinnis, Dmitry Fyodorov, Vassili Alexiadis, Buyung Santoso, Scott Iyama, Tammy Juven-Gershon, Mark Levenstein, and Tom Boulay for critical reading of the manuscript. We are grateful to Bill McGinnis, Nadine McGinnis, Alexey Veraksa, Mark Siegal, Ethan Bier, Brian Biehs, Annabel Guichard, and Jim Posakony for invaluable advice and assistance with Drosophila techniques; Mark Siegal and Daniel Hartl for the waffle vector and fly lines that express FLP and Cre recombinases; Hong Tran and Cristina Ramones for technical assistance; and Trish Willy, Tom Burke, and Beth Blackwood for insightful discussions and advice. This work was supported by a grant from the NIH to J.T.K. (GM41249). J.E.F.B. is a recipient of an ARCS Graduate Fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jkadonaga@ucsd.edu; FAX (858) 534-0555.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.924301.
References
- Bellen HJ, O’Kane CJ, Wilson C, Grossniklaus U, Pearson RK, Gehring WJ. P-element mediated enhancer detection: A versatile method to study development in Drosophila. Genes & Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- Bergson C, McGinnis W. An autoregulatory enhancer element of the Drosophila homeotic gene Deformed. EMBO J. 1990;9:4287–4297. doi: 10.1002/j.1460-2075.1990.tb07877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E, Vaessin H, Shepherd S, Lee K, McCall K, Barbel S, Ackerman L, Carretto R, Uemura T, Grell E, et al. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes & Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- Blackwood EM, Kadonaga JT. Going the distance: A current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- Burke TW, Kadonaga JT. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes & Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- ————— The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes & Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami KH, Navarre WW, Smale ST. Core promoter specificities of the Sp1 and VP16 transcriptional activation domains. Mol Cell Biol. 1995;15:5906–5916. doi: 10.1128/mcb.15.11.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway IP, Semple K, Smale ST. Transcription of the lymphocyte-specific terminal deoxynucleotidyltransferase gene requires a specific core promoter structure. Proc Natl Acad Sci. 1996;93:4336–4341. doi: 10.1073/pnas.93.9.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutach AK, Kadonaga JT. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol Cell Biol. 2000;20:4754–4764. doi: 10.1128/mcb.20.13.4754-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH. New core promoter element in RNA polymerase II-dependent transcription: Sequence-specific DNA binding by transcription factor IIB. Genes & Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- Lemon B, Tjian R. Orchestrated response: A symphony of transcription factors for gene control. Genes & Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- Li X, Noll M. Compatibility between enhancers and promoters determines the transcriptional specificity of gooseberry and gooseberry neuro in the Drosophila embryo. EMBO J. 1994;13:400–406. doi: 10.1002/j.1460-2075.1994.tb06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci. 2000;25:277–283. doi: 10.1016/s0968-0004(00)01596-6. [DOI] [PubMed] [Google Scholar]
- Merli C, Bergstrom DE, Cygan JA, Blackman RK. Promoter specificity mediates the independent regulation of neighboring genes. Genes & Dev. 1996;10:1260–1270. doi: 10.1101/gad.10.10.1260. [DOI] [PubMed] [Google Scholar]
- Mizobuchi M, Frohman LA. Rapid amplification of genomic DNA ends. Biotechniques. 1993;15:215–216. [PubMed] [Google Scholar]
- Ohtsuki S, Levine M, Cai HN. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes & Dev. 1998;12:547–556. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Kane CJ, Gehring WJ. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes & Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Siegal ML, Hartl DL. Transgene coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics. 1996;144:715–726. doi: 10.1093/genetics/144.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— An experimental test for lineage-specific position effects on alcohol dehydrogenase (Adh) genes in Drosophila. Proc Natl Acad Sci. 1997;95:15513–15518. doi: 10.1073/pnas.95.26.15513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- Smale ST, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- White RJ. Gene transcription: Mechanisms and control. Oxford, UK: Blackwell Science, Ltd.; 2001. [Google Scholar]
- Willy PJ, Kobayashi R, Kadonaga JT. A basal transcription factor that activates or represses transcription. Science. 2000;290:982–984. doi: 10.1126/science.290.5493.982. [DOI] [PubMed] [Google Scholar]