Abstract
Background
Glioblastoma is a complex multifactorial disorder that has swift and devastating consequences. Few genes have been consistently identified as prognostic biomarkers of glioblastoma survival. The goal of this study was to identify general and clinical-dependent biomarker genes and biological processes of three complementary events: lifetime, overall and progression-free glioblastoma survival.
Methods
A novel analytical strategy was developed to identify general associations between the biomarkers and glioblastoma, and associations that depend on cohort groups, such as race, gender, and therapy. Gene network inference, cross-validation and functional analyses further supported the identified biomarkers.
Results
A total of 61, 47 and 60 gene expression profiles were significantly associated with lifetime, overall, and progression-free survival, respectively. The vast majority of these genes have been previously reported to be associated with glioblastoma (35, 24, and 35 genes, respectively) or with other cancers (10, 19, and 15 genes, respectively) and the rest (16, 4, and 10 genes, respectively) are novel associations. Pik3r1, E2f3, Akr1c3, Csf1, Jag2, Plcg1, Rpl37a, Sod2, Topors, Hras, Mdm2, Camk2g, Fstl1, Il13ra1, Mtap and Tp53 were associated with multiple survival events.
Most genes (from 90 to 96%) were associated with survival in a general or cohort-independent manner and thus the same trend is observed across all clinical levels studied. The most extreme associations between profiles and survival were observed for Syne1, Pdcd4, Ighg1, Tgfa, Pla2g7, and Paics. Several genes were found to have a cohort-dependent association with survival and these associations are the basis for individualized prognostic and gene-based therapies. C2, Egfr, Prkcb, Igf2bp3, and Gdf10 had gender-dependent associations; Sox10, Rps20, Rab31, and Vav3 had race-dependent associations; Chi3l1, Prkcb, Polr2d, and Apool had therapy-dependent associations. Biological processes associated glioblastoma survival included morphogenesis, cell cycle, aging, response to stimuli, and programmed cell death.
Conclusions
Known biomarkers of glioblastoma survival were confirmed, and new general and clinical-dependent gene profiles were uncovered. The comparison of biomarkers across glioblastoma phases and functional analyses offered insights into the role of genes. These findings support the development of more accurate and personalized prognostic tools and gene-based therapies that improve the survival and quality of life of individuals afflicted by glioblastoma multiforme.
Background
Glioblastoma multiforme (glioblastoma, World Health Organization grade IV astrocytoma) accounts for 15%-20% of all intracranial tumors and 50% of all brain malignancies [1]. This aggressive malignant type of primary brain tumor has swift and devastating consequences resulting in a median survival after diagnosis of one year [2,3,2]. Primary glioblastoma has a higher incidence in Caucasian men than in other racial and gender groups [4] although these differences may be confounded with differences in access to health care or diagnostic practices [5]. Also, the variation in response to glioblastoma therapies and similar median survival across therapies has prevented the identification of a therapy or therapies directly associated with glioblastoma survival [6-9].
Numerous studies have proposed biomarker genes that can be used to accurately predict the clinical course of glioblastoma [10-16]. Although some genes have been associated with the presence of glioblastoma, few have been identified as prognostic biomarkers of glioblastoma survival and fewer have been confirmed in independent reports. The limited reproducibility of gene-glioblastoma associations may be, in part, due to limited or no consideration of the clinical characteristics of the individuals studied, such as gender and therapy subject [17-19]. Another reason for the lack of confirmation of biomarker genes of glioblastoma may be the consideration of the association between glioblastoma and individual genes independently, although multiple genes acting in unison are known to influence this disease. Statistical reasons for this lack of confirmation include the analysis of gene expression levels in glioblastoma versus non-glioblastoma samples instead of analyzing survival, and the failure to correctly model the censored nature of the observations that may not exhibit the progression or death event by the end of the period considered. For example, The Cancer Genome Atlas Research Network (TCGA [20]) identified gene expression aberrations among the 206 glioblastoma cases considered but did not consider the age at glioblastoma death or progression, nor the clinical characteristics of the individuals studied.
The goal of this study was to identify general and clinical-dependent biomarker genes and biological processes of three complementary events: lifetime, overall and progression-free glioblastoma survival. A novel analytical strategy was developed to identify general and cohort-dependent associations between the biomarkers and the three glioblastoma events. Cross-validation and functional analysis further supported the identified biomarkers. The identification of gene biomarkers of glioblastoma survival supports the efficient follow-up studies using in vitro and in vivo experiments and augments the molecular toolbox that can be used to classify patients across and within cohort groups with respect to prognosis and the development of targeted treatments.
Methods
Data
Clinical and gene expression information from 320 individuals diagnosed with glioblastoma was obtained from the TCGA repository (September 2009 data freeze [21]. Protocols for specimen preparation and gene expression measurements are described in detail in the report by The Cancer Genome Atlas Research Network [20]. Briefly, a retrospective search in glioblastoma sample banks identified newly diagnosed glioblastoma cases based on surgical pathology reports and clinical records. Only samples that had demographic, clinical and pathological information, a minimum of 80% tumor nuclei, and a maximum of 50% necrosis, qualified for gene expression analyses.
The data included glioblastoma diagnostic and death records between the years 1989 and 2009. Clinical factors used to classify individuals into cohort groups were Gender (Male or Female), Race (White Caucasian or Other), Therapy received (R = radiation alone; CRnoT = chemo, radiation and not targeted therapy plus other therapy if present; CRT = chemo plus radiation and targeted therapy only; Other = any other combination of radiation, chemo, targeted, immune and hormonal therapy; or None = no therapy), and detection of glioblastoma progression or recurrence (ProgRec - Yes/No).
Three glioblastoma time-to-event variables were considered: lifetime survival (encompassing the period from birth to death), overall or post-diagnosis survival (encompassing the period from glioblastoma diagnosis to death) and post-diagnosis progression-free survival (encompassing the period from glioblastoma diagnosis to progression of glioblastoma or to recurrence of glioblastoma). The distribution of the observations among the levels of the clinical or cohort variables is presented in Table 1. There were 287 individuals with sufficient survival information for analysis. Three individuals were excluded from the progression-free survival analysis because of inconsistency in the dates for diagnosis and progression or recurrence.
Table 1.
Median length of the hazard period and (relative) frequency of the individuals across clinical cohort levels
| Lifetime survival (n = 287)1 |
Overall survival (n = 287)2 |
Progression-free survival (n = 284) 2 |
||||
|---|---|---|---|---|---|---|
| Censored | (0.12) | (0.12) | (0.23) | |||
| Lifetime | 59.2 | 13.1 | 6.5 | |||
| Gender | ||||||
| Male | 58.8 | (0.63) | 13.6 | (0.63) | 6.6 | (0.63) |
| Female | 61.1 | (0.37) | 12.1 | (0.37) | 6.4 | (0.37) |
| Race | ||||||
| White | 59.7 | (0.77) | 13.1 | (0.77) | 6.8 | (0.77) |
| Other | 57.7 | (0.23) | 12.6 | (0.23) | 5.3 | (0.23) |
| Therapy | ||||||
| CRnoT | 57.7 | (0.40) | 15.7 | (0.40) | 8.0 | (0.34) |
| R | 60.7 | (0.35) | 12.3 | (0.35) | 5.3 | (0.41) |
| CRT | 53.4 | (0.10) | 15.4 | (0.10) | 6.8 | (0.10) |
| Other | 64.8 | (0.08) | 14.2 | (0.08) | 7.9 | (0.08) |
| None | 70.5 | (0.07) | 2.9 | (0.07) | 1.4 | (0.07) |
| ProgRec | ||||||
| Yes | 57.6 | (0.77) | 15.1 | (0.77) | - | (0.77) |
| No | 64.8 | (0.23) | 5.9 | (0.23) | - | (0.23) |
1 expressed in years;
2 expressed in months.
Gene expression measurements were obtained using the Affymetrix HT HG-U133A platform, comprising 22,277 probe sets. The gene expression measurements were obtained in ten experimental batches, in which the percentage of individuals per batch ranged from 4.35% to 21.25%. For samples with multiple gene expression measurements, the correlation between measurements across microarrays was higher than 0.98 and, thus, the average expression was used to represent the sample. Raw expression data was log2 transformed and normalized using quantile normalization and GC-RMA [22] approaches implemented in Beehive [23].
In addition to detecting genes in the microarray platform associated with the glioblastoma survival, particular attention was given to genes known to be associated with glioblastoma and the association detected in this study. A list, including 123 genes known to be associated with glioblastoma were identified from the literature [20,24-27] and 51 genes in the KEGG glioma pathway [28], was compiled (see Additional file 1).
Statistical Analysis
A five-step approach was used to reduce the dimensionality of the data set caused by the large number of probes and few records of the individuals in this experiment. First, a Cox proportional hazards survival analysis [29] was undertaken for each non-control probe in the microarray platform. The model included all the clinical variables with the profile of only one probe. This step allowed the selection of probes associated with each of the three survival variables at P-value < 0.01. This mild threshold was used to minimize the chances of false negative associations and evaluate in subsequent steps probes with strong or moderate associations with glioblastoma per se. The number of probes identified for lifetime survival, overall, and progression-free survival was 963, 839, and 1048 respectively. Second, for each one of the three glioblastoma time-to-event variables, the clinical variables and all remaining probes identified in the first step were included simultaneously in a Cox survival model. In this manner, the clinical variables were fixed component of the model and the probes associated with the survival variables were selected using a combination of forward and stepwise model selection methods. The forward selection method was used to add one probe at a time to the model containing the clinical variables using a significance level for entry of 30%. In the complementary stepwise selection method, the inclusion of probes followed the same rules as in the forward method but a probe only remained in the model if its P-value was lower than the significance levels for stay of 10%. Since these two selection methods could result in different models, a second stepwise selection was performed using the significant probes. This step allowed identifying broad or general associations between probe profiles and glioblastoma survival. Third, the interaction between the remaining probes and clinical variables was evaluated using the stepwise approach. This step permitted the detection of clinical or cohort-dependent associations between probe profiles and glioblastoma survival. The fourth stage of our approach aimed to select the significant probes from our list of 174 known genes associated with glioblastoma (see Additional file 1) fitting the probes and interaction with the clinical variables using the stepwise selection method. The consideration of the known probes alone aimed at minimizing the potential masking of associations by other probes in the model. Similarly to the previous step, in the final step the probes identified from both sets of analyses were combined and further streamlined using the stepwise method. This final step allowed the confirmation of prior probes associations reported in previous studies as well as the identification of novel associations. With respect to P-value threshold selection at each stage, a lenient first-stage threshold was used to capture most true positive associations at the expense of some false positives. The more stringent threshold used in the subsequent steps and repeated selection process minimized the number of false positives remaining in the index. Thus, this approach would have the same effect than reducing the threshold in the first step with the added benefit of minimizing the loss of true positives. Likewise, extending the first-stage threshold would have resulted in more false positives being considered in the second stage and higher risks of overparameterization.
In addition to a P-value, each probe had a hazard ratio (HR) estimate and associated 95% confidence interval limits. Hazard ratios below 1 indicate that the hazard under consideration decreases as the level of the gene increases. The proportional hazards assumption was assessed for the final predictive models corresponding to each survival variable based on the residuals. There was no evidence of departure from the assumptions for all the models reported. The association between survival and clinical and probe expression profiles was visualized by plotting the probability of survival predicted by the Cox model against time. For depiction purposes, individuals were divided into low and high probe expression groups that corresponded to the 25th and 75th percentile respectively given the median expression for all other probes in the predictive model. The survival curves were computed based on the information used to identify the significant gene associations. Biomarker genes resulting from the multi-stage approach were compared to previous reports of genes associated with glioblastoma or other cancers. The protein interaction resource at the NCBI Gene data base [30] was used to check that biomarkers not previously associated with cancer were also not indirectly associated with cancer through intermediate genes.
The genes identified by the five-step approach were compared to those resulting from a more conventional analysis using a one-step Cox survival analysis with a stringent cut-off (P-value < 0.0001).
Functional and Gene Network Analyses
Identification of Gene Ontology (GO) categories (molecular function and biological process) and KEGG pathways represented among the significant genes associated with each glioblastoma survival variable was undertaken [31,32]. The representation of genes in the GO and KEGG pathway classes was evaluated using Fisher's exact (two-tailed) test and False Discovery Rate multiple test adjustment [33]. The relationships between the biomarker genes were further studied for the three glioblastoma survival variables and significant functional categories. The BisoGenet plug-in [34] from the Cytoscape software [35] was used to build and visualize the networks for each one of the three glioblastoma survival variables using the respective list of significant genes from the GO categories. All the available data sources in BisoGenet (including BIOGRID, DIP, BIND and others) were selected to generate the interactions. To facilitate the visualization of the networks, only interactions (edges) connecting two significant genes (nodes) directly or through an intermediate gene were depicted.
Cross-validation
The associations between gene profiles and survival detected in this study were confirmed using a three-fold approach. First, a leave-one-out cross-validation (LOOCV) approach [36-38]. LOOCV is specially recommended in data sets of limited size, providing an almost unbiased estimator and identifying the same best classifiers as other X-fold training-test data partitions [38,39]. Validation of the predictive survival equation and biomarkers detected in a training data set on an independent test data set is desirable, followed by X-fold cross-validation on a particular data set. The representation of all cohort factors on both the training and test sets is necessary for unbiased evaluation of the biomarkers and to ensure that the detected biomarkers were not a spurious artifact of ignored cohort effects and for a fair evaluation of the training estimates. Consideration of race is particular critical for the validation of biomarkers detected in this study because lack of adjustment for this cohort factor could result in the identification of associations that are due to genetic background and not the particular gene expression profile.
For the X-fold validation approach, the specification of suitable training and testing data sets would have required at least 200 patients in each data set (5 individuals × 2 races × 2 genders × 5 therapies × 2 recurrence groups) and only 287individuals were available. The minimum of 5 individuals per group minimizes the risk of confounding between individual variation and cohort variation. Use of smaller data sets would have lead to low power and biased findings because of the ill-representation of individuals across cohort groups. Thus, the X-fold cross-validation could not be implemented. Likewise, the test of the predictive hazard equations (that include cohort factors) on an independent data set could not be implemented due to the lack of dataset with comparable cohort information or adequate structure that would minimize the risk of confounding between factors.
Accurate validation of associations between biomarkers and survival was attained using LOOCV discriminant analysis[40] that allows the assessment of the performance of biomarkers to classify individuals into high and low hazard (low and high survival, respectively). The same cohort information was used to obtain parameter estimates and to train the predictive hazard equations. For each survival variable, the median length of the period considered (age at death for lifetime survival; months from diagnosis to death for overall survival and; months from diagnosis to progression/recurrence for progression free survival) was calculated, and individuals were classified into either a high or low hazard group based on the median. The 20% of the individuals that had a length of period closest (higher or lower) to the median were not considered in order to minimize borderline cases that could affect the assessment of the model performance. Only non-censored records were used in the cross-validation analysis to favor unbiased classification. All individuals except for one were used to develop a new hazard index using the biomarkers previously detected and the new index was used to classify the remaining individuals. This leave-one-individual-out analysis was repeated for all individuals and the observed and predicted affiliations of the individuals to the high and low groups for each hazard were compared in order to assess the correct assignment rate.
Second, in addition to LOOCV, confirmation of the genes associated with the three glioblastoma hazards was investigated on the independent database REMBRANDT (REpository for Molecular BRAin Neoplasia DaTa) [41,42]. This database includes gene expression and survival information on 181 individuals diagnosed with glioblastoma. Third, a literature review was undertaken to identify independent studies that have reported associations between the genes associated with survival detected in this study and glioblastoma or other cancer types.
Results
Confirmed and Novel Biomarkers of Glioblastoma
The median length of the periods associated with lifetime, overall, and progression-free survival across and within clinical or cohort group are presented in Table 1. The age of the individuals at death or at the end of the considered period ranged from 14 to 87 years with a median age of 60 years. The median survival length was 59 years, 13 months and 7 months for lifetime, overall, and progression-free survival, respectively.
A total of 168 significant associations between expression profiles and glioblastoma survival (61, 47 and 60 associations for lifetime, overall, and progression-free survival, respectively) from 139 genes were identified. Among these, 10 associations are borderline significant (0.1 < P-value < 0.05) and are included in the tables in support of other more significant associations.
The vast majority of the genes associated with glioblastoma survival have been previously reported to be associated with glioblastoma (35, 24, and 35 genes, respectively) or with another cancer (10, 19, and 15 genes, respectively) and the rest (16, 4, and 10 genes, respectively) exhibited novel associations with glioblastoma. Table 2 presents the distribution of genes and probes associated with more than one hazard. Cohort-independent and cohort-dependent associations, respectively, were uncovered for lifetime (Tables 3 and 4), overall (Tables 5 and 6), and progression-free (Tables 7 and 8) survival. Cohort-independent associations represented 90%, 96% and 92% of the significant gene associations for lifetime, overall, and progression-free survival, respectively.
Table 2.
Genes and probes represented more than one time within or across the glioblastoma survival events
| Gene | Lifetime Survival | Overall Survival | Progression-free survival |
|---|---|---|---|
| Actr2 | 200727_s_at | ||
| 200729_s_at | |||
| Akr1c3 | 209160_at | 209160_at | |
| App | 211277_x_at | ||
| 214953_s_at | |||
| Camk2b | 211483_x_at | ||
| 209956_s_at | |||
| Camk2g | 212757_s_at | 214322_at | |
| 214322_at | |||
| Cdc42 | 208727_s_at | ||
| 208728_s_at | |||
| 214230_at | |||
| Chi3l1 | 216546_s_at | ||
| 209396_s_at | |||
| Csf1 | 207082_at | 209716_at | |
| E2f3 | 203692_s_at | 203693_s_at | 203693_s_at |
| Egfr | 211551_at | ||
| 211607_x_at | |||
| Fstl1 | 208782_at | 208782_at | |
| Hras | 212983_at | 212983_at | |
| Ighg1 | 211908_x_at | ||
| 211693_at | |||
| Il13ra1 | 210904_s_at | 211612_s_at | |
| Jag2 | 32137_at | 209784_s_at | |
| Mdm2 | 217373_x_at | 217373_x_at | |
| Mtap | 204956_at | 204956_at | |
| Pik3r1 | 212240_s_at | 212249_at | 212239_at |
| Plcg1 | 216551_x_at | 216551_x_at | |
| Prkcb | 207957_s_at | ||
| 209685_s_at | |||
| Rpl37a | 213459_at | 213459_at | |
| Sod2 | 221477_s_at | 215078_at | |
| Timp3 | 201148_s_at | ||
| 201150_s_at | |||
| Topors | 204071_s_at | 204071_s_at | |
| Tp53 | 211300_s_at | 211300_s_at |
Table 3.
Genes that have a general association (P-value < 0.05) with the lifetime glioblastoma survival
| Gene Symbol | Probe Identifier | P-value | Hazard Ratio1 | Relevant literature references |
|---|---|---|---|---|
| Syne1 | 209447_at | <.0001 | 0.17 (0.10-0.32) | [60]O |
| E2f3 | 203692_s_at | <.0001 | 0.26 (0.15-0.44) | [28]G |
| Fstl1 | 208782_at | <.0001 | 0.31 (0.22-0.42) | [25]G |
| Ep300 | 213579_s_at | <.0001 | 0.34 (0.29-0.57) | [26]G |
| Gigyf2 | 212261_at | <.0001 | 0.39 (0.26-0.58) | n/a |
| Topors | 204071_s_at | <.0001 | 0.41 (0.29-0.59) | [26]G |
| Chst4 | 220446_s_at | 0.09892 | 0.44 (0.17-1.16) | [89]O |
| Sar1a | 201543_s_at | <.0001 | 0.44 (0.29-0.66) | n/a |
| Il13ra1 | 210904_s_at | <.0001 | 0.47 (0.36-0.60) | [24]G |
| Sod2 | 221477_s_at | <.0001 | 0.47 (0.37-0.59) | [25]G |
| Rab15 | 221810_at | <.0001 | 0.48 (0.34-0.69) | n/a |
| Timm23 | 218118_s_at | 0.0239 | 0.50 (0.27-0.91) | n/a |
| Kcnj4 | 208359_s_at | <.0001 | 0.50 (0.38-0.66) | n/a |
| Rpl37a | 213459_at | 0.0023 | 0.51 (0.33-0.79) | [90]G |
| Camk2g | 214322_at | 0.0135 | 0.53 (0.32-0.88) | [56]G |
| Plcg1 | 216551_x_at | 0.0068 | 0.55 (0.35-0.85) | [26]G |
| Slc43a3 | 213113_s_at | 0.0004 | 0.56 (0.40-0.77) | n/a |
| Cdc42 | 208727_s_at | <.0001 | 0.57 (0.45-0.73) | [26]G |
| Csf1 | 207082_at | 0.0092 | 0.58 (0.38-0.88) | [26]G |
| Ccnb2 | 202705_at | 0.0118 | 0.60 (0.40-0.89) | [91]G |
| Tlk2 | 212997_s_at | 0.0004 | 0.64 (0.49-0.82) | n/a |
| Mtap | 204956_at | 0.0091 | 0.67 (0.49-0.91) | [26]G |
| Egfr | 211551_at | 0.07432 | 0.68 (0.45-1.04) | [24]G |
| Akt2 | 211453_s_at | 0.0292 | 0.68 (0.48-0.96) | [86]G |
| Akr1c3 | 209160_at | <.0001 | 0.70 (0.62-0.81) | [26]G |
| Tp53 | 211300_s_at | 0.0215 | 0.76 (0.60-0.96) | [25]G |
| Igf1 | 209541_at | 0.0183 | 0.76 (0.61-0.95) | [26]G |
| Rpl10 | 221989_at | 0.0392 | 0.80 (0.64-0.99) | [24]G |
| Arhgef4 | 205109_s_at | 0.06472 | 0.80 (0.64-1.01) | n/a |
| Cdc42 | 214230_at | 0.05542 | 0.82 (0.67-1.00) | [26]G |
| Chi3l1 | 216546_s_at | 0.061 | 0.87 (0.75-1.00) | [70]G |
| Ppbp | 214146_s_at | 0.012 | 1.16 (1.03-1.30) | n/a |
| Cdkn2a | 209644_x_at | 0.0003 | 1.18 (1.08-1.29) | [92]G |
| Wdr67 | 214061_at | 0.0237 | 1.30 (1.03-1.63) | [93]O |
| Tspyl5 | 213122_at | 0.0003 | 1.34 (1.14-1.56) | n/a |
| Usf2 | 215737_x_at | <.0001 | 1.42 (1.19-1.69) | [94]O |
| Camk2g | 212757_s_at | 0.0078 | 1.54 (1.12-2.13) | [56]G |
| Pik3r1 | 212240_s_at | 0.0022 | 1.67 (1.20-2.32) | [20]G |
| Akt1 | 207163_s_at | 0.0005 | 1.70 (1.26-2.30) | [84]O |
| Rac2 | 213603_s_at | 0.0001 | 1.74 (1.31-2.31) | [95]G |
| Six6 | 207250_at | <.0001 | 1.82 (1.45-2.28) | [96]O |
| Spg21 | 217827_s_at | 0.0387 | 1.91 (1.03-3.52) | n/a |
| Wdyhv1 | 219060_at | 0.0015 | 1.95 (1.29-2.94) | n/a |
| Uros | 203031_s_at | 0.0067 | 2.37 (1.27-4.42) | n/a |
| Lin7c | 219399_at | 0.0002 | 2.40 (1.51-3.80) | [97]O |
| Ros1 | 207569_at | <.0001 | 2.58 (1.73-3.85) | [98]O |
| Cdk2 | 204252_at | <.0001 | 2.74 (1.78-4.21) | [91]G |
| Jag2 | 32137_at | <.0001 | 2.78 (1.86-4.14) | [24]G |
| Kiaa0090 | 212395_s_at | <.0001 | 2.89 (1.87-4.47) | n/a |
| Ccnb1 | 214710_s_at | <.0001 | 3.16 (2.00-4.98) | [91]G |
| Scn5a | 207413_s_at | <.0001 | 3.21 (1.79-5.74) | n/a |
| Col14a1 | 212865_s_at | <.0001 | 3.30 (1.93-5.63) | [99]O |
| Hoxa10 | 213147_at | <.0001 | 3.30 (1.93-5.65) | [100]O |
| Cdc42 | 208728_s_at | <.0001 | 3.94 (2.12-7.32) | [26]G |
| Pdcd4 | 202731_at | <.0001 | 4.68 (3.01-7.28) | [26]G |
n/a, No association with any type of cancer found in literature;
1 Hazard ratio estimate (95% confidence interval);
2 Borderline significant (P-value < 0.1) included for completeness;
G Gene confirmed in an independent glioblastoma multiforme study; the number between square brackets denotes the corresponding literature reference;
O Gene confirmed in an independent study on any other type of cancer; the number between square brackets denotes the corresponding literature reference.
Table 4.
Genes that have a cohort-dependent association (P-value < 0.05) with the lifetime glioblastoma survival
| Gene Symbol | Probe Identifier | Clinical Cohort | P-value | Level of Clinical Cohort | Hazard Ratio1 | Relevant literature references |
|---|---|---|---|---|---|---|
| Prkcb2 | 207957_s_at | Gender | <.0001 | Male | 0.36 (0.24-0.55) | [28]G |
| Female | 1.27 (0.84-1.93) | |||||
| Therapy | 0.0006 | None | 0.38 (0.25-0.60) | |||
| CRnoT | 0.51 (0.36-0.73) | |||||
| R | 0.64 (0.46-0.88) | |||||
| CRT | 0.71 (0.44-1.13) | |||||
| Other | 0.75 (0.43-1.32) | |||||
| Sox10 | 209843_s_at | Race | 0.0018 | White | 0.55 (0.44-0.68) | [10]G |
| Other | 1.08 (0.72-1.62) | |||||
| Egfr | 211607_x_at | Gender | <.0001 | Male | 0.60 (0.50-0.72) | [24]G |
| Female | 0.88 (0.74-1.04) | |||||
| Chi3l1 | 209396_s_at | Therapy | 0.0006 | CRT | 1.27 (0.96-1.70) | [70]G |
| R | 1.28 (1.07-1.52) | |||||
| Other | 1.31 (1.04-1.66) | |||||
| CRnoT | 1.53 (1.31-1.79) | |||||
| None | 2.42 (1.56-3.75) | |||||
| C2 | 203052_at | Gender | 0.0033 | Female | 1.30 (1.03-1.65) | n/a |
| Male | 1.93 (1.56-2.39) | |||||
| Prkcb | 209685_s_at | Gender | <.0001 | Female | 1.31 (0.79-2.14) | [28]G |
| Male | 5.21 (3.16-8.61) |
n/a, No association with any type of cancer found in literature;
1 Hazard ratio estimate (95% confidence interval);
2 Interaction with a single clinical cohort factor;
G Gene confirmed in an independent glioblastoma multiforme study; the number between square brackets denotes the corresponding literature reference;
O Gene confirmed in an independent study on any other type of cancer; the number between square brackets denotes the corresponding literature reference.
Table 5.
Genes that have a general association (P-value < 0.05) with the overall glioblastoma survival
| Gene Symbol | Probe Identifier | P-value | Hazard Ratio1 | Relevant literature references |
|---|---|---|---|---|
| Tgfa | 205015_s_at | 0.0002 | 0.12 (0.04-0.37) | [28]G |
| Sirpa | 202895_s_at | <.0001 | 0.24 (0.14-0.41) | [26]G |
| Ctbp2 | 210835_s_at | <.0001 | 0.28 (0.16-0.48) | [101]O |
| Eef1e1 | 213907_at | <.0001 | 0.37 (0.23-0.61) | [102]O |
| Mapk3 | 212046_x_at | 0.0041 | 0.43 (0.24-0.76) | [103]O |
| Actr2 | 200727_s_at | <.0001 | 0.43 (0.29-0.63) | [24]G |
| Igh@ | 211637_x_at | 0.0167 | 0.44 (0.23-0.86) | n/a |
| Plcg1 | 216551_x_at | <.0001 | 0.46 (0.31-0.68) | [26]G |
| Mgat3 | 209764_at | <.0001 | 0.51 (0.37-0.71) | [26]G |
| Lrp10 | 201412_at | 0.0041 | 0.60 (0.42-0.85) | [24]G |
| Idh1 | 201193_at | 0.0051 | 0.60 (0.42-0.86) | [24]G |
| Tmem8b | 207839_s_at | <.0001 | 0.60 (0.46-0.77) | n/a |
| Ccna2 | 203418_at | <.0001 | 0.60 (0.49-0.75) | [104]O |
| Topors | 204071_s_at | 0.0007 | 0.61 (0.46-0.81) | [26]G |
| Rpl37a | 213459_at | 0.0164 | 0.66 (0.47-0.93) | [90]G |
| Mdm2 | 217373_x_at | <.0001 | 0.69 (0.61-0.78) | [26]G |
| E2f3 | 203693_s_at | 0.06722 | 0.75 (0.55-1.02) | [28]G |
| Mdfic | 211675_s_at | 0.0006 | 0.78 (0.68-0.90) | [105]O |
| Sod2 | 215078_at | <.0001 | 0.80 (0.73-0.88) | [25]G |
| Akr1c3 | 209160_at | 0.0014 | 0.83 (0.73-0.93) | [26]G |
| Thbs4 | 204776_at | 0.0007 | 1.18 (1.07-1.30) | [106]O |
| Shc3 | 206330_s_at | 0.0031 | 1.32 (1.10-1.59) | [28]G |
| Pik3r1 | 212249_at | 0.0145 | 1.34 (1.06-1.69) | [20]G |
| Nkx2-5 | 206578_at | 0.0027 | 1.38 (1.12-1.70) | [107]O |
| Hras | 212983_at | 0.0187 | 1.42 (1.06-1.90) | [85]G |
| Bhlhb9 | 213709_at | 0.0192 | 1.42 (1.06-1.92) | [108]O |
| C9orf95 | 219147_s_at | 0.0004 | 1.43 (1.17-1.73) | [109]O |
| C17orf101 | 219254_at | 0.0085 | 1.46 (1.10-1.94) | [110]O |
| Nol3 | 59625_at | <.0001 | 1.46 (1.21-1.76) | [111]O |
| Rangap1 | 212125_at | 0.0225 | 1.47 (1.06-2.05) | [26]G |
| Ftsj2 | 222130_s_at | 0.017 | 1.48 (1.07-2.05) | [112]O |
| Rrm1 | 201476_s_at | 0.0006 | 1.49 (1.19-1.87) | [27]G |
| Jag2 | 209784_s_at | 0.0351 | 1.63 (1.03-2.57) | [24]G |
| Tnpo1 | 212635_at | 0.0054 | 1.89 (1.20-2.96) | n/a |
| Myo7a | 211103_at | 0.0033 | 1.97 (1.25-3.10) | [113]O |
| Actr2 | 200729_s_at | 0.0001 | 2.18 (1.47-3.23) | [24]G |
| Csf1 | 209716_at | <.0001 | 2.33 (1.65-3.27) | [26]G |
| Ank1 | 208352_x_at | 0.0003 | 2.38 (1.49-3.82) | [24]G |
| B3galnt1 | 211379_x_at | <.0001 | 2.40 (1.76-3.28) | [114]O |
| Kras | 214352_s_at | 0.002 | 2.44 (1.38-4.31) | [85]G |
| Ewsr1 | 210012_s_at | 0.0005 | 2.49 (1.49-4.15) | [26]G |
| Sec24c | 202361_at | <.0001 | 2.84 (1.76-4.60) | n/a |
| Rpl10l | 217559_at | <.0001 | 2.95 (1.83-4.74) | [115]O |
| Ighg1 | 211908_x_at | 0.0007 | 3.41 (1.68-6.93) | [116]O |
| Ighg1 | 211693_at | 0.0007 | 4.33 (1.86-10.04) | [116]O |
n/a, No association with any type of cancer found in literature;
1 Hazard ratio estimate (95% confidence interval);
2 Borderline significant (P-value < 0.1) included for completeness;
G Gene confirmed in an independent glioblastoma multiforme study; the number between square brackets denotes the corresponding literature reference;
O Gene confirmed in an independent study on any other type of cancer; the number between square brackets denotes the corresponding literature reference.
Table 6.
Genes that have a cohort-dependent association (P-value < 0.05) with the overall glioblastoma survival
| Gene Symbol | Probe Identifier | Clinical Cohort | P-value | Level of Clinical Cohort | Hazard Ratio1 | Relevant literature references |
|---|---|---|---|---|---|---|
| Polr2d | 214144_at | Therapy | 0.0044 | Other | 0.35 (0.18-0.70) | [117]O |
| R | 0.50 (0.31-0.81) | |||||
| CRT | 0.68 (0.38-1.21) | |||||
| None | 0.77 (0.43-1.39) | |||||
| CRnoT | 0.93 (0.58-1.5) | |||||
| Igf2bp3 | 203820_s_at | Gender | 0.0146 | Female | 1.02 (0.84-1.24) | [118]O |
| Male | 1.29 (1.12-1.49) |
n/a, No association with any type of cancer found in literature;
1 Hazard ratio estimate (95% confidence interval);
G Gene confirmed in an independent glioblastoma multiforme study; the number between square brackets denotes the corresponding literature reference;
O Gene confirmed in an independent study on any other type of cancer; the number between square brackets denotes the corresponding literature reference.
Table 7.
Genes that have a general association (P-value < 0.05) with the progression-free glioblastoma survival
| Gene Symbol | Probe Identifier | P-value | Hazard Ratio1 | Relevant literature references |
|---|---|---|---|---|
| Pla2g7 | 206214_at | <.0001 | 0.11 (0.05-0.23) | [119]O |
| Pdgfb | 216061_x_at | <.0001 | 0.18 (0.09-0.35) | [28]G |
| Calm2 | 207243_s_at | 0.0011 | 0.22 (0.09-0.54) | [28]G |
| Timp3 | 201148_s_at | <.0001 | 0.26 (0.16-0.41) | [26]G |
| Agpat1 | 215535_s_at | <.0001 | 0.27 (0.15-0.51) | n/a |
| Ifngr1 | 202727_s_at | <.0001 | 0.32 (0.19-0.53) | [27]G |
| Pvr | 214444_s_at | <.0001 | 0.32 (0.19-0.54) | [26]G |
| Ndufv1 | 208714_at | 0.0002 | 0.33 (0.19-0.59) | n/a |
| Fgfr2 | 211401_s_at | <.0001 | 0.39 (0.26-0.57) | [26]G |
| E2f3 | 203693_s_at | <.0001 | 0.42 (0.29-0.60) | [28]G |
| Pold2 | 201115_at | 0.0007 | 0.43 (0.27-0.70) | [26]G |
| Calm3 | 200622_x_at | 0.0001 | 0.43 (0.28-0.67) | [28]G |
| Tp53 | 211300_s_at | <.0001 | 0.43 (0.29-0.64) | [25]G |
| Raf1 | 201244_s_at | 0.0141 | 0.46 (0.25-0.85) | [26]G |
| Pknox2 | 219046_s_at | <.0001 | 0.46 (0.33-0.62) | [120]O |
| App | 214953_s_at | 0.0016 | 0.47 (0.30-0.75) | [24]G |
| Fstl1 | 208782_at | <.0001 | 0.47 (0.35-0.62) | [25]G |
| Camk2b | 211483_x_at | 0.0008 | 0.54 (0.38-0.77) | [28]G |
| Pten | 204053_x_at | 0.0003 | 0.60 (0.46-0.79) | [26]G |
| Mdm2 | 217373_x_at | <.0001 | 0.66 (0.57-0.76) | [26]G |
| Ccnd1 | 208711_s_at | 0.0273 | 0.80 (0.66-0.98) | [28]G |
| Hspa1a/Hspa1b | 202581_at | 0.0529 | 0.82 (0.67-1. 00) | [24]G |
| Cd24 | 208650_s_at | 0.0069 | 0.85 (0.75-0.95) | [121]O |
| Clec2b | 209732_at | 0.06452 | 1.19 (0.99-1.44) | [122]O |
| Cav2 | 203324_s_at | 0.0024 | 1.25 (1.08-1.44) | [123]O |
| Snx10 | 218404_at | <.0001 | 1.34 (1.16-1.57) | [124]O |
| Wee1 | 215711_s_at | 0.0083 | 1.37 (1.08-1.74) | [27]G |
| Hras | 212983_at | 0.08022 | 1.49 (0.95-2.33) | [85]G |
| Mns1 | 219703_at | 0.0053 | 1.51 (1.13-2.02) | n/a |
| Ppp1r15a | 37028_at | 0.011 | 1.54 (1.10-2.15) | n/a |
| App | 211277_x_at | 0.08112 | 1.56 (0.95-2.56) | [24]G |
| Fadd | 202535_at | 0.09342 | 1.57 (0.93-2.65) | [125]O |
| Pik3r1 | 212239_at | 0.018 | 1.60 (1.08-2.36) | [20]G |
| Mmp14 | 217279_x_at | 0.0182 | 1.66 (1.09-2.52) | [24]G |
| Mtap | 204956_at | 0.0016 | 1.66 (1.21-2.27) | [26]G |
| Il13ra1 | 211612_s_at | 0.0003 | 1.72 (1.28-2.32) | [24]G |
| Kcnj13 | 210179_at | 0.0235 | 1.74 (1.08-2.82) | [126]O |
| Clip3 | 212358_at | 0.0022 | 1.75 (1.22-2.50) | n/a |
| Aanat | 207225_at | 0.0114 | 1.78 (1.14-2.79) | [127]O |
| Camk2g | 214322_at | 0.0024 | 1.86 (1.24-2.78) | [56]G |
| Prkca | 215195_at | 0.0005 | 1.90 (1.32-2.73) | [28]G |
| Kdm6b | 41386_i_at | 0.0003 | 2.03 (1.39-2.96) | n/a |
| Zfy | 207246_at | 0.0016 | 2.06 (1.31-3.22) | [128]O |
| Smarcb1 | 212167_s_at | 0.0004 | 2.06 (1.38-3.07) | [26]G |
| Utp20 | 209725_at | <.0001 | 2.08 (1.46-2.98) | n/a |
| Igl@ | 211655_at | 0.0209 | 2.22 (1.13-4.38) | [129]O |
| Atf5 | 204998_s_at | <.0001 | 2.31 (1.72-3.11) | [130]G |
| Shox | 207570_at | <.0001 | 2.66 (1.73-4.07) | [24]G |
| Loc283079 | 215929_at | 0.0071 | 2.73 (1.31-5.69) | n/a |
| Ung | 202330_s_at | 0.0001 | 2.79 (1.66-4.68) | [27]G |
| Hnrnpd | 213359_at | <.0001 | 2.94 (1.91-4.52) | n/a |
| Camk2b | 209956_s_at | <.0001 | 3.02 (2.13-4.29) | [28]G |
| Timp3 | 201150_s_at | <.0001 | 3.10 (1.88-5.11) | [26]G |
| Nras | 202647_s_at | <.0001 | 3.93 (2.60-5.95) | [28]G |
| Paics | 214664_at | <.0001 | 5.28 (3.13-8.91) | [66]O |
n/a, No association with any type of cancer found in literature;
1 Hazard ratio estimate (95% confidence interval);
2 Borderline significant (P-value < 0.1) included for completeness;
G Gene confirmed in an independent glioblastoma multiforme study; the number between square brackets denotes the corresponding literature reference;
O Gene confirmed in an independent study on any other type of cancer; the number between square brackets denotes the corresponding literature reference.
Table 8.
Genes that have a cohort-dependent association (P-value < 0.05) with progression-free glioblastoma survival
| Gene Symbol | Probe Identifier | Clinical Cohort | P-value | Level of Clinical Cohort | Hazard Ratio1 | Relevant literature references |
|---|---|---|---|---|---|---|
| Gdf10 | 206159_at | Gender | 0.0317 | Male | 0.37 (0.23-0.60) | n/a |
| Female | 0.80 (0.45-1.42) | |||||
| Vav3 | 218807_at | Race | 0.008 | Other | 0.41 (0.29-0.59) | [81]G |
| White | 0.68 (0.55-0.85) | |||||
| Rps20 | 216246_at | Race | 0.0003 | Other | 0.75 (0.39-1.44) | [75]O |
| White | 1.83 (1.03-3.24) | |||||
| Rab31 | 217764_s_at | Race | <.0001 | White | 1.47 (0.93-2.30) | [74]O |
| Other | 7.72 (3.71-16.07) | |||||
| Apool | 213289_at | Therapy | 0.0026 | R | 1.64 (1.13-2.38) | [131]O |
| None | 1.93 (0.63-5.98) | |||||
| CRnoT | 2.23 (1.55-3.20) | |||||
| Other | 3.86 (1.83-8.13) | |||||
| CRT | 4.82 (2.69-8.63) |
n/a, No association with any type of cancer found in literature;
1 Hazard ratio estimate (95% confidence interval);
G Gene confirmed in an independent glioblastoma multiforme study; the number between square brackets denotes the corresponding literature reference
O Gene confirmed in an independent study on any other type of cancer; the number between square brackets denotes the corresponding literature reference.
The five-step approach was consistently superior to a one-step Cox analysis with more stringent P-value < 0.001 on all three survival indicators. For the three variables studied; lifetime, overall, and progression-free survival, the simpler approach identified 60, 71 and 67 probes of which 19, 17, and 23 respectively overlapped with the corresponding 61, 47, and 60 probes identified in the five-step analyses of the three survival indicators. Of the 139 probes identified by the simpler approach and not identified by our approach, the vast majority (123 probes across all three variables) have not been associated with glioblastoma and could not be confirmed.
Genes Associated with Lifetime Death Hazard
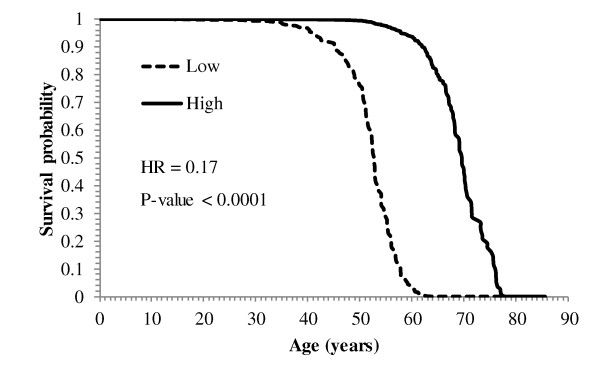
Sixty-one gene profiles, representing 55 genes, were associated with lifetime survival. An increase in the level of expression of 31 genes was associated with a decrease in HR, with estimates ranging from 0.17 (Syne1) to 0.87 (Chi3l1). The changes in survival across levels of gene expression and clinical variables for the population under consideration were visualized using survival plots. The decline on the probability of lifetime survival across age (in years) for individuals with high (75th percentile) and low (25th percentile) levels of Syne1 is depicted in Figure 1. Consistent with the hazard ratio estimate (HR = 0.17, P-value < 0.0001), the probability of survival of individuals with high levels of Syne1 remains higher across age. Individuals with high and low levels of Syne1 have a survival probability of 50% at 69 and 52 years of age, respectively. The opposite trend was observed in the remaining 24 profiles that have hazard ratio estimates ranging from 1.16 (Ppbp) to 4.7 (Pdcd4).
Figure 1.
Probability of lifetime glioblastoma survival across age for spectrin repeat containing, nuclear envelope 1 (Syne1). Probability of glioblastoma survival across age for individuals with Low (25th percentile) and High (75th percentile) expression level of spectrin repeat containing, nuclear envelope 1 (Syne1). With a lower hazard estimate (HR = 0.17), the probability of survival of individuals with high levels of Syne1 remains higher across age.
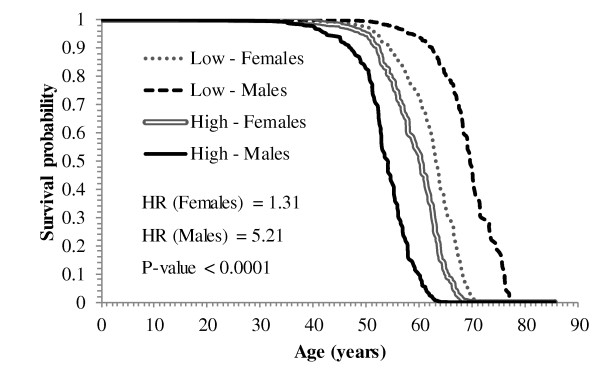
Among the genes exhibiting cohort-dependent associations with lifetime survival (Table 4), the drop in the probability of lifetime survival across age (in years) for females and males with high (75th percentile) and low (25th percentile) levels of Prkcb 209685_s_at is portrayed in Figure 2. Consistent with the hazard ratio estimates for females (HR = 1.31) and males (HR = 5.21), the probability of survival declines faster in males with high levels of Prkcb than females with low levels of this gene.
Figure 2.
Probability of lifetime glioblastoma survival across age in females and males for protein kinase, C beta (Prkcb). Probability of glioblastoma survival across age for Females and Males with Low (25th percentile) and High (75th percentile) expression level of protein kinase, C beta (Prkcb). Consistent with the hazard ratio estimates for females (HR = 1.31) and males (HR = 5.21), the probability of survival in individuals with high levels of Prkcb declines before than in individuals with lower levels of Prkcb. Due to the significant interaction between the expression of Prkcb and gender, the probability of survival for females with high level of the gene declines faster than the probability of survival for males with low level of the gene.
Genes Associated with Overall Survival
A total of 45 genes were associated with overall survival (Tables 5 and 6). Among the cohort-independent associations, an increase in the levels of 20 genes was associated with a decrease in overall hazard with HR ranging from 0.12 (Tgfa) to 0.83 (Akr1c3). On the other hand, an increase in the level of 25 genes was associated with an increase in overall hazard with HR ranging from 1.18 (Thbs4) to 4.33 (Ighg1). Among the cohort-dependent associations, the hazard increased more in males (HR = 1.29) than in females (HR = 1.02) per unit increase in the levels of Igf2bp3.
Genes Associated with Progression-free Survival
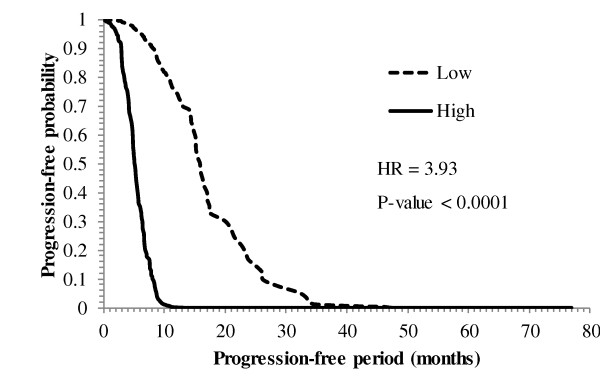
Of the 60 probes (corresponding to 57 genes) associated with progression-free survival, 55 had general associations and 5 had cohort-dependent associations (Tables 7 and 8). Among the genes that have cohort-independent associations, an increase in the level of 23 genes was associated with a decrease in HR, ranging from 0.11 (Pla2g7) to 0.85 (Cd24). For the remaining 32 genes, an increase in the level of expression was associated with an increase in the progression-free HR ranging from 1.19 (Clec2b) to 5.28 (Paics). The decline in the progression-free survival probability across time (in months) for individuals with high (75th percentile) and low (25th percentile) levels of neuroblastoma RAS viral (v-RAS) oncogene homolog (Nras) is depicted in Figure 3. Consistent with the hazard ratio estimate (HR = 3.93, P-value < 0.0001), the progression-free survival probability falls faster in individuals with high expression levels of Nras. With regard to the cohort-dependent association with progression-free survival, an increase in the expression of Gdf10 was associated with a higher decrease of the hazard ratio in males (HR = 0.37) than in females (HR = 0.80).
Figure 3.
Progression-free probability across post-diagnosis months for neuroblastoma RAS viral (v-RAS) oncogene homolog (Nras). Progression-free probability across post-diagnosis months for individuals with Low (25th percentile) and High (75th percentile) expression level of neuroblastoma RAS viral (v-RAS) oncogene homolog (Nras). With a high hazard estimate (HR = 3.93), the progression-free probability falls faster in individuals with high expression levels of Nras.
Gene That Have Multiple Probes and Hazard Ratios
When multiple probes of the same gene had opposite associations with the glioblastoma hazard (e.g. HR > 1 for probe 1 and HR < 1 for probe 2), the disagreements were resolved by assessing the dependability of each probe. Information on dissenting probes is briefly summarized here. Probe 214322_at, of Cam2kg, was obtained from an ovary EST and thus is less reliable in respect to brain cancer than probe 212757_s_at. Probe 208728_s_at, of Cdc42, is expected to be more reliable than 208727_s_at because the former was obtained from an mRNA sequence that has double the length than the later. Probe 200729_s_at, of Actr2, corresponds to an mRNA and is more reliable than 200727_s_at, which corresponds to an EST from mixed tissues. Probe 210904_s_at, of Il13ra1, corresponds to a cluster of mRNA assigned to this gene in the NCBI-Gene database; meanwhile, probe 211612_s_at pertains to a single mRNA not assigned to the gene and is considered less reliable. Other probes include 201148_s_at of Timp3, which corresponds to a pancreatic EST, 200727_s_at of Actr2, which corresponds to a mixed tissue EST, and 209956_s_at of Camk2b, which corresponds to a proline rich sequence.
Functional and Gene Network Analyses
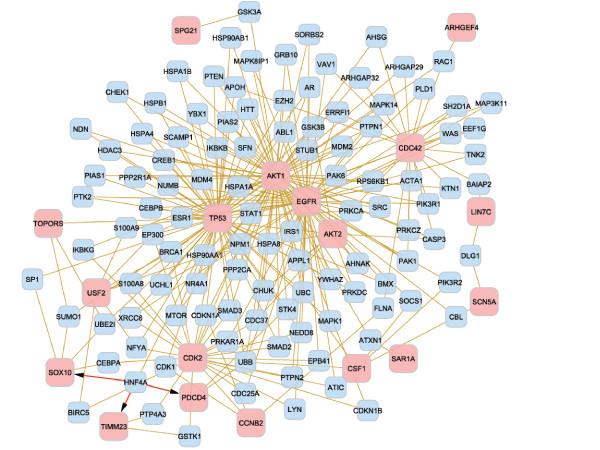
The GO categories enriched (FDR adjusted P-value < 0.1, ≥ 3 genes/category) among the genes associated with each of the three glioblastoma survival variables are summarized in Tables 9, 10, and 11. The functional analysis revealed nine, two and ten biological processes enriched among the genes associated with lifetime, overall, and progression-free survival respectively, and three molecular functions enriched among the genes associated with progression-free survival. The biological processes of cell cycle (GO:0007049) and death (GO:0016265) were over-represented among the genes associated with the lifetime and progression-free survivals. The gene networks for the significant genes from the functional analyses associated with lifetime, overall, and progression-free survival are depicted in Figures 4, 5 and 6, respectively.
Table 9.
Gene Ontology categories enriched among the genes associated with lifetime glioblastoma survival1
| Gene Ontology | Level | Term | P-value | FDR adjusted P-value | Number of genes | Genes |
|---|---|---|---|---|---|---|
| Biological process | 3 | aging (GO:0007568) | 2.13E-05 | 1.62E-03 | 3 | Pdcd4, Cdkn2a, Tp53 |
| regulation of biological process (GO:0050789) | 2.77E-04 | 1.50E-02 | 20 | Usf2, Cdkn2a, Ccnb2, Akt2, Tp53, Cdc42, Six6, Jag2, Lin7c, Pdcd4, Csf1, Topors, Spg21, Akt1, Egfr, Sox10, C2, Scn5a, Arhgef4, Cdk2 | ||
| protein localization (GO:0008104) | 1.49E-03 | 3.77E-02 | 7 | Topors, Akt1, Sar1a, Egfr, Timm23, Tp53, Lin7c | ||
| cell division (GO:0051301) | 2.04E-03 | 3.88E-02 | 3 | Ccnb2, Cdc42, Cdk2 | ||
| cell cycle (GO:0007049) | 3.75E-03 | 5.14E-02 | 7 | Pdcd4, Egfr, Cdk2, Cdkn2a, Ccnb2, Tp53, Jag2 | ||
| nitrogen compound metabolic process (GO:0006807) | 4.06E-03 | 5.14E-02 | 4 | Chst4, Akt1, Egfr, Chi3l1 | ||
| cell proliferation (GO:0008283) | 8.32E-03 | 8.99E-02 | 6 | Csf1, Topors, Egfr, Cdk2, Tp53, Jag2 | ||
| death (GO:0016265) | 9.46E-03 | 8.99E-02 | 6 | Akt1, Cdkn2a, Tp53, Jag2, Pdcd4, Topors | ||
| 4 | cell aging (GO:0007569) | 4.38E-06 | 9.42E-04 | 3 | Pdcd4, Cdkn2a, Tp53 | |
1 Only GO categories with False Discovery Rate (FDR) adjusted P-value < 0.1 and represented by three or more genes.
Table 10.
Gene Ontology categories enriched among the genes associated with overall glioblastoma survival
| Gene Ontology | Level | Term | P-value | FDR Adjusted P-value | Number of genes | Genes |
|---|---|---|---|---|---|---|
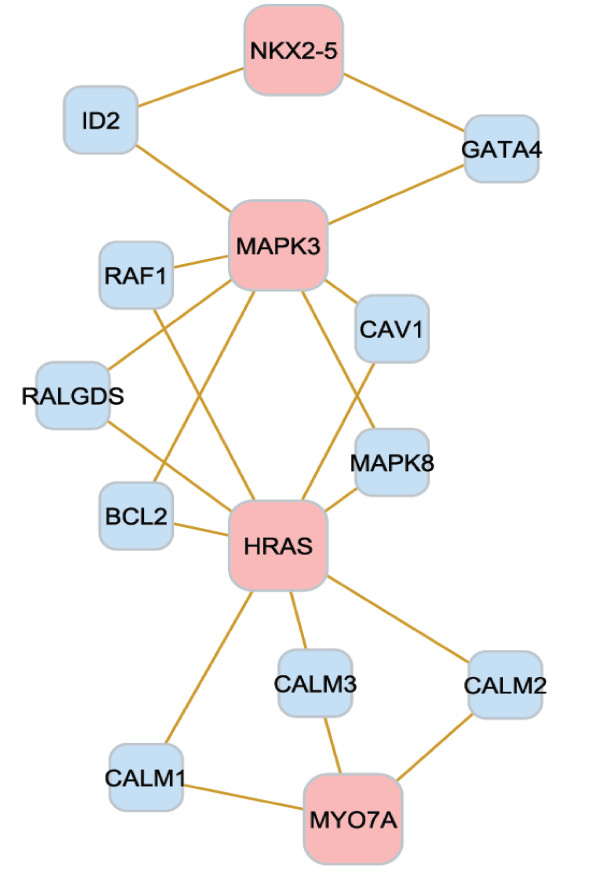
| Biological process | 4 | anatomical structure morphogenesis (GO:0009653) | 5.39E-05 | 1.16E-02 | 9 | Nkx2-5, Csf1, Mapk3, Tgfa, Thbs4, Jag2, Igf2bp3, Myo7a, Hras |
| 6 | organ morphogenesis (GO:0009887) | 3.83E-06 | 2.40E-03 | 7 | Nkx2-5, Csf1, Mapk3, Tgfa, Jag2, Myo7a, Hras | |
1 Only GO categories with False Discovery Rate (FDR) adjusted P-value < 0.1 and represented by three or more genes.
Table 11.
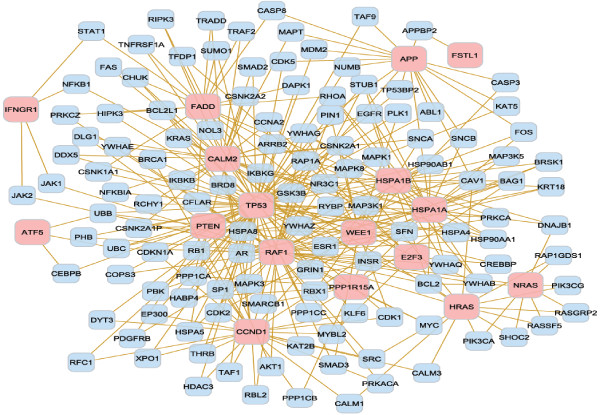
Gene Ontology categories enriched among the genes associated with progression-free survival
| Gene Ontology | Level | Term | P-value | FDR Adjusted P-value | Number of genes | Genes |
|---|---|---|---|---|---|---|
| Biological process | 3 | cell cycle (GO:0007049) | 3.89E-06 | 2.96E-04 | 11 | Hras, Ppp1r15a, App, Calm2, Atf5, Pten, E2f3, Wee1, Tp53, Ccnd1, Nras |
| death (GO:0016265) | 3.18E-04 | 1.21E-02 | 92 | App, Raf1, Atf5, Pten, Fadd, Hspa1a/Hspa1b, Tp53, Ppp1r15a | ||
| response to biotic stimulus (GO:0009607) | 4.11E-03 | 5.13E-02 | 62 | Fadd, Hspa1a/Hspa1b, Clec2b, Ccnd1, Ifngr1 | ||
| response to abiotic stimulus (GO:0009628) | 8.17E-03 | 6.90E-02 | 62 | Fadd, Hspa1a/Hspa1b, Clec2b, Ccnd1, Ifngr1 | ||
| 4 | cell cycle process (GO:0022402) | 4.26E-06 | 9.16E-04 | 10 | App, Atf5, Pten, E2f3, Wee1, Tp53, Ccnd1, Nras, Hras, Ppp1r15a | |
| 5 | regulation of cell cycle (GO:0051726) | 2.90E-07 | 1.22E-04 | 10 | Wee1, Tp53, Ccnd1, App, Nras, Hras, Ppp1r15a, Atf5, Pten, E2f3 | |
| 6 | regulation of progression through cell cycle (GO:0000074) | 1.47E-07 | 9.24E-05 | 10 | Tp53, Ccnd1, Nras, Hras, Ppp1r15a, App, Atf5, Pten, E2f3, Wee1 | |
| cell death (GO:0008219) | 2.13E-04 | 6.18E-02 | 92 | Ppp1r15a, App, Raf1, Atf5, Pten, Fadd, Hspa1a/Hspa1b, Tp53 | ||
| 7 | programmed cell death (GO:0012501) | 1.19E-04 | 2.23E-02 | 92 | Ppp1r15a, App, Raf1, Atf5, Pten, Fadd, Hspa1a/Hspa1b, Tp53 | |
| 8 | apoptosis (GO:0006915) | 1.41E-04 | 5.21E-02 | 92 | Ppp1r15a, App, Raf1, Atf5, Pten, Fadd, Hspa1a/Hspa1b, Tp53 | |
| Molecular function | 3 | pattern binding (GO:0001871) | 3.26E-04 | 3.55E-02 | 3 | Fstl1, App, Fgfr2 |
| carbohydrate binding (GO:0030246) | 1.26E-03 | 6.89E-02 | 4 | Fstl1, App, Fgfr2, Clec2b | ||
| 4 | polysaccharide binding (GO:0030247) | 3.01E-04 | 8.70E-02 | 3 | Fstl1, App, Fgfr2 | |
1 Only GO categories with False Discovery Rate (FDR) adjusted P-value < 0.1 and represented by three or more genes;
2 Although Hspa1a/Hspa1b are represented by the same probe (202581_at), these isoforms are counted as two units.
Figure 4.
Gene network from the functional analysis of lifetime glioblastoma survival. Interaction between the significant genes from the functional analysis of lifetime glioblastoma death. The gold edges represent protein interactions whereas the red edges represent interaction of the HNF4A protein with the DNA of the genes Pdcd4, Sox10 and Timm23. Of the 24 genes from Table 9, 18 (pink nodes) interact among each other in a direct way or through an intermediate gene (blue nodes).
Figure 5.
Gene network from the functional analysis of overall glioblastoma survival. Relationship between the significant genes from the functional analysis of overall survival. Of the nine genes from Table 10, four (pink nodes) interact among each other in a direct way or through an intermediate gene (blue nodes).
Figure 6.
Gene network from the functional analysis of progression-free survival. Relationship between the significant genes from the functional analysis of progression-free survival. Of the 19 genes from Table 11, 17 (pink nodes) interact among each other in a direct way or through an intermediate gene (blue nodes).
Cross-validation
The performance of the gene sets as reliable prognosticators of the three glioblastoma survival variables was evaluated. The generalization capability of the biomarker index was tested in individuals other than those used to develop a hazard index using a leave-one-individual-out discriminant analysis. Individuals were predicted to pertain to the high or low glioblastoma hazard groups for each event using the predictive biomarker index, and the prediction was compared to the observed classification based on the length of the period corresponding to each event. For both, lifetime and overall hazard, the number of observed high and low individuals was 100, and the number of predicted high and low individuals was 97 and 103, respectively. For the progression-free hazard, the number of observed high and low individuals was 87 and 88, respectively, and the number of predicted high and low individuals was 83 and 92, respectively.
Additional evaluation of the results was pursued by comparing the genes associated with the three glioblastoma hazards identified in this study and the target genes of microRNAs associated with the glioblastoma hazard reported by Delfino et al. [43]. One third of the sequences identified in this study are putative targets of microRNAs associated with glioblastoma. A hypergeometric test confirmed that the overlap between the genes uncovered in this study and the target genes was significant (P-value ≤ 0.005).
Discussion
The data set analyzed offered a suitable representation of the general population of glioblastoma cases. The median overall survival was 13 months, and the probabilities of survival at 12, 24, 36, 48 and 60 months post-diagnosis were 0.59, 0.25, 0.15, 0.11 and 0.07 respectively, in this study. The median survival is similar to that reported by Krex et al. [14], and the 60 month survival probability is comparable to the 5-year survival rate of 0.13 estimated for grade IV brain cancer reported by the National Cancer Institute Surveillance Epidemiology and End Results [44]. The similarity between the survival rate in this study and that reported for primary glioblastoma suggests an insignificant fraction of secondary glioblastoma samples among the samples analyzed [20].
Comparing findings against a literature review confirmed that the Cox survival analysis of multiple gene expression profiles and clinical variables simultaneously was an effective tool to detect an integrated set of gene expression profiles exhibiting general and cohort-dependent associations with the three glioblastoma survival variables. The majority of the genes associated with lifetime, overall, and progression-free survival, in this study, have been previously reported to be associated with glioblastoma (35, 24, and 35 genes, respectively) or with another cancer (10, 19, and 15, respectively). In addition, the multi-factor analysis and data used in this study allowed the uncovering several novel associations between gene profiles and glioblastoma survival. Specifically, 16, 4, and 10 previously unreported genes were associated with lifetime, overall, and progression-free survival, respectively in the present work. The discussion of the findings from our study is divided into genes associated with multiple survival variables, genes associated with glioblastoma in a cohort-independent or cohort-dependent manner, and further investigation of complex associations.
Pik3r1 and E2f3 were associated with all three glioblastoma survival variables (Tables 2, 3, 5 and 7). The higher glioblastoma hazards associated with higher levels of Pik3r1 observed in this study are supported by previous work showing that over-expression of this gene plays a role in the activation of the PI3K/Akt pathway resulting in cell proliferation and tumor invasion [45]. Likewise, a link between E2f3 and glioblastoma has been reported [28,46]. Among the 15 genes associated with two glioblastoma events (Table 2), Akr1c3, Csf1, Jag2, Plcg1, Rpl37a, Sod2, and Topors were associated with lifetime and overall survival (Tables 3 and 5). Jag2 has been associated with adenomas [47], pancreatic [48] and breast cancer [49], Rpl37a with nasopharyngeal carcinoma cell lines [50], and the rest with glioblastoma [26,51-54]. The consistent findings across both glioblastoma survival events suggest that these genes may have specific roles in death. Likewise, the association between Hras and overall and progression-free survival (Tables 2, 5 and 7), is consistent with previous glioblastoma studies [55] and suggests that this gene may have a role in aggressive glioblastoma growth. Fstl1, Mtap, Tp53, Camk2g 214322_at, and Il13ra1 probe 210904_s_at, were associated with lifetime and progression-free survival (Tables 2, 3 and 7) and these associations are supported by previous studies [20,24,25,28,56-58].
Most genes (lifetime survival, 55 out of 61 genes; overall survival, 45 out of 47 genes; and progression-free survival, 55 out of 60 genes) were associated with survival in a general or cohort-independent manner. The most extreme cohort-independent changes in lifetime survival were observed in Syne1 (HR = 0.17) and Pdcd4 (HR = 4.68), and the former profile has been found in lung [59], ovarian [60], colon, and breast cancers [61]; while, the second has been associated with glioma [62]. The most extreme cohort-independent changes in overall survival were observed in Ighg1 (HR = 4.33) and Tgfa (HR = 0.12), and the former trend has been found in cancer cell lines [63]; meanwhile the later is present in the KEGG glioma pathway [28]. Lastly, the genes that presented extreme hazard ratio values and general association with progression-free survival are Pla2g7 (HR = 0.11) and Paics (HR = 5.28). The Pla2g7 and Paics trends identified in this study are consistent with those reported for breast cancer in mice [64] and in non-glioma types of cancer [65,66], respectively.
Several genes (lifetime survival, 6 out of 61 genes; overall survival, 2 out of 47 genes; and progression-free survival, 5 out of 60 genes) were associated with glioblastoma survival in a cohort-dependent manner. These findings indicate that effective use of these genes in prognostic indices or in therapy development must consider the personal characteristics of the individual. Higher levels of C2 and Prkcb (probe 209685_s_at) were associated with a higher lifetime death hazard in males (HR = 1.93 and 5.22, respectively) than in females (HR = 1.30 and 1.31, respectively) and the profile of the latter gene has been observed in colon cancer cell lines [67]. The lifetime hazard estimate decreased with increased levels of Sox10 in Caucasian individuals (HR = 0.55) compared to non-Caucasian individuals, and this pattern is concordant with broad distribution of Sox10 in high grade gliomas [68]. Increases in the level of Chi3l1 were associated with significant increases in lifetime hazard estimates across all therapies with the highest hazard ratio observed in individuals receiving no therapy (None, HR = 2.42). This trend is consistent with reports that Chi3l1/Ykl-40 was highly overexpressed in glioblastoma relative to nonneoplastic brain [69] and that Ykl-40 is associated with poorer response to radiation and shorter lifetime survival in glioblastoma [70]. Males (HR = 0.36) and individuals receiving no therapy (HR = 0.38) have the lowest hazard ratio per increase in Prkcb (probe 207957_s_at). These trends are consistent with those reported for other cancer types [67] and with observations of protein kinase C activation in gamma-irradiated proliferating and confluent human lung fibroblast cells [71].
The cohort-dependent associations between overall survival and both Polr2d and Igf2bp3 have been observed in colorectal cancer [72] and glioblastoma [73], respectively. Three genes (Rab31, Rps20 and Apool) exhibited a cohort-dependent association with overall survival that is consistent with previously reported trends [74-76]. Lastly, the gender-dependent association between Gdf10 and progression-free survival is in agreement with reports of copy number loss of Gdf10 in mesothelioma [77].
Further analyses of the association between individual genes (with or without clinical variables) and hazards were undertaken when the trend estimated from the multi-gene index was opposite to that previously reported. Nine genes and survival events were re-analyzed individually and compared to previous reports including: E2f3 and all three survival variables [28,46], Egfr and lifetime survival [78], Cfs1 and lifetime survival, Mdm2 with overall hazard [79], Fstl1 and lifetime and progression-free survival [25], Mtap and progression-free hazard [57], Pdcd4 and lifetime survival [62], Tgfa and overall survival [80], and race-dependent Vav3 and overall survival [81]. In the first six cases, the consideration of the gene alone as predictor of glioblastoma survival as standard in previous reports resulted in non-significant associations, in this study. These results indicate that the accurate identification of biomarkers and precise characterization of the trend requires the study of the genes in concert with other genes in a systems biology framework, such as the approach implemented, in this study. Re-analysis of Pdcd4 and Vav3 confirmed the significant trend detected in the multi-gene analysis, suggesting that further studies are needed to precisely characterize the trend.
The LOOCV confirmed the adequacy of the set of genes and clinical variables identified to predict the glioblastoma hazards. The minor differences between the observed and predicted numbers in each group may be due to the discretization of the survival length into high and low groups required by the discriminant analysis; whereas, the Cox survival analysis models continuous time to the glioblastoma event. The significant number of genes prognostic of glioblastoma survival identified in this study that are also targets of microRNAs associated with glioblastoma [43] further confirms our results.
In addition to literature review and LOOCV, the gene-survival associations detected in this study were confirmed using the information from the REMBRANDT database. The associations between survival and the 10 gene probes with the most extreme hazard ratio estimate for each of the three survival variables studied that did not interact with cohort variables (Tables 3, 5, 7) were investigated in REMBRANDT. The query was performed using the Kaplan-Meier survival plot for Gene Expression Data. Of these, eight genes had the same significant trend observed in our study (Syne1, Gigyf2/Tnrc15, Scn5a, Hoxa10, Pdcd4, Tgfa, Pla2g7 and Agpat1), two did not have information on the REMBRANDT database (Ighg1 and Hnrnpd), Fstl1 had an opposite trend than the one observed in our study and in previous independent studies (Table 3) and most of the remaining genes, although non-significant, had the same trend observed in our analysis. The latter results are consistent with the simpler analytical approach based on Kaplan-Meier curves available in REMBRANDT, when compared to the more flexible Cox survival analysis used in our study. The Kaplan-Meier approach relies on non-parametric rank-based test to compare the survival between individuals with high and low gene expression. These groups are obtained by setting up an arbitrary expression threshold. Non-parametric rank-based approaches tend to have lower power to detect significant variation than semi- and parametric approaches such as the Cox survival analysis. In addition, the Kaplan-Meier analysis only allows the consideration of one explanatory variable at a time, and this variable has to be discrete (thus, the reason for comparing high and low expression groups in REMBRANDT). This approach does not allow considering multiple continuous covariates (i.e. gene expression) and factors (e.g. race, gender, therapy and progression) or interactions simultaneously. The Cox-survival analysis implemented in our study allows the simultaneous consideration of multiple factors (such as possible population stratification due to race), covariates (e.g. other gene expression profiles) and interactions, and it does not require the discretization of the gene expression values that could result in potential loss of information. Thus, the Cox approach used in our study is able to capture the association between continuous gene expression values and survival conditional on all other model terms and is able to detect associations that are likely not to reach statistical significance using the Kaplan-Meier comparison of survival between high and low gene expression groups.
Among the GO categories, 19 biological processes and three molecular functions were over-represented (FDR adjusted P-value < 0.1, ≥ 3 genes per category) in the genes associated with the three glioblastoma events studied (Tables 9, 10 and 11). Two biological processes, cell cycle (GO:0007049) and death (GO:0016265), were over-represented in the lifetime and progression-free survival (Tables 9 and 11), and several biological processes have been previously associated with glioblastoma [17,62,68,70,79,82-86]. These processes included: aging, morphogenesis, cell cycle and proliferation, and death for lifetime survival; morphogenesis for overall survival; and cell cycle, death and recognition, death, response to biotic and abiotic stimuli, programmed cell death, and apoptosis for progression-free survival.
The study of complementary glioblastoma survival variables allowed to confirm that the gene profiles associated with lifetime survival resulting in the enriched functional category of aging are clearly associated with cancer initiation and progression and are not a simply reflection of the natural aging process. Two results confirm that the biomarkers are not mere confounding with aging. First, the genes in the GO terms "aging (GO:0007568)" and "cell aging (GO:0007569)", Pdcd4, Cdkn2a, and Tp53, have all been associated with GBM in previous independent studies (Table 3). In addition, Tp53 was associated with progression-free survival (Table 7). Second, other functional terms enriched among the genes associated with lifetime glioblastoma survival were also identified on the other glioblastoma survival variables studied. The biological processes of cell death and cell cycle were enriched both for lifetime and progression-free survival.
The biological processes, molecular functions and gene networks particular to a glioblastoma survival event offered insights into the processes particular to the initiation and progression of this cancer. For instance, eight biological processes associated with lifetime survival were level 3, and one was level 4, indicating that the differentially expressed genes associated with lifetime survival participate on broad or general biological mechanisms. The interconnection between the genes pertaining to aging further confirms the significance of this gene network on lifetime survival (Figure 4). Although only two biological processes were associated with overall survival, these processes correspond to levels 4 and 6. This result indicates that the genes associated with overall survival correspond to more specific mechanisms. Moreover, both biological processes are related to generation and organization of anatomical structures, such as organs, and this finding may be associated to the dispersion and development of malignant cells after diagnosis and resection. The close relationship between biomarker genes in this network supports this finding (Figure 5). Albeit the study of progression-free survival encompassed a shorter period than lifetime and overall survival, the functional analysis showed several biological processes and molecular functions over-represented among the genes associated with this survival. Four of the biological processes are from level 6 to 8, indicating that specific gene networks and roles are associated with progression-free survival. The biological processes associated with progression-free survival include regulation of progression through cell cycle, programmed cell death, and apoptosis. Extensive relationships between the biomarker genes in the cell cycle were identified further, supporting the major role of this network on glioblastoma progression (Figure 6). In addition, three molecular functions were enriched among the genes associated with progression-free survival. Therefore, many biological and molecular events occur in the period between the diagnosis of malignancy and progression or recurrence, probably due to response to numerous treatments, surgery, and cancer progression. Two genes were highly represented across the categories (Tables 9 to 11). Tp53 has an important role as a tumor repressor [83], and App is highly expressed in individuals with short-term glioblastoma survival [24].
Conclusions
An innovative approach to identify simultaneously multiple biomarkers of lifetime, overall and progression-free glioblastoma survival in a systems biology framework was presented. Furthermore, the inclusion of clinical information allowed the uncovering of general and individualized associations between gene expression profiles and three complementary survival metrics. This study demonstrated the pre-eminence of developing multi-gene prognostic indices of glioblastoma survival through the integration of variable selection and survival models relative to the simple-yet- simplistic single-gene analysis. Known biomarker gene profiles were confirmed, and new general and clinical-dependent gene profiles were uncovered. The present study looked at glioblastoma in general and complements work on the identification of genes associated with specific glioblastoma types [42,87,88]. Empirically confirmed findings will be the basis for improved prognostic tools and individualized treatments that improve the survival and quality of life of individuals suffering glioblastoma multiforme.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
NVLS compiled the data; performed the normalization, survival, and functional analyses; contributed to the interpretation of results; and drafted the manuscript. KRD participated in the analyses. BRS participated in the analyses and contributed in drafting the manuscript. JEB obtained funding for the study and helped in the drafting of the manuscript. SRZ obtained funding for the study and permission to use the data; participated in its conception, coordination, and interpretation of results; and helped to draft the manuscript. All authors have read and approved the final version of this manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
List of genes associated in glioblastoma from the literature Table containing the list of 174 genes previously reported in the literature.
Contributor Information
Nicola VL Serão, Email: serao1@illinois.edu.
Kristin R Delfino, Email: delfino2@illinois.edu.
Bruce R Southey, Email: southey@illinois.edu.
Jonathan E Beever, Email: jbeever@illinois.edu.
Sandra L Rodriguez-Zas, Email: rodrgzzs@uiuc.edu.
Acknowledgements
We thank Dr. Kun Yu and Dr. Jen-Tsan Chi for suggestions and advice on analysis and interpretation. The support of USDA NIFA (Number: ILLU-538-554) to NSV and JEB, NCI (Grant Number: 1R03CA143975) to SRZ and KRD, and NIH/NIDA (Grant Number: R21DA027548 and P30DA018310) to BRS and SRZ are greatly appreciated.
References
- Dai C, Holland E. In: Glioblastoma Multiforme. 1. Markert J,DeVita Jr VT,Rosenberg SA,Hellman S.Sudbury, MA, editor. Jones and Barlett Publishers; 2005. Astrocyte Differentiation States and Glioma Formation; p. 1. [Google Scholar]
- Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL, Burkhard C, Schuler D, Probst-Hensch NM, Maiorka PC, Baeza N, Pisani P, Yonekawa Y, Yasargil MG, Lutolf UM, Kleihues P. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64(19):6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- Novakova J, Slaby O, Vyzula R, Michalek J. MicroRNA involvement in glioblastoma pathogenesis. Biochem Biophys Res Commun. 2009;386(1):1–5. doi: 10.1016/j.bbrc.2009.06.034. [DOI] [PubMed] [Google Scholar]
- Wrensch M, Minn Y, Chew T, Bondy M, Berger MS. Epidemiology of primary brain tumors: current concepts and review of the literature. Neuro Oncol. 2002;4(4):278–299. doi: 10.1093/neuonc/4.4.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butowski N, Chang S. In: High-Grade Gliomas Diagnosis and Treatment. 1. Barnett GH. Danvers, MA, editor. Humana Press; 2006. Adult High-Grade Glioma; p. 59. [Google Scholar]
- Kesari S, Schiff D, Henson JW, Muzikansky A, Gigas DC, Doherty L, Batchelor TT, Longtine JA, Ligon KL, Weaver S, Laforme A, Ramakrishna N, Black PM, Drappatz J, Ciampa A, Folkman J, Kieran M, Wen PY. Phase II study of temozolomide, thalidomide, and celecoxib for newly diagnosed glioblastoma in adults. Neuro Oncol. 2008;10(3):300–308. doi: 10.1215/15228517-2008-005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGirt MJ, Than KD, Weingart JD, Chaichana KL, Attenello FJ, Olivi A, Laterra J, Kleinberg LR, Grossman SA, Brem H, Quinones-Hinojosa A. Gliadel (BCNU) wafer plus concomitant temozolomide therapy after primary resection of glioblastoma multiforme. J Neurosurg. 2009;110(3):583–588. doi: 10.3171/2008.5.17557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minniti G, Muni R, Lanzetta G, Marchetti P, Enrici RM. Chemotherapy for glioblastoma: current treatment and future perspectives for cytotoxic and targeted agents. Anticancer Res. 2009;29(12):5171–5184. [PubMed] [Google Scholar]
- Maucort-Boulch D, Baron MH, Pommier P, Weber DC, Mizoe JE, Rochat J, Boissel JP, Balosso J, Tsujii H, Amsallem E. Rationale for carbon ion therapy in high-grade glioma based on a review and a meta-analysis of neutron beam trials. Cancer Radiother. 2010;14(1):34–41. doi: 10.1016/j.canrad.2009.08.141. [DOI] [PubMed] [Google Scholar]
- Mischel PS, Shai R, Shi T, Horvath S, Lu KV, Choe G, Seligson D, Kremen TJ, Palotie A, Liau LM, Cloughesy TF, Nelson SF. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene. 2003;22(15):2361–2373. doi: 10.1038/sj.onc.1206344. [DOI] [PubMed] [Google Scholar]
- Rich JN, Hans C, Jones B, Iversen ES, McLendon RE, Rasheed BK, Dobra A, Dressman HK, Bigner DD, Nevins JR, West M. Gene expression profiling and genetic markers in glioblastoma survival. Cancer Res. 2005;65(10):4051–4058. doi: 10.1158/0008-5472.CAN-04-3936. [DOI] [PubMed] [Google Scholar]
- Houillier C, Lejeune J, Benouaich-Amiel A, Laigle-Donadey F, Criniere E, Mokhtari K, Thillet J, Delattre JY, Hoang-Xuan K, Sanson M. Prognostic impact of molecular markers in a series of 220 primary glioblastomas. Cancer. 2006;106(10):2218–2223. doi: 10.1002/cncr.21819. [DOI] [PubMed] [Google Scholar]
- Wang L, Wei Q, Wang LE, Aldape KD, Cao Y, Okcu MF, Hess KR, El-Zein R, Gilbert MR, Woo SY, Prabhu SS, Fuller GN, Bondy ML. Survival prediction in patients with glioblastoma multiforme by human telomerase genetic variation. J Clin Oncol. 2006;24(10):1627–1632. doi: 10.1200/JCO.2005.04.0402. [DOI] [PubMed] [Google Scholar]
- Krex D, Klink B, Hartmann C, von Deimling A, Pietsch T, Simon M, Sabel M, Steinbach JP, Heese O, Reifenberger G, Weller M, Schackert G. German Glioma Network. Long-term survival with glioblastoma multiforme. Brain. 2007;130(Pt 10):2596–2606. doi: 10.1093/brain/awm204. [DOI] [PubMed] [Google Scholar]
- Dreyfuss JM, Johnson MD, Park PJ. Meta-analysis of glioblastoma multiforme versus anaplastic astrocytoma identifies robust gene markers. Mol Cancer. 2009;8:71. doi: 10.1186/1476-4598-8-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat; 2009. [DOI] [PubMed] [Google Scholar]
- Comincini S, Paolillo M, Barbieri G, Palumbo S, Sbalchiero E, Azzalin A, Russo MA, Schinelli S. Gene expression analysis of an EGFR indirectly related pathway identified PTEN and MMP9 as reliable diagnostic markers for human glial tumor specimens. J Biomed Biotechnol. 2009;2009:924565. doi: 10.1155/2009/924565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castells X, Acebes JJ, Boluda S, Moreno-Torres A, Pujol J, Julia-Sape M, Candiota AP, Arino J, Barcelo A, Arus C. Development of a predictor for human brain tumors based on gene expression values obtained from two types of microarray technologies. OMICS. 2010;14(2):157–164. doi: 10.1089/omi.2009.0093. [DOI] [PubMed] [Google Scholar]
- Wei KC, Huang CY, Chen PY, Feng LY, Wu TW, Chen SM, Tsai HC, Lu YJ, Tsang NM, Tseng CK, Pai PC, Shin JW. Evaluation of the prognostic value of CD44 in glioblastoma multiforme. Anticancer Res. 2010;30(1):253–259. [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Data Portal. http://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp
- Irizarry RA, Gautier L, Bolstad BM, Miller C. Methods for Affymetrix Oligonucleotide Arrays. http://bioconductor.org/packages/2.5/bioc/html/affy.html
- Beehive. http://stagbeetle.animal.uiuc.edu/Beehive
- Marko NF, Toms SA, Barnett GH, Weil R. Genomic expression patterns distinguish long-term from short-term glioblastoma survivors: a preliminary feasibility study. Genomics. 2008;91(5):395–406. doi: 10.1016/j.ygeno.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Reddy SP, Britto R, Vinnakota K, Aparna H, Sreepathi HK, Thota B, Kumari A, Shilpa BM, Vrinda M, Umesh S, Samuel C, Shetty M, Tandon A, Pandey P, Hegde S, Hegde AS, Balasubramaniam A, Chandramouli BA, Santosh V, Kondaiah P, Somasundaram K, Rao MR. Novel glioblastoma markers with diagnostic and prognostic value identified through transcriptome analysis. Clin Cancer Res. 2008;14(10):2978–2987. doi: 10.1158/1078-0432.CCR-07-4821. [DOI] [PubMed] [Google Scholar]
- Bredel M, Scholtens DM, Harsh GR, Bredel C, Chandler JP, Renfrow JJ, Yadav AK, Vogel H, Scheck AC, Tibshirani R, Sikic BI. A network model of a cooperative genetic landscape in brain tumors. JAMA. 2009;302(3):261–275. doi: 10.1001/jama.2009.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu B, Jiang X, Zhao H, Fan M, Fan Z, Lee JJ, Jiang T, Jiang T, Song SW. A systems biology-based gene expression classifier of glioblastoma predicts survival with solid tumors. PLoS One. 2009;4(7):e6274. doi: 10.1371/journal.pone.0006274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KEGG PATHWAY: Glioma - Homo sapiens (humans) http://www.genome.jp/kegg/pathway/hsa/hsa05214.html
- Cox DR, Oakes D. Analysis of survival data. 1. London: Chapman and Hall; 1984. [Google Scholar]
- NCBI-Gene database. http://www.ncbi.nlm.nih.gov/gene
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F, Minguez P, Tarraga J, Montaner D, Alloza E, Vaquerizas JM, Conde L, Blaschke C, Vera J, Dopazo J. BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res. 2006;34(Web Server issue):W472–6. doi: 10.1093/nar/gkl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Ochagavia ME, Rabasa LC, Miranda J, Fernandez-de-Cossio J, Bringas R. BisoGenet: a new tool for gene network building, visualization and analysis. BMC Bioinformatics. 2010;11:91. doi: 10.1186/1471-2105-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumur CI, Ladd AC, Wright HV, Penberthy LT, Wilkinson DS, Powers CN, Garrett CT, DiNardo LJ. Genes involved in radiation therapy response in head and neck cancers. Laryngoscope. 2009;119(1):91–101. doi: 10.1002/lary.20005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LY, Yang CH, Wu KC, Yang CH. A hybrid feature selection method for DNA microarray data. Comput Biol Med. 2011;41(4):228–237. doi: 10.1016/j.compbiomed.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Chuang LY, Yang CS, Li JC, Yang CH. Chaotic genetic algorithm for gene selection and classification problems. OMICS. 2009;13(5):407–420. doi: 10.1089/omi.2009.0007. [DOI] [PubMed] [Google Scholar]
- Petrausch U, Martus P, Tonnies H, Bechrakis NE, Lenze D, Wansel S, Hummel M, Bornfeld N, Thiel E, Foerster MH, Keilholz U. Significance of gene expression analysis in uveal melanoma in comparison to standard risk factors for risk assessment of subsequent metastases. Eye (Lond) 2008;22(8):997–1007. doi: 10.1038/sj.eye.6702779. [DOI] [PubMed] [Google Scholar]
- Lachenbruch PA, Mickey MR. Estimation of error rates in discriminant analysis. Technometrics. 1968;10(1):1–11. doi: 10.2307/1266219. [DOI] [Google Scholar]
- National Cancer Institute. REMBRANDT home page. http://rembrandt.nci.nih.gov
- Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7(2):157–167. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfino KR, Rodriguez-Zas SL, Serão NVL, Southey BR. A Computational Strategy To Identify General And Individualized Genomic Biomarkers. Proceedings of the 6th International Symposium on Bioinformatics Research and Applications: 23-26 May 2010; Storrs, Connecticut. 2010. pp. 31–33.
- National Cancer Institute. Surveillance Epidemiology and End Results. http://seer.cancer.gov/publications/survival
- Fu Y, Zhang Q, Kang C, Zhang J, Zhang K, Pu P, Wang G, Wang T. Inhibitory effects of adenovirus mediated Akt1 and PIK3R1 shRNA on the growth of malignant tumor cells in vitro and in vivo. Cancer Biol Ther. 2009;8(11):1002–1009. doi: 10.4161/cbt.8.11.8285. [DOI] [PubMed] [Google Scholar]
- Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26(34):5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- Fre S, Pallavi SK, Huyghe M, Lae M, Janssen KP, Robine S, Artavanis-Tsakonas S, Louvard D. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proc Natl Acad Sci USA. 2009;106(15):6309–6314. doi: 10.1073/pnas.0900427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullendore ME, Koorstra JB, Li YM, Offerhaus GJ, Fan X, Henderson CM, Matsui W, Eberhart CG, Maitra A, Feldmann G. Ligand-dependent Notch signaling is involved in tumor initiation and tumor maintenance in pancreatic cancer. Clin Cancer Res. 2009;15(7):2291–2301. doi: 10.1158/1078-0432.CCR-08-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal S, Subramanyam D, Dey D, Kumar RV, Rangarajan A. Cooperation of Notch and Ras/MAPK signaling pathways in human breast carcinogenesis. Mol Cancer. 2009;8:128. doi: 10.1186/1476-4598-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim EU, Ang CH, Ng CC, Lee CW, Narayanan K. Differential expression of a subset of ribosomal protein genes in cell lines derived from human nasopharyngeal epithelium. J Hum Genet. 2010;55(2):118–120. doi: 10.1038/jhg.2009.124. [DOI] [PubMed] [Google Scholar]
- Alterman RL, Stanley ER. Colony stimulating factor-1 expression in human glioma. Mol Chem Neuropathol. 1994;21(2-3):177–188. doi: 10.1007/BF02815350. [DOI] [PubMed] [Google Scholar]
- Sala G, Dituri F, Raimondi C, Previdi S, Maffucci T, Mazzoletti M, Rossi C, Iezzi M, Lattanzio R, Piantelli M, Iacobelli S, Broggini M, Falasca M. Phospholipase Cgamma1 is required for metastasis development and progression. Cancer Res. 2008;68(24):10187–10196. doi: 10.1158/0008-5472.CAN-08-1181. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Yang Z, Xu JG, Yang MS, Zeng ZX, You C. Differentially expressed genes from the glioblastoma cell line SHG-44 treated with all-trans retinoic acid in vitro. J Clin Neurosci. 2009;16(2):285–294. doi: 10.1016/j.jocn.2007.11.014. [DOI] [PubMed] [Google Scholar]
- Gene Expression Atlas. Summary for AKR1C3 (Homo sapiens) http://www.ebi.ac.uk/gxa/gene/ENSG00000196139
- Rich JN, Guo C, McLendon RE, Bigner DD, Wang XF, Counter CM. A genetically tractable model of human glioma formation. Cancer Res. 2001;61(9):3556–3560. [PubMed] [Google Scholar]
- van den Boom J, Wolter M, Kuick R, Misek DE, Youkilis AS, Wechsler DS, Sommer C, Reifenberger G, Hanash SM. Characterization of gene expression profiles associated with glioma progression using oligonucleotide-based microarray analysis and real-time reverse transcription-polymerase chain reaction. Am J Pathol. 2003;163(3):1033–1043. doi: 10.1016/S0002-9440(10)63463-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Maruno M, Wada K, Kagawa N, Fujimoto Y, Hashimoto N, Izumoto S, Yoshimine T. Genetic analysis of human glioblastomas using a genomic microarray system. Brain Tumor Pathol. 2004;21(1):27–34. doi: 10.1007/BF02482174. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. P53 in Health and Disease. Nat Rev Mol Cell Biol. 2007;8(4):275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Tessema M, Belinsky SA. Mining the epigenome for methylated genes in lung cancer. Proc Am Thorac Soc. 2008;5(8):806–810. doi: 10.1513/pats.200805-045TH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty JA. et al. ESR1/SYNE1 polymorphism and invasive epithelial ovarian cancer risk: an Ovarian Cancer Association Consortium study. Cancer Epidemiol Biomarkers Prev. 2010;19(1):245–250. doi: 10.1158/1055-9965.EPI-09-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TA, Glockner S, Yi JM, Chen W, Van Neste L, Cope L, Herman JG, Velculescu V, Schuebel KE, Ahuja N, Baylin SB. Convergence of mutation and epigenetic alterations identifies common genes in cancer that predict for poor prognosis. PLoS Med. 2008;5(5):e114. doi: 10.1371/journal.pmed.0050114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Zhang P, Zhou C, Li J, Wang Q, Zhu F, Ma C, Sun W, Zhang L. Frequent loss of PDCD4 expression in human glioma: possible role in the tumorigenesis of glioma. Oncol Rep. 2007;17(1):123–128. [PubMed] [Google Scholar]
- Chen Z, Gu J. Immunoglobulin G expression in carcinomas and cancer cell lines. FASEB J. 2007;21(11):2931–2938. doi: 10.1096/fj.07-8073com. [DOI] [PubMed] [Google Scholar]
- Calogero RA, Cordero F, Forni G, Cavallo F. Inflammation and breast cancer. Inflammatory component of mammary carcinogenesis in ErbB2 transgenic mice. Breast Cancer Res. 2007;9(4):211. doi: 10.1186/bcr1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Zhang K, Zhang X, Lei W, Xiao T, Ma J, Guo S, Shao S, Zhang H, Liu Y, Yuan J, Hu Z, Ma Y, Feng X, Hu S, Zhou J, Cheng S, Gao Y. Identification of differentially expressed genes in human lung squamous cell carcinoma using suppression subtractive hybridization. Cancer Lett. 2004;212(1):83–93. doi: 10.1016/j.canlet.2004.03.023. [DOI] [PubMed] [Google Scholar]
- Martin DN, Boersma BJ, Yi M, Reimers M, Howe TM, Yfantis HG, Tsai YC, Williams EH, Lee DH, Stephens RM, Weissman AM, Ambs S. Differences in the tumor microenvironment between African-American and European-American breast cancer patients. PLoS One. 2009;4(2):e4531. doi: 10.1371/journal.pone.0004531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Su W, Thompson EA, Leitges M, Murray NR, Fields AP. Protein kinase CbetaII regulates its own expression in rat intestinal epithelial cells and the colonic epithelium in vivo. J Biol Chem. 2004;279(44):45556–45563. doi: 10.1074/jbc.M407701200. [DOI] [PubMed] [Google Scholar]
- Ferletta M, Uhrbom L, Olofsson T, Ponten F, Westermark B. Sox10 has a broad expression pattern in gliomas and enhances platelet-derived growth factor-B--induced gliomagenesis. Mol Cancer Res. 2007;5(9):891–897. doi: 10.1158/1541-7786.MCR-07-0113. [DOI] [PubMed] [Google Scholar]
- Nigro JM, Misra A, Zhang L, Smirnov I, Colman H, Griffin C, Ozburn N, Chen M, Pan E, Koul D, Yung WK, Feuerstein BG, Aldape KD. Integrated array-comparative genomic hybridization and expression array profiles identify clinically relevant molecular subtypes of glioblastoma. Cancer Res. 2005;65(5):1678–1686. doi: 10.1158/0008-5472.CAN-04-2921. [DOI] [PubMed] [Google Scholar]
- Pelloski CE, Mahajan A, Maor M, Chang EL, Woo S, Gilbert M, Colman H, Yang H, Ledoux A, Blair H, Passe S, Jenkins RB, Aldape KD. YKL-40 expression is associated with poorer response to radiation and shorter overall survival in glioblastoma. Clin Cancer Res. 2005;11(9):3326–3334. doi: 10.1158/1078-0432.CCR-04-1765. [DOI] [PubMed] [Google Scholar]
- Baskar R, Hande MP. A comparative study of protein kinase C activation in gamma-irradiated proliferating and confluent human lung fibroblast cells. J Radiat Res (Tokyo) 2009;50(5):415–423. doi: 10.1269/jrr.08125. [DOI] [PubMed] [Google Scholar]
- Nibbe RK. Systems biology of human colorectal cancer. 2010.
- Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, Holland E. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4(11):e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nindl I, Dang C, Forschner T, Kuban RJ, Meyer T, Sterry W, Stockfleth E. Identification of differentially expressed genes in cutaneous squamous cell carcinoma by microarray expression profiling. Mol Cancer. 2006;5:30. doi: 10.1186/1476-4598-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt CL, Mani DR, Betensky RA, Tamayo P, Cairncross JG, Ladd C, Pohl U, Hartmann C, McLaughlin ME, Batchelor TT, Black PM, von Deimling A, Pomeroy SL, Golub TR, Louis DN. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63(7):1602–1607. [PubMed] [Google Scholar]
- Trougakos IP, Gonos ES. Clusterin/apolipoprotein J in human aging and cancer. Int J Biochem Cell Biol. 2002;34(11):1430–1448. doi: 10.1016/S1357-2725(02)00041-9. [DOI] [PubMed] [Google Scholar]
- Christensen BC, Houseman EA, Poage GM, Godleski JJ, Bueno R, Sugarbaker DJ, Wiencke JK, Nelson HH, Marsit CJ, Kelsey KT. Integrated profiling reveals a global correlation between epigenetic and genetic alterations in mesothelioma. Cancer Res. 2010;70(14):5686–5694. doi: 10.1158/0008-5472.CAN-10-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60(5):1383–1387. [PubMed] [Google Scholar]
- Halatsch ME, Schmidt U, Unterberg A, Vougioukas VI. Uniform MDM2 overexpression in a panel of glioblastoma multiforme cell lines with divergent EGFR and p53 expression status. Anticancer Res. 2006;26(6B):4191–4194. [PubMed] [Google Scholar]
- Razis E, Selviaridis P, Labropoulos S, Norris JL, Zhu MJ, Song DD, Kalebic T, Torrens M, Kalogera-Fountzila A, Karkavelas G, Karanastasi S, Fletcher JA, Fountzilas G. Phase II study of neoadjuvant imatinib in glioblastoma: evaluation of clinical and molecular effects of the treatment. Clin Cancer Res. 2009;15(19):6258–6266. doi: 10.1158/1078-0432.CCR-08-1867. [DOI] [PubMed] [Google Scholar]
- Salhia B, Tran NL, Chan A, Wolf A, Nakada M, Rutka F, Ennis M, McDonough WS, Berens ME, Symons M, Rutka JT. The guanine nucleotide exchange factors trio, Ect2, and Vav3 mediate the invasive behavior of glioblastoma. Am J Pathol. 2008;173(6):1828–1838. doi: 10.2353/ajpath.2008.080043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Li W, Jiang W, Humphrey LE, Howell GM, Brattain MG. Autocrine TGFalpha expression in the regulation of initiation of human colon carcinoma growth. J Cell Physiol. 1998;177(3):387–395. doi: 10.1002/(SICI)1097-4652(199812)177:3<387::AID-JCP2>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol. 2003;13(1):49–58. doi: 10.1016/S1044-579X(02)00099-8. [DOI] [PubMed] [Google Scholar]
- Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, Uhlik M, Lin A, Du J, Qian YW, Zeckner DJ, Tucker-Kellogg G, Touchman J, Patel K, Mousses S, Bittner M, Schevitz R, Lai MH, Blanchard KL, Thomas JE. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448(7152):439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- Lymbouridou R, Soufla G, Chatzinikola AM, Vakis A, Spandidos DA. Down-regulation of K-ras and H-ras in human brain gliomas. Eur J Cancer. 2009;45(7):1294–1303. doi: 10.1016/j.ejca.2008.12.028. [DOI] [PubMed] [Google Scholar]
- Wang G, Kang C, Pu P. Increased expression of Akt2 and activity of PI3K and cell proliferation with the ascending of tumor grade of human gliomas. Clin Neurol Neurosurg. 2010;112(4):324–327. doi: 10.1016/j.clineuro.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O'Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN. Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John T, Black MA, Toro TT, Leader D, Gedye CA, Davis ID, Guilford PJ, Cebon JS. Predicting clinical outcome through molecular profiling in stage III melanoma. Clin Cancer Res. 2008;14(16):5173–5180. doi: 10.1158/1078-0432.CCR-07-4170. [DOI] [PubMed] [Google Scholar]
- MacDonald TJ, Pollack IF, Okada H, Bhattacharya S, Lyons-Weiler J. Progression-associated genes in astrocytoma identified by novel microarray gene expression data reanalysis. Methods Mol Biol. 2007;377:203–222. doi: 10.1007/978-1-59745-390-5_13. [DOI] [PubMed] [Google Scholar]
- Hodgson JG, Yeh RF, Ray A, Wang NJ, Smirnov I, Yu M, Hariono S, Silber J, Feiler HS, Gray JW, Spellman PT, Vandenberg SR, Berger MS, James CD. Comparative analyses of gene copy number and mRNA expression in glioblastoma multiforme tumors and xenografts. Neuro Oncol. 2009;11(5):477–487. doi: 10.1215/15228517-2008-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller M, Felsberg J, Hartmann C, Berger H, Steinbach JP, Schramm J, Westphal M, Schackert G, Simon M, Tonn JC, Heese O, Krex D, Nikkhah G, Pietsch T, Wiestler O, Reifenberger G, von Deimling A, Loeffler M. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27(34):5743–5750. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- Lagerstedt KK, Kristiansson E, Lonnroth C, Andersson M, Iresjo BM, Gustafsson A, Hansson E, Kressner U, Nordgren S, Enlund F, Lundholm K. Genes with relevance for early to late progression of colon carcinoma based on combined genomic and transcriptomic information from the same patients. Cancer Inform. 2010;9:79–91. doi: 10.4137/cin.s4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocejo-Garcia M, Baokbah TA, Ashurst HL, Cowlishaw D, Soomro I, Coulson JM, Woll PJ. Roles for USF-2 in lung cancer proliferation and bronchial carcinogenesis. J Pathol. 2005;206(2):151–159. doi: 10.1002/path.1775. [DOI] [PubMed] [Google Scholar]
- Bhatti P, Stewart PA, Hutchinson A, Rothman N, Linet MS, Inskip PD, Rajaraman P. Lead exposure, polymorphisms in genes related to oxidative stress, and risk of adult brain tumors. Cancer Epidemiol Biomarkers Prev. 2009;18(6):1841–1848. doi: 10.1158/1055-9965.EPI-09-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulier J, Clappier E, Cayuela JM, Regnault A, Garcia-Peydro M, Dombret H, Baruchel A, Toribio ML, Sigaux F. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL) Blood. 2005;106(1):274–286. doi: 10.1182/blood-2004-10-3900. [DOI] [PubMed] [Google Scholar]
- Onda T, Uzawa K, Nakashima D, Saito K, Iwadate Y, Seki N, Shibahara T, Tanzawa H. Lin-7C/VELI3/MALS-3: an essential component in metastasis of human squamous cell carcinoma. Cancer Res. 2007;67(20):9643–9648. doi: 10.1158/0008-5472.CAN-07-1911. [DOI] [PubMed] [Google Scholar]
- Jenssen TK, Kuo WP, Stokke T, Hovig E. Associations between gene expressions in breast cancer and patient survival. Hum Genet. 2002;111(4-5):411–420. doi: 10.1007/s00439-002-0804-5. [DOI] [PubMed] [Google Scholar]
- Pavlides S, Tsirigos A, Vera I, Flomenberg N, Frank PG, Casimiro MC, Wang C, Pestell RG, Martinez-Outschoorn UE, Howell A, Sotgia F, Lisanti MP. Transcriptional evidence for the "Reverse Warburg Effect" in human breast cancer tumor stroma and metastasis: similarities with oxidative stress, inflammation, Alzheimer's disease, and "Neuron-Glia Metabolic Coupling". Aging (Albany NY) 2010;2(4):185–199. doi: 10.18632/aging.100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Jiang Y, Liu C, Shen O, Tang W, Wang X. Identification of aberrant promoter hypomethylation of HOXA10 in ovarian cancer. J Cancer Res Clin Oncol. 2010;136(8):1221–1227. doi: 10.1007/s00432-010-0772-4. [DOI] [PubMed] [Google Scholar]
- Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, Crenshaw A, Cancel-Tassin G, Staats BJ, Wang Z, Gonzalez-Bosquet J, Fang J, Deng X, Berndt SI, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cussenot O, Valeri A, Andriole GL, Crawford ED, Tucker M, Gerhard DS, Fraumeni JF Jr, Hoover R, Hayes RB, Hunter DJ, Chanock SJ. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40(3):310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- Hasegawa S, Furukawa Y, Li M, Satoh S, Kato T, Watanabe T, Katagiri T, Tsunoda T, Yamaoka Y, Nakamura Y. Genome-wide analysis of gene expression in intestinal-type gastric cancers using a complementary DNA microarray representing 23,040 genes. Cancer Res. 2002;62(23):7012–7017. [PubMed] [Google Scholar]
- Madoz-Gurpide J, Canamero M, Sanchez L, Solano J, Alfonso P, Casal JI. A proteomics analysis of cell signaling alterations in colorectal cancer. Mol Cell Proteomics. 2007;6(12):2150–2164. doi: 10.1074/mcp.M700006-MCP200. [DOI] [PubMed] [Google Scholar]
- Li HP, Ji JF, Hou KY, Lei YT, Zhao HM, Wang J, Zheng J, Liu JY, Wang MP, Xiao Y, Wang YF, Lu YY, Sun Y. Prediction of recurrence risk in early breast cancer using human epidermal growth factor 2 and cyclin A2. Chin Med J (Engl) 2010;123(4):431–437. [PubMed] [Google Scholar]
- Cigognini D, Corneo G, Fermo E, Zanella A, Tripputi P. HIC gene, a candidate suppressor gene within a minimal region of loss at 7q31.1 in myeloid neoplasms. Leuk Res. 2007;31(4):477–482. doi: 10.1016/j.leukres.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Rorive S, Maris C, Debeir O, Sandras F, Vidaud M, Bieche I, Salmon I, Decaestecker C. Exploring the distinctive biological characteristics of pilocytic and low-grade diffuse astrocytomas using microarray gene expression profiles. J Neuropathol Exp Neurol. 2006;65(8):794–807. doi: 10.1097/01.jnen.0000228203.12292.a1. [DOI] [PubMed] [Google Scholar]
- Kwabi-Addo B, Wang S, Chung W, Jelinek J, Patierno SR, Wang BD, Andrawis R, Lee NH, Apprey V, Issa JP, Ittmann M. Identification of differentially methylated genes in normal prostate tissues from African American and Caucasian men. Clin Cancer Res. 2010;16(14):3539–3547. doi: 10.1158/1078-0432.CCR-09-3342. [DOI] [PubMed] [Google Scholar]
- Jacinto FV, Ballestar E, Ropero S, Esteller M. Discovery of epigenetically silenced genes by methylated DNA immunoprecipitation in colon cancer cells. Cancer Res. 2007;67(24):11481–11486. doi: 10.1158/0008-5472.CAN-07-2687. [DOI] [PubMed] [Google Scholar]
- Lae M, Ahn EH, Mercado GE, Chuai S, Edgar M, Pawel BR, Olshen A, Barr FG, Ladanyi M. Global gene expression profiling of PAX-FKHR fusion-positive alveolar and PAX-FKHR fusion-negative embryonal rhabdomyosarcomas. J Pathol. 2007;212(2):143–151. doi: 10.1002/path.2170. [DOI] [PubMed] [Google Scholar]
- Graham K, de las Morenas A, Tripathi A, King C, Kavanah M, Mendez J, Stone M, Slama J, Miller M, Antoine G, Willers H, Sebastiani P, Rosenberg CL. Gene expression in histologically normal epithelium from breast cancer patients and from cancer-free prophylactic mastectomy patients shares a similar profile. Br J Cancer. 2010;102(8):1284–1293. doi: 10.1038/sj.bjc.6605576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Kobunai T, Toda E, Kanazawa T, Kazama Y, Tanaka J, Tanaka T, Yamamoto Y, Hata K, Kojima T, Yokoyama T, Konishi T, Okayama Y, Sugimoto Y, Oka T, Sasaki S, Ajioka Y, Muto T, Nagawa H. Gene expression signature and the prediction of ulcerative colitis-associated colorectal cancer by DNA microarray. Clin Cancer Res. 2007;13(2 Pt 1):415–420. doi: 10.1158/1078-0432.CCR-06-0753. [DOI] [PubMed] [Google Scholar]
- Campbell JM, Lockwood WW, Buys TP, Chari R, Coe BP, Lam S, Lam WL. Integrative genomic and gene expression analysis of chromosome 7 identified novel oncogene loci in non-small cell lung cancer. Genome. 2008;51(12):1032–1039. doi: 10.1139/G08-086. [DOI] [PubMed] [Google Scholar]
- Fernandez LP, Milne RL, Pita G, Floristan U, Sendagorta E, Feito M, Aviles JA, Martin-Gonzalez M, Lazaro P, Benitez J, Ribas G. Pigmentation-related genes and their implication in malignant melanoma susceptibility. Exp Dermatol. 2009;18(7):634–642. doi: 10.1111/j.1600-0625.2009.00846.x. [DOI] [PubMed] [Google Scholar]
- Schraders M, Jares P, Bea S, Schoenmakers EF, van Krieken JH, Campo E, Groenen PJ. Integrated genomic and expression profiling in mantle cell lymphoma: identification of gene-dosage regulated candidate genes. Br J Haematol. 2008;143(2):210–221. doi: 10.1111/j.1365-2141.2008.07334.x. [DOI] [PubMed] [Google Scholar]
- Rohozinski J, Anderson ML, Broaddus RE, Edwards CL, Bishop CE. Spermatogenesis associated retrogenes are expressed in the human ovary and ovarian cancers. PLoS One. 2009;4(3):e5064. doi: 10.1371/journal.pone.0005064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grade M, Hormann P, Becker S, Hummon AB, Wangsa D, Varma S, Simon R, Liersch T, Becker H, Difilippantonio MJ, Ghadimi BM, Ried T. Gene expression profiling reveals a massive, aneuploidy-dependent transcriptional deregulation and distinct differences between lymph node-negative and lymph node-positive colon carcinomas. Cancer Res. 2007;67(1):41–56. doi: 10.1158/0008-5472.CAN-06-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Shields CL. Differential gene expression profile of retinoblastoma compared to normal retina. Mol Vis. 2010;16:1292–1303. [PMC free article] [PubMed] [Google Scholar]
- Schaeffer DF, Owen DR, Lim HJ, Buczkowski AK, Chung SW, Scudamore CH, Huntsman DG, Ng SS, Owen DA. Insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) overexpression in pancreatic ductal adenocarcinoma correlates with poor survival. BMC Cancer. 2010;10:59. doi: 10.1186/1471-2407-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MW, Yue ZN, Geiss GK, Sadovnikova NY, Carter VS, Boix L, Lazaro CA, Rosenberg GB, Bumgarner RE, Fausto N, Bruix J, Katze MG. Identification of novel tumor markers in hepatitis C virus-associated hepatocellular carcinoma. Cancer Res. 2003;63(4):859–864. [PubMed] [Google Scholar]
- Shirahata M, Iwao-Koizumi K, Saito S, Ueno N, Oda M, Hashimoto N, Takahashi JA, Kato K. Gene expression-based molecular diagnostic system for malignant gliomas is superior to histological diagnosis. Clin Cancer Res. 2007;13(24):7341–7356. doi: 10.1158/1078-0432.CCR-06-2789. [DOI] [PubMed] [Google Scholar]
- Ikenaga N, Ohuchida K, Mizumoto K, Yu J, Kayashima T, Hayashi A, Nakata K, Tanaka M. Characterization of CD24 expression in intraductal papillary mucinous neoplasms and ductal carcinoma of the pancreas. Hum Pathol; 2010. [DOI] [PubMed] [Google Scholar]
- Takeno A, Takemasa I, Doki Y, Yamasaki M, Miyata H, Takiguchi S, Fujiwara Y, Matsubara K, Monden M. Integrative approach for differentially overexpressed genes in gastric cancer by combining large-scale gene expression profiling and network analysis. Br J Cancer. 2008;99(8):1307–1315. doi: 10.1038/sj.bjc.6604682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim HN, Lee KT, Lee JK, Choi SH, Paik SW, Rhee JC, Lowe AW. Gene expression profiles in gallbladder cancer: the close genetic similarity seen for early and advanced gallbladder cancers may explain the poor prognosis. Tumour Biol. 2008;29(1):41–49. doi: 10.1159/000132570. [DOI] [PubMed] [Google Scholar]
- Chen C, Mendez E, Houck J, Fan W, Lohavanichbutr P, Doody D, Yueh B, Futran ND, Upton M, Farwell DG, Schwartz SM, Zhao LP. Gene expression profiling identifies genes predictive of oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17(8):2152–2162. doi: 10.1158/1055-9965.EPI-07-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prapinjumrune C, Morita K, Kuribayashi Y, Hanabata Y, Shi Q, Nakajima Y, Inazawa J, Omura K. DNA amplification and expression of FADD in oral squamous cell carcinoma. J Oral Pathol Med. 2010;39(7):525–532. doi: 10.1111/j.1600-0714.2009.00847.x. [DOI] [PubMed] [Google Scholar]
- Blaveri E, Brewer JL, Roydasgupta R, Fridlyand J, DeVries S, Koppie T, Pejavar S, Mehta K, Carroll P, Simko JP, Waldman FM. Bladder cancer stage and outcome by array-based comparative genomic hybridization. Clin Cancer Res. 2005;11(19 Pt 1):7012–7022. doi: 10.1158/1078-0432.CCR-05-0177. [DOI] [PubMed] [Google Scholar]
- Chevillard S, Ugolin N, Vielh P, Ory K, Levalois C, Elliott D, Clayman GL, El-Naggar AK. Gene expression profiling of differentiated thyroid neoplasms: diagnostic and clinical implications. Clin Cancer Res. 2004;10(19):6586–6597. doi: 10.1158/1078-0432.CCR-04-0053. [DOI] [PubMed] [Google Scholar]
- Lau YF, Zhang J. Expression analysis of thirty one Y chromosome genes in human prostate cancer. Mol Carcinog. 2000;27(4):308–321. doi: 10.1002/(SICI)1098-2744(200004)27:4<308::AID-MC9>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Garcia-Munoz R, Panizo C, Bendandi M, Llorente L. Autoimmunity and lymphoma: is mantle cell lymphoma a mistake of the receptor editing mechanism? Leuk Res. 2009;33(11):1437–1439. doi: 10.1016/j.leukres.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Dong S, Nutt CL, Betensky RA, Stemmer-Rachamimov AO, Denko NC, Ligon KL, Rowitch DH, Louis DN. Histology-based expression profiling yields novel prognostic markers in human glioblastoma. J Neuropathol Exp Neurol. 2005;64(11):948–955. doi: 10.1097/01.jnen.0000186940.14779.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorringe KL, Ramakrishna M, Williams LH, Sridhar A, Boyle SE, Bearfoot JL, Li J, Anglesio MS, Campbell IG. Are there any more ovarian tumor suppressor genes? A new perspective using ultra high-resolution copy number and loss of heterozygosity analysis. Genes Chromosomes Cancer. 2009;48(10):931–942. doi: 10.1002/gcc.20694. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of genes associated in glioblastoma from the literature Table containing the list of 174 genes previously reported in the literature.