Abstract
When Chlamydomonas reinhardtii cells mate, a zygotic maturation program is activated, part of which leads to destruction of chloroplast DNA (cpDNA) from the mating type minus (mt−) parent, and, therefore, to uniparental inheritance of mating type plus (mt+) cpDNA. A long-standing model that explains the selective destruction of mt− cpDNA in zygotes invokes a methylation-restriction system. We tested this model by using the potent methylation inhibitor 5-aza-2‘-deoxycytidine (5adc) to hypomethylate parental cpDNA and found that the pattern of cpDNA inheritance is altered by 5adc in a manner that is consistent with the model. Surprisingly, however, hypomethylated mt+ cpDNA is not destroyed in zygotes as the methylation-restriction model predicts it should be. Destruction of mt− cpDNA is also unaffected when the parental mt+ cpDNA is hypomethylated. Instead, loss of methylation affects the relative rates of replication of residual mt− cpDNA and mt+ cpDNA in germinating zygotes. The mode of action for 5adc on cpDNA replication in germinating zygotes may be via hypomethylation of mt+ cpDNA, but is also consistent with its action as a DNA-damaging agent. Interestingly, 5adc causes reduced cpDNA replication only in germinating zygotes, not in vegetatively grown cells, indicating that cpDNA replication is qualitatively different in these two stages of the life cycle. Our results demonstrate that methylation is not necessary for protection of the mt+ cpDNA in early zygotes and uncover a novel stage of the Chlamydomonas life cycle when replication of cpDNA is highly susceptible to perturbation. Our data support a model in which differential cpDNA replication in germinating zygotes is used as a mechanism to selectively amplify intact and properly methylated cpDNA molecules.
Keywords: uniparental inheritance, 5-aza-2‘-deoxycytidine, ethionine, organelle, replication
A ubiquitous feature of sexual eukaryotes is the non-Mendelian inheritance pattern of their organellar genomes. Meiotic progeny typically receive most or all of their mitochondrial and chloroplast DNA from a single parent (Birky 1995). The diverse strategies employed by various lineages to achieve uniparental organelle inheritance is itself a testament to the strong selective pressure that must exist to ensure this result and the broad importance of uniparental inheritance for sexual eukaryotes. For example, in plants, there are various methods by which pollen plastids are prevented from entering oocytes, but there are also exceptions, notably in gymnosperms, in which pollen plastid DNA not only enters oocytes but is preferentially retained or propagated, resulting in paternal (rather than the usual maternal) inheritance (Whatley 1982; Corriveau and Coleman 1991; Gillham et al. 1991). Despite a wealth of interesting descriptive knowledge, the molecular bases for uniparental inheritance in various taxa remain poorly understood.
One of the best-characterized systems for investigating uniparental chloroplast inheritance is the haploid soil alga, Chlamydomonas reinhardtii (Armbrust 1998). C. reinhardtii is a unicellular flagellate that exists as two mating types, termed plus (mt+) and minus (mt−). Each C. reinhardtii cell has a single large chloroplast that contains ∼80 copies of a circular 200-kb genome. The chloroplast DNA (cpDNA) is organized into ∼10 discrete nucleoprotein bodies in the stroma termed nucleoids and is replicated continuously during the mitotic cell cycle (Turmel et al. 1980).
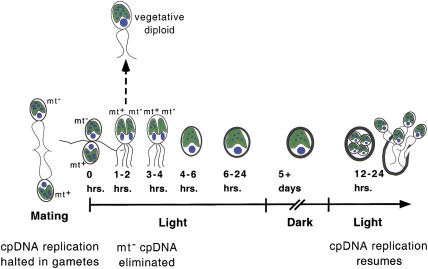
In nutrient-replete conditions, vegetative cells divide mitotically, but when starved of nitrogen, they differentiate into mating-competent gametes. When gametes of opposite mating type are mixed, they fuse rapidly to form diploid zygotes and almost immediately begin a differentiation program that leads to major structural and cytological changes (Fig. 1). These changes include deposition of a thick cell wall, retraction of flagella, nuclear fusion, and fusion of the chloroplasts contributed by the two parents. A key event with respect to chloroplast inheritance is the degradation of mt−-derived cpDNA within the first few hours of zygote formation, prior to chloroplast fusion (Burton et al. 1979; Kuroiwa et al. 1982). After a maturation period in the dark (∼5 d), and reintroduction to light and nutrients, zygotes undergo meiosis and germinate to produce four haploid progeny that can reenter the vegetative growth cycle (Fig. 1; Harris 1989; Goodenough 1991).
Figure 1.
Schematic representation of zygote-maturation time course. Cells are pictured with the nucleus as a blue circle and the chloroplast as a green cup-shaped structure containing nucleoids that are represented by small blue dots. cpDNA is not replicated in gametes or maturing zygotes. Replication resumes in germinating zygotes. Zygotic maturation is diagrammed as a time course with cell fusion occurring at 0 h. Nucleoids from the mt− parent are shown disappearing ∼3–4 h after mating. In the next several hours, the flagella are withdrawn, the parental chloroplasts and nuclei fuse, and the zygotic cell wall begins forming. After 5 d of dark incubation, zygotes that are returned to light and nutrients will undergo meiosis and hatch within 12 to 24 h. Rare vegetative diploids can form prior to mt− cpDNA destruction.
Whereas nuclear markers segregate in the typical 2:2 fashion, elimination of mt− cpDNA from zygotes results in uniparental inheritance of chloroplast genes from the mt+ parent by most progeny. The system is not, however, invariant: A small number (∼0.1%–5%) of zygotes in a cross fail to eliminate all the mt− cpDNA. The resulting meiotic progeny, termed exceptionals, are usually heteroplasmic initially, but random segregation subsequently generates clones that have become homoplasmic for one of the two parental inputs (Birky 1994).
Experiments with mt+/mt− diploids indicate that the mt+ locus controls inheritance of cpDNA. Such diploids can be produced in two ways: (1) genetic selection of rare zygotes that forgo zygotic maturation and instead divide mitotically as diploid vegetative cells (Ebersold 1963, 1967; Fig. 1), or (2) genetic selection of diploids produced by polyethylene glycol-induced fusion of haploid cells (Matagne et al. 1979). mt+/mt− diploids produced by either method differentiate as mt− gametes because of dominance of the mt− locus (Ebersold 1967; Ferris and Goodenough 1997) and, thus, they can be mated to either mt+ haploids or mt+/mt+ diploids. In either case, the progeny of such crosses inherit cpDNA markers biparentally (Matagne and Beckers 1983). This observation is consistent with a simple protection/destruction model for uniparental chloroplast inheritance in which the mt+ locus encodes the ability to protect cpDNA from later destruction by a zygotic nuclease. To date, no mt+-encoded protection gene(s) or zygotic nucleases that participate in uniparental cpDNA inheritance have been identified.
The most prominent model for uniparental cpDNA inheritance in C. reinhardtii, first proposed by Sager and Lane (1972), invokes a methylation-restriction system. The model specifically states that mt+ cpDNA is protected by methylation, and that the zygotic nuclease is a methylation-sensitive enzyme, analogous to bacterial restriction/modification systems.
The methylation-restriction model is most robustly supported by the finding of differential cpDNA methylation in gametes and zygotes. Whereas vegetative cells of both mating types have barely detectable levels of cpDNA methylation, mt+ cpDNA becomes more heavily methylated on cytosine residues than mt− cpDNA (12% vs. 4%) during gametogenesis (Feng and Chiang 1984), where 5-methylcytosine is the only identified modification in C. reinhardtii (Burton et al. 1979). During zygote maturation, mt+ cpDNA undergoes extensive further methylation until ∼50% of all cytosines are converted to 5-methylcytosine (Feng and Chiang 1984). Interestingly, the residual mt− cpDNA of zygotes is reported to be far less methylated than the mt+ cpDNA (Burton et al. 1979). Once zygotes germinate, the methylation pattern is soon lost, as vegetatively growing cells do not remethylate their cpDNA (Sano et al. 1984). Other experiments have established that the presence of a mt+ locus in mt+/mt− diploids induces levels of gametic methylation similar to those seen in mt+ cells (Sager et al. 1981), a result that would be predicted from the methylation-restriction model.
The model was challenged when a spontaneous mutation, me-1, was found to cause up to 35% cytosine methylation of cpDNA in vegetative cells and gametes of both mating types. Although increased methylation of mt− cpDNA in me-1 cells does not spare it from destruction in zygotes (Bolen et al. 1982), it was later determined that some of the sites that are methylated in mt+ gametic and zygotic cpDNA are not methylated in me-1 mutants (Sager and Grabowy 1983). Thus, although there are several observations that support the methylation-restriction model of cpDNA inheritance, the model still awaits rigorous testing.
In this work, we used the potent and specific methylation inhibitor, 5-aza-2‘-deoxycytidine (5adc), to test the methylation-restriction model. We first found that 5adc treatment of parental strains affects the outcome of crosses in a manner that is predicted by the model. Surprisingly, however, we subsequently found that destruction of hypomethylated mt+ cpDNA does not occur in zygotes as the methylation-restriction model predicts that it should. 5adc treatment has no effect on the early program of zygotic maturation during which mt− cpDNA is destroyed and mt+ cpDNA is retained. Instead, 5adc treatment reduces the ability of cpDNA to replicate during zygote germination. Zygote germination was found to be a unique and critical stage of the life cycle when cpDNA replication is highly sensitive to perturbation, and we suggest that it may also be a time when residual unmethylated and/or damaged cpDNA is eliminated.
Results
Genetic test of the methylation-restriction model
To test the methylation-restriction model of cpDNA inheritance for Chlamydomonas, we used 5adc to block cytosine methylation in either mt+ or mt− gametes. The model predicts that if the mt+ parent loses cpDNA methylation, its cpDNA should be partially or completely destroyed in zygotes. Therefore, the progeny should either be inviable, because of a loss of cpDNA, or have a mt+:mt− cpDNA ratio that is skewed toward mt−, resulting in a higher proportion of exceptional progeny that inherit mt− cpDNA. For example, if 90% of the mt+ cpDNA were destroyed, and the residual mt− cpDNA stayed constant, there would be a 10-fold increase in the frequency of exceptional progeny. Correspondingly, if the mt− cpDNA were hypomethylated, then it might be eliminated more efficiently, resulting in fewer exceptional progeny compared with control crosses.
Cells plated on 5adc up to the highest concentration tested (250 μM) grew at normal rates and did not show any obvious phenotypic differences from control cells (data not shown). Moreover, mt+ or mt− gametes produced from cells grown on 5adc-containing plates mated normally and underwent zygotic maturation and germination like untreated cells. Table 1 and Figure 2 show the result of crosses in which either the mt+ or mt− parent was grown in the presence of 5adc before mating. In the first set of crosses, (Table 1, crosses 1–3; Fig. 2) we scored germinated zygote colonies for the presence of both parental cpDNA markers. This method of scoring gives an indication of overall changes in cpDNA inheritance patterns but is nonquantitative, as a biparental zygote could contain a wide range of distributions of mt+ and mt− cpDNA among its ∼80 copies of the chloroplast genome.
Table 1.
Effect of 5adc on chloroplast inheritance
| Parents
|
Progeny
|
||||||
|---|---|---|---|---|---|---|---|
|
|
Cross
|
5adc treatment
|
Total scored
|
%Uniparental (mt+)
|
%Biparental
|
%Uniparental (mt−)
|
%Exceptional
|
| 1 | CC86 mt+ kanR × CC2663 mt− erR | none | 192 | 95 | 5 | 0 | 5 |
| 2 | CC86 mt+ kanR × CC2663 mt− erR | mt+ parent | 192 | 49 | 30 | 21 | 51 |
| 3 | CC86 mt+ kanR × CC2663 mt− erR | mt− parent | 192 | 99 | 0.5 | 0 | 0.5 |
| 4 | R3 mt+ × CJU10 mt− SpecR | none | 340 | na | na | na | 1.8 |
| 5 | R3 mt+ × CJU10 mt− SpecR | mt+ parent | 289 | na | na | na | 22 |
| 6 | R3 mt+ × CJU10 mt− SpecR | mt− parent | 276 | na | na | na | 0 |
Each cross was carried out using strains with the indicated chloroplast antibiotic resistance markers (erR, erythromycin resistance; kanR, kanamycin resistance; specR, spectinomycin resistance). Parental strains were grown on 200 μM 5adc as indicated. For crosses 1–3, zygote colonies were scored for chloroplast markers with uniparental (mt+) indicating that only the mt+ parental marker was detected, uniparental (mt−) indicating that only the mt− marker was detected and biparental indicating that both markers were detected in the zygote colony. The value %exceptionals reflects the sum of biparental and uniparental (mt−) progeny. For crosses 4–6, progeny were propagated to homoplasmy and subcloned; individuals were then scored for the presence of the mt− cpDNA marker, and those that were spectinomycin resistant were scored as exceptionals.
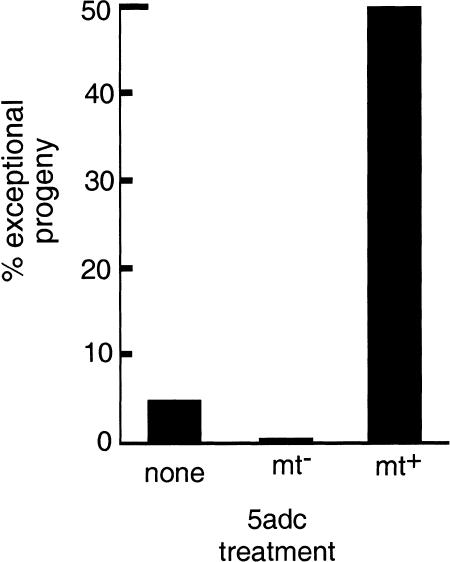
Figure 2.
Bar graph showing the effect of 5adc on chloroplast DNA inheritance. (Horizontal axis) Parental strain that was grown on 5adc (200 μM); (vertical axis) number of exceptional progeny produced (those carrying the mt− parental chloroplast marker). A cross with a relatively high frequency of exceptional progeny (∼5%) in the control mating (no 5adc treatment) was used to illustrate the ability of 5adc to increase or decrease the frequency of exceptional progeny depending on which parent is treated with the drug.
In all subsequent crosses (Table 1, crosses 4–6), we allowed zygote progeny to grow for ∼10–12 generations and become homoplasmic before scoring for the mt− cpDNA marker. The frequency of exceptional progeny determined by this method is a direct reflection of the frequency of mt− cpDNA genomes in the population after zygote germination.
When chloroplast genetic markers were scored by either of the above methods, the cpDNA from the hypomethylated parental strain was always observed to be under-represented in the progeny compared with its frequency in control crosses. 5adc treatment typically caused at least a 10- to 20-fold increase in exceptional progeny when the mt+ parent was treated and a 10- to 20-fold decrease in exceptional progeny when the mt− parent was treated (Table 1; Fig. 2). The example we chose for the first set of crosses (Table 1, crosses 1–3; Fig. 2) had a higher than usual frequency of exceptional progeny in the control cross and illustrates the ability of 5adc to either increase or decrease the frequency of exceptional progeny depending on whether it is administered to the mt+ or mt− parent.
Dose-dependent responses to 5adc
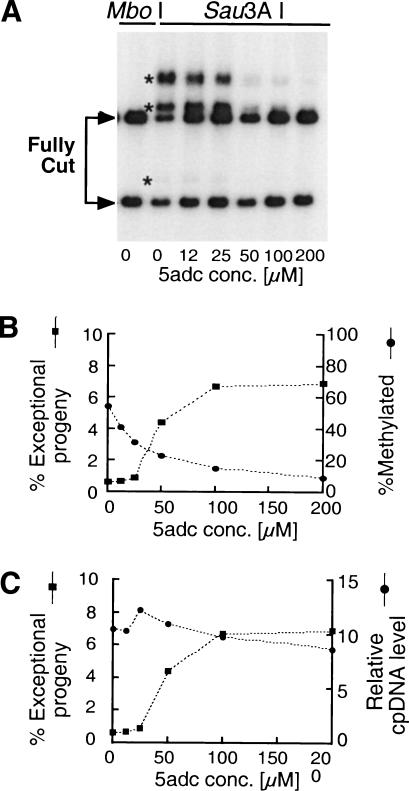
To determine whether decreased cpDNA methylation is correlated with changes in cpDNA inheritance, we looked at the dose response of each to different concentrations of 5adc. The level of mt+ cpDNA methylation in gametes (Fig. 3A,B) was measured by cutting cpDNA with the methylcytosine-sensitive enzyme, Sau3AI, or its methylcytosine-insensitive isoschizomer, MboI as a control (Fig. 3A). Similar results were obtained with other methylcytosine-sensitive restriction enzymes, such as HpaII and MspI, and also when looking at methylation of the chloroplast 16S rDNA genes (data not shown). Decreased methylation and increased frequencies of exceptional progeny are correlated and appear to reach limit values at 100–250 μM 5adc (Fig. 3B).
Figure 3.
Dose-dependent responses of methylation levels, chloroplast DNA levels, and exceptional progeny frequencies to 5adc concentration. (A) mt+ cells were grown on varying concentrations of 5adc, and gametic DNA was prepared and cut with either MboI (methylcytosine insensitive) or its isoschizomer Sau3AI (methylcytosine sensitive). The DNA was used to prepare a Southern blot that was probed with atpB sequences. (Arrows) Positions of fully cut DNA fragments; (asterisks) methylated bands. (B) Gametes used in A were mated, and the frequency of exceptional progeny and level of gametic cpDNA methylation (vertical axes) were plotted against 5adc dosage (horizontal axis). Methylation was quantitated from data in A. (C) Same experiment as in B except the total amount of chloroplast DNA in gametes rather than methylation levels is plotted on the right vertical axis.
Decreased cpDNA copy number in mt+ gametes is known to affect cpDNA inheritance (Wurtz et al. 1977; Armbrust et al. 1995). Therefore, we also measured the levels of cpDNA in cells treated with 5adc to determine whether the drug blocks cpDNA replication and decreases cpDNA copy number in vegetative cells and gametes. We found, however, that 5adc treatment has no significant effect on levels of cpDNA in gametes (Fig. 3C).
Next, we examined the effect of 5adc treatment on cpDNA methylation in zygotes. We mated mt+ gametes grown with and without 200 μM 5adc to untreated mt− gametes and assayed zygotic cpDNA methylation at intervals after mating. In an untreated control mating mt− cpDNA is destroyed during the first several hours (see Figs. 5 and 6, below) whereas the remaining mt+ cpDNA continues to be methylated (Fig. 4A). Pretreatment of mt+ gametes with 5adc strongly inhibits but does not completely prevent cpDNA methylation in zygotes (Fig. 4B).
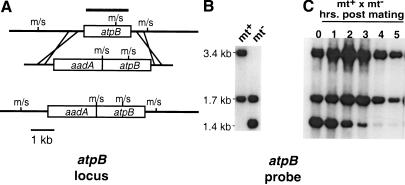
Figure 5.
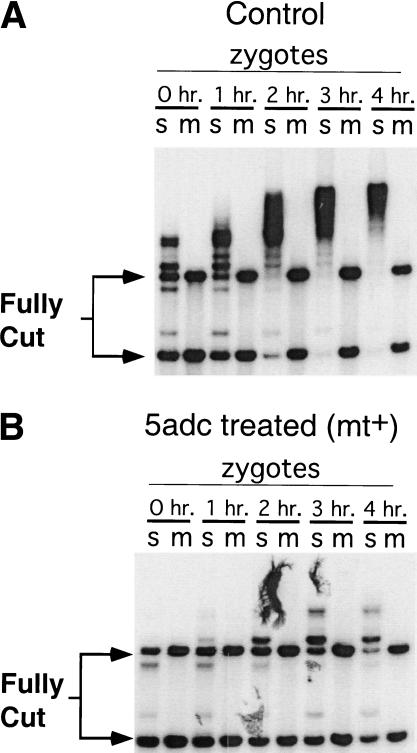
Molecularly tagging the atpB locus. (A) Schematic of the recombination event that generated the tagged atpB locus in mt− cpDNA. (Top line) Wild-type atpB locus with the coding region boxed. The locations of MboI/Sau3AI recognition sites are marked m/s. The atpB probe used in all these experiments is indicated by the thick shaded line above the locus. The recombining plasmid sequence is shown below the atpB locus with aadA (spectinomycin resistance) and atpB coding sequences boxed. (Bottom) Recombination product. (B) DNA from wild-type mt+ or tagged mt− gametes was cut with MboI, Southern blotted, and probed with the atpB fragment indicated in A. (C) Gametes from B were mated, and DNA prepared from zygotes at the indicated time points was cut with MboI, and blotted as in B. Residual unmated gamete DNA was not removed from the zygote DNA preparations in this experiment.
Figure 6.
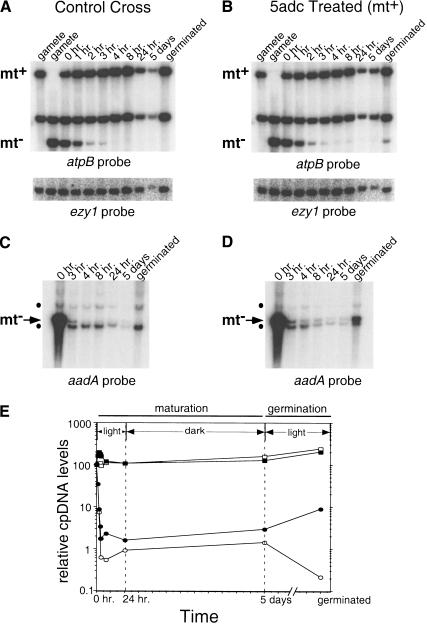
Effect of 5adc on the fate of cpDNA. (A) DNA from gametes or zygotes formed from a wild-type mt+ strain mated to a tagged mt− strain was cut with MboI, Southern blotted, and probed with chloroplast atpB sequences (top) or nuclear ezy1 sequences (bottom). The time course of zygote maturation extends to the end of the dark period (5 d). The last lane contains DNA from progeny that have germinated and grown vegetatively for several generations (4–5 d). Lanes from 24-h and 5-d samples contain less total DNA than the other lanes. (B) Same experiment as in A except the mt+ parent was grown on 200 μM 5adc before mating. (C) Same experiment as in A except that the DNA was cut with the methylcytosine-insensitive enzyme EcoRV, which releases a single ∼4.2-kb band containing the aadA transgene, and was probed with aadA sequences. The dots indicate the locations of weakly cross-hybridizing bands that are unrelated to the presence of the aadA transgene and serve as useful internal controls for levels of the authentic aadA band (marked as mt−). (D) Same experiment as in C except that the mt+ parent was grown on 200 μM 5adc before mating. (E) cpDNA signals from A and C were normalized to the nuclear signal and plotted on a logarithmic scale against the same time course of zygote maturation and germination. The symbols indicate control cross mt+ cpDNA (□), control cross mt− cpDNA (○), 5adc-treated cross mt+ cpDNA (▪), and 5adc-treated cross mt− cpDNA (●). Starting cpDNA for all samples was set at 100. The light and dark periods during which zygotes mature and germinate are also indicated.
Figure 4.
Effect of 5adc treatment on zygotic chloroplast DNA methylation. (A) Control mating. DNA from zygotes was prepared at the indicated times after mating, cut with either MboI (M) or Sau3AI (S), Southern blotted, and probed with atpB sequences. (B) Same experiment as in A except the mt+ parent was grown in the presence of 200 μM 5adc before mating.
Determining the fate of hypomethylated cpDNA
The above results are consistent with the methylation-restriction model, but they do not reveal the actual fate of mt+ and mt− cpDNA in developing zygotes. The methylation-restriction model specifically predicts that hypomethylated mt+ cpDNA should be eliminated in developing zygotes: A 10-fold increase in the frequency of exceptional progeny that inherit mt− cpDNA would require that ∼90% of the mt+ cpDNA be destroyed.
The individual fates of mt+ and mt− zygotic cpDNA in the above experiment (Fig. 4A,B) could not be determined because the mt+ and the mt− cpDNA could not be distinguished. To follow the fates of mt+ and mt− cpDNA in zygotes, we constructed a strain, CJU10, by transforming an mt− wild-type strain with a construct that integrates at the chloroplast atpB locus and places the bacterial aadA gene next to atpB (Fig. 5A; Zerges and Rochaix 1994). The aadA gene confers resistance to spectinomycin, allowing us to follow cpDNA inheritance genetically, and provides a unique sequence tag that allowed us to follow the fate of mt− cpDNA on Southern blots. The inserted transgene also generates a useful RFLP at the atpB locus when DNA is cut with MboI or Sau3AI: When CJU10 DNA is probed with atpB sequences, the 3.4-kb MboI fragment found in wild-type strains is replaced by a 1.4-kb fragment (Fig. 5B). When CJU10 was mated to a wild-type mt+ strain, we observed elimination of mt− cpDNA during the first several hours of zygote maturation (Fig. 5C). Interestingly, putative degradation intermediates were never observed on blots probed with atpB or aadA sequences, regardless of exposure time (Figs. 5C and 6A,C, plus other data not shown), indicating that once initiated, cpDNA degradation takes place rapidly. As expected, very few progeny from the above cross inherited the spectinomycin resistance marker carried by CJU10 (Table 1, cross 4). When the spectinomycin resistance transgene marker was carried instead by the mt+ parent, >99% of the progeny were spectinomycin resistant, indicating that the transgene is inherited in the same manner as endogenous chloroplast genes (data not shown).
Next, we used CJU10 as the mt− strain in matings with wild-type gametes, with and without 5adc pretreatment. To ensure the most sensitive and quantitative results possible, we developed procedures to eliminate unmated gametes prior to zygotic DNA preparation and to efficiently break open the walls of zygotic cells, resulting in high yields of pure zygotic DNA (Materials and Methods). In Southern blots prepared with DNA from the control mating and probed with atpB sequences, the mt−-specific cpDNA band disappeared during the first several hours after mating and could only be detected with long exposures in 24-h zygotes, in 5-day-old zygotes (just prior to germination), or in the germinated progeny (Fig. 6A). When Southern blots were prepared with the same DNA preparations and then probed with aadA sequences to specifically detect mt− cpDNA, a very low signal could be observed in DNA prepared from mature zygotes and from zygotes just prior to and after germination, confirming the presence of a small fraction of exceptional zygotes and progeny (Fig. 6C). Strikingly, in the cross where the cpDNA of the mt+ parent was hypomethylated, there was no discernable loss of mt+ cpDNA during zygotic development as the methylation-restriction model would predict (Fig. 6B,D). Even more surprising, the process of mt− cpDNA destruction also appeared completely normal: The amount of residual mt− cpDNA at the end of zygote maturation (Fig. 6A–E, 5 d) was not significantly different than in the control cross (less than twofold; Fig. 6E). However, during germination, the relative amount of mt− cpDNA in the control cross decreased, whereas in the 5adc-treated cross it increased, generating an ∼40-fold difference between the two sets of progeny in the final amounts of mt− cpDNA (Fig. 6E, 5 d). The ratios of mt+:mt− cpDNA in the germinated progeny of the 5adc-treated and control crosses were consistent with our genetic results in this experiment in which we observed ∼7% exceptional progeny in the 5adc-treated cross and ∼0.4% exceptional progeny in the control cross (data not shown).
The increased ratio of mt−:mt+ cpDNA observed in progeny from the 5adc-treated cross (Fig. 6E; 5 d vs. germinated) could result either from: (1) slower replication of mt+ cpDNA (relative to residual mt− cpDNA) during germination or (2) active destruction of 5adc-treated mt+ cpDNA during germination. To test these possibilities, we used a quantitative PCR assay to monitor levels of nuclear and cpDNA during zygote maturation. This assay is more sensitive and accurate than Southern hybridization for detecting the low-abundance residual mt− cpDNA in mature zygotes and progeny. Zygotes from crosses with and without 5adc treatment of the mt+ parent were germinated in liquid medium, and DNA was prepared from aliquots of cells taken at intervals. Then, primers that are specific for nuclear DNA, mt− cpDNA, and mt+ cpDNA were used to amplify the three types of DNA from each sample and follow their relative abundance over time (Fig. 7). In this time course, the first progeny cells appeared between 16 and 24 h, with the majority of zygotes hatching by 32 h. The germination efficiency of the cultures was 83% for the control and 67% for the 5adc-treated cells. The nuclear DNA in the cultures increased by approximately threefold for the control sample and ∼twofold for the 5adc-treated sample with the 5adc-treated culture showing a slight delay (Fig. 7A). Although not all the zygotes germinated, following meiosis, some meiotic progeny immediately underwent one or two additional mitotic divisions following meiosis (Chiang and Sueoka 1967; Coleman 1984; J.G. Umen and U.W. Goodenough, unpubl.) allowing the nuclear DNA content of the culture to increase over the expected meiotic doubling.
Figure 7.
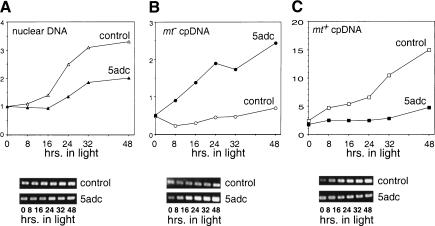
Nuclear and chloroplast DNA levels during germination. (A, top) Graph of nuclear DNA levels during germination from a control (▵) and 5adc-treated (mt+) (▴) cross. (y axis) Relative nuclear DNA levels normalized to the starting concentration (0 h.); (x axis) time course of germination, starting with the transfer to light and liquid growth media at 0 hrs. (Bottom) PCR-amplified fragments from the nuclear gene fus1 used for the quantitation. (B, top) Graph of mt− cpDNA levels from control (○) and 5adc-treated (mt+) (●) samples. (y axis) Relative amounts of mt− cpDNA normalized to the starting nuclear DNA content. The x axis is labeled as in A. (Bottom) PCR-amplified fragments from the aadA gene (mt− specific) used for the quantitation. (C, top) Graph of mt+ cpDNA levels from control (□) and 5adc-treated (mt+) (▪) samples. (y axis) Relative amounts of mt+ cpDNA normalized to the starting nuclear DNA content. The x axis is labeled as in A. (Bottom) PCR-amplified fragments from the 3′ region of the chloroplast atpB gene (mt+ specific) used for the quantitation.
The patterns of cpDNA replication were as anticipated from Figure 6. The mt− cpDNA from each culture showed very different replication patterns: In the control sample, the residual mt− cpDNA replicated very slowly, and was eventually outpaced by the nuclear DNA, a result that we also observed by Southern hybridization in the previous experiment where we saw a decrease in the mt− cpDNA:nuclear DNA ratio (Fig. 6E). In contrast, the mt− cpDNA began immediate and rapid replication in the sample in which the mt+ cells had been 5adc-treated, increasing in abundance ∼fivefold during the course of germination (Fig. 7B). A reciprocal result was obtained when we examined mt+ cpDNA: In the control sample, mt+ cpDNA replicated rapidly, increasing ∼sixfold, whereas in the 5adc-treated sample, mt+ cpDNA replicated more slowly, increasing by only ∼threefold (Fig. 7C). It should be noted that it would be difficult to see the replication effect on mt+ cpDNA in Figure 6 in which germinated cells had grown for several generations and, therefore, had time to rectify any cpDNA deficit by increasing the total cpDNA copy number back to normal levels. In summary, 5adc treatment of mt+ cells had two effects on germinating zygotes: It caused increased replication of mt− cpDNA and decreased replication of mt+ cpDNA compared with cpDNA in control samples.
Testing the DNA damage hypothesis
Because 5adc is known to cause DNA damage as well as hypomethylation (Jackson-Grusby et al. 1997), we sought to test whether DNA damage could be partly responsible for the effect of 5adc on apparent cpDNA replication rates. We used the DNA-alkylating agent methanesulfonic acid ethyl ester (EMS) to treat gametes because the bulky lesions caused by EMS are similar to those caused by 5adc when it forms an irreversible adduct with the methyltransferase enzyme (Santi et al. 1984; Gabbara and Bhagwat 1995), but EMS had no measurable effect on cpDNA methylation (data not shown). We compared the relative efficacies of EMS and 5adc as mutagens and as agents affecting cpDNA inheritance. As shown in Table 2 (experiments I–III), we measured dose-dependent or time-dependent effects of both agents for generation of spectinomycin-resistant mutations (which are most commonly the result of mutations in Chlamydomonas chloroplast 16S rDNA; Harris et al. 1987). Comparable results were obtained when scoring for erythromycin resistance, another common chloroplast rDNA mutation (data not shown). Although there was variability between experiments, in all cases there was a rough correspondence between the fold increase in mutation frequency observed in cpDNA and the fold increase in frequency of exceptional progeny caused by each compound when compared with controls (Table 2, experiments I–III). It should be noted that the mutagenic efficiency of 5adc is probably an overestimate: 5adc treatment requires incorporation of the drug into replicating DNA, meaning that some of the antibiotic-resistant colonies may be mitotic clones, whereas EMS can be administered to stationary phase cells, meaning that antibiotic-resistant colonies arise from independent mutagenic events. Therefore, the effect of 5adc on cpDNA inheritance is likely to stem both from DNA damage and loss of cpDNA methylation, a conclusion supported by our results with L-ethionine (below). We were not able to follow the fate of cpDNA in these experiments because EMS-treated cells mate poorly.
Table 2.
Effect of 5adc, EMS, and L-ethionine on chloroplast DNA mutation frequency and inheritance
| Experiment
|
Treatment of mt+ parent
|
Mutagenesis
|
Progeny
|
|||
|---|---|---|---|---|---|---|
| SpecR frequency
|
Fold increase
|
Number scored
|
%exceptional
|
Fold increase
|
||
| I | none | 1.3 × 10−9 | 1× | 484 | 0.21 | 1× |
| 12.5 μm 5adc | 1.3 × 10−8 | 10× | 389 | 0.51 | 2.4× | |
| 25 μm 5adc | 3.7 × 10−8 | 30× | 541 | 1.1 | 5× | |
| 50 μm 5adc | 9.3 × 10−8 | 74× | 474 | 3.0 | 14× | |
| 100 μm 5adc | 9.7 × 10−8 | 78× | 458 | 6.5 | 31× | |
| 250 μm 5adc | 1.8 × 10−7 | 144× | 554 | 18 | 86× | |
| II | none | 1.3 × 10−9 | 1× | 578 | 0.17 | 1× |
| 200 μm 5adc | 1.9 × 10−7 | 152× | 550 | 6.7 | 39× | |
| 60′ EMS | 3.5 × 10−8 | 28× | 625 | 1.8 | 11× | |
| 120′ EMS | 9.5 × 10−8 | 76× | 688 | 6.7 | 39× | |
| III | none | 1.3 × 10−9 | 1× | 426 | 0.70 | 1× |
| 120′ EMS | 1.8 × 10−7 | 140× | 483 | 9.9 | 14× | |
| IVa | none | n.d. | n.d. | 859 | 0.93 | 1× |
| 1 mM L-ethionine | n.d. | n.d. | 605 | 5.8 | 6× | |
| IVb | none | n.d. | n.d. | 567 | 0.53 | 1× |
| 1 mM L-ethionine | n.d. | n.d. | 506 | 3.8 | 7× | |
In each cross the mt+ parent was wild type (R3) and was either plated directly on selective media, to score mutagenesis, or was mated to an untreated mt− strain, CJU10, so that exceptional progeny could be scored. For 5adc and L-ethionine, cells were grown on plates with the indicated concentration. For EMS treatment, gametes were incubated with EMS for the indicated amount of time prior to plating or mating. Exceptional progeny were scored as in Table 1.
n.d., Not determined.
We also evaluated a second methylation inhibitor, L-ethionine. L-ethionine does not incorporate into DNA or cause DNA damage like 5adc, but is instead a competitive inhibitor of S-adenosyl-methonine, the methyl donor used by methyltransferases (Cox 1986; Ahmad and Rao 1996). The effects of L-ethionine were similar to 5adc, consistent with methylation playing a direct role in cpDNA inheritance (Table 2, experiments IVa and IVb). Again, cells plated on L-ethionine grew and mated poorly, meaning that we could not follow the fate of their cpDNA during zygote development as we did for 5adc-treated cells. Taken together, our results with 5adc, EMS, and L-ethionine suggest that both loss of methylation and DNA damage affect cpDNA inheritance.
Effect of 5adc on vegetative diploids versus germinating zygotes
Our results indicate that 5adc exerts a specific effect on replication rates of cpDNA in germinating zygotes without affecting growth, viability, or cpDNA copy number in vegetative cells or gametes (Fig. 3C). Zygotes differ from vegetative and gametic cells in that they are diploid and have gone through a mating and a unique differentiation program. To ask whether either of these factors has an influence on the 5adc response, we created heteroplasmic vegetative diploid strains by crossing marked haploid strains and selecting for nuclear complementation (Ebersold 1967; see Materials and Methods; Table 3). We followed inheritance of mt+ versus mt− cpDNA in the diploids by following the segregation of cpDNA markers in these strains during their subsequent mitotic growth. Vegetative diploids have been shown to inherit far more mt− cpDNA than do meiotic progeny (Matagne 1981), possibly because the destruction process is incomplete, and as expected, 20%–25% of the cpDNA in the diploids derived from the mt− parent (Table 3). Of interest is the observation that when either parental haploid strain was treated with 5adc before performing the cross and selecting for vegetative diploids, this inheritance pattern was not affected (Table 3). Therefore, neither DNA methylation levels nor DNA damage has the same effect on cpDNA replication in vegetative diploids as it has in germinating zygotes, supporting the conclusion that the 5adc effect is exerted during the germination process, perhaps in concert with meiosis. As a control, cpDNA inheritance in normal meiotic progeny was scored in the same crosses, and the result of 5adc treatment was similar to what we have observed previously (Table 3).
Table 3.
Effect of 5adc on vegetative diploids versus meiotic progeny
| Parents
|
Vegetative diploids
|
Meiotic progeny
|
|||
|---|---|---|---|---|---|
| Cross
|
5adc treatment
|
Total
|
%erR
|
Total
|
%erR
|
| CC425 mt+ arg7 srR × CC2663 mt− nic7 erR | none | 252 | 21 | ∼1100 | 0.18 |
| CC425 mt+ arg7 srR × CC2663 mt− nic7 erR | mt+ | 171 | 25 | ∼1400 | 8.7 |
| CC425 mt+ arg7 srR × CC2663 mt− nic7 erR | mt− | 141 | 19 | ∼1200 | 0 |
The indicated parental strains with nuclear markers (arg7, arginine-requiring; nic7, nicotinamide-requiring) and chloroplast markers (srR, streptomycin resistant; erR, erythromycin resistant) were mated and either plated directly on selective media in the light, to generate vegetative diploids, or allowed to undergo zygotic maturation and germination, to generate meiotic progeny. Growth on 200 μM 5adc prior to mating is indicated. For both the vegetative diploids and meiotic progeny, cells were propagated to homoplasmy and then scored for the mt− derived cpDNA marker.
Discussion
Evidence against the methylation-restriction model of cpDNA inheritance
Although circumstantial evidence has accumulated supporting the methylation-restriction model of chloroplast inheritance in Chlamydomonas as originally proposed by Sager and Lane (1972), critical testing of the model has been limited. Our strategy for testing the model combined use of a potent methylation inhibitor, 5adc, and molecularly tagged strains for monitoring the fate of chloroplast DNA in developing zygotes.
A previous attempt to test the methylation-restriction model using 5adc and L-ethionine had yielded negative results (Feng and Chiang 1984). Because the inhibitors were added just prior to gametogenesis or mating, however, they presumably did not have time to act before critical methylation patterns were established. In our experiments, methylation inhibitors did affect the outcome of crosses and did so in a way that might be predicted if methylation protects cpDNA from digestion in zygotes (i.e., we observed reduced transmission of cpDNA from the parent with hypomethylated cpDNA).
However, molecular analysis of crosses in which the mt+ parent contained hypomethylated cpDNA was not consistent with the methylation-restriction model. According to the model, at least 90%–95% of the 5adc-treated mt+ cpDNA would have to be eliminated in zygotes to account for the 10- to 20-fold increase in the frequency of mt− cpDNA that was seen in the progeny. Contrary to this prediction, we could not detect any loss of mt+ cpDNA in zygotes when the mt+ parent’s cpDNA had been hypomethylated. Although it might be argued that there are some special methylation sites that are resistant to 5adc, the mechanism of this inhibitor is such that any cytosine methyltransferase should be blocked (Santi et al. 1984; Gabbara and Bhagwat 1995). To overcome such a block, a large excess of enzyme that is specific for a single or small number of critical sites would have to be produced in mt+ gametes. Although such a scenario cannot be completely ruled out, it seems highly unlikely as we have shown here that the methyltransferase(s) that carries out the vast majority of chloroplast cytosine methylation is very effectively inhibited by 5adc.
DNA damage versus hypomethylation
We realized that it was important to establish whether the effect of 5adc on cpDNA inheritance is due to loss of DNA methylation per se, or due to DNA damage. The DNA damaging effects of 5adc are well known (Jackson-Grusby et al. 1997) but seldom taken into account when interpreting its biological effects. Our results using EMS, which damages cpDNA without affecting cpDNA methylation, suggest that DNA damage might be partly responsible for the effect we see with 5adc. However, L-ethionine , a non-DNA-damaging methylation inhibitor, caused altered cpDNA inheritance in a manner similar to that of 5adc. Most importantly, we see differential replication of mt+ and mt− cpDNA even in control crosses where the cpDNA has not been subject to DNA-damaging agents, meaning that methylation differences alone can influence cpDNA replication rates in germinating zygotes. Taken together, our data are consistent with the possibility that both hypomethylation and DNA damage feed into a common pathway that is designed to eliminate undesirable cpDNA molecules. The rationale for such a pathway is discussed in the next section.
5adc affects a novel step of the uniparental inheritance process
An unexpected finding using our tagged chloroplast strains in mating reactions was that destruction of mt− cpDNA proceeded normally when the mt+ parent had hypomethylated cpDNA. Other treatments of mt+ gametes that block uniparental cpDNA inheritance, such as UV light or 5-fluordeoxyuridine (5fdu), act to abolish or attenuate the destruction of mt− cpDNA in early zygotes (Munaut et al. 1990; Rosen et al. 1991; Uchida et al. 1992). In the case of 5adc, it was only after progeny had germinated that enhanced transmission of mt− cpDNA could be observed in 5adc-treated versus control crosses. The novel effect on cpDNA inheritance uncovered by 5adc is further underscored by the fact that the drug elicits this effect only in germinating zygotes, not in vegetatively grown cells, as demonstrated by our comparison of vegetative diploids and meiotic progeny from 5adc-treated crosses. Importantly, even in control crosses, the relative amounts of mt+ and mt− cpDNA underwent changes during zygote germination, meaning that the effect of 5adc is to perturb a normal process of differential cpDNA replication that occurs at this time. Therefore, our experiments with 5adc have led us to the discovery of a novel mechanism controlling cpDNA inheritance: elimination of residual mt− cpDNA in germinating zygotes by restricted replication. In a normal cross, this secondary mechanism results in a ∼5- to 10-fold further reduction in residual mt− cpDNA during zygote germination and subsequent progeny outgrowth (Fig. 6E). The unusual nature of cpDNA replication during zygote germination has been hinted at previously, where a rapid increase in cpDNA was seen by DAPI staining just prior to meiosis (Coleman 1984). Our results with 5adc and EMS suggest that differential cpDNA replication may function not only to eliminate improperly methylated cpDNA but also damaged cpDNA, thus ensuring that meiotic progeny inherit an intact and uniform population of cpDNA.
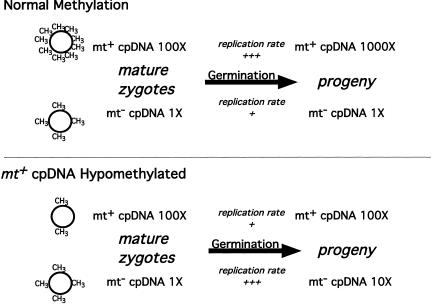
Why does 5adc treatment of mt+ cpDNA cause a change in the replication rates of both mt+ and mt− cpDNA in germinating zygotes? The simplest explanation is that both parental cpDNAs are competing for a limited pool of replication resources (e.g., initiator proteins or initiation sites). In a normal cross, heavily methylated mt+ cpDNA would outcompete the undermethylated mt− cpDNA (Fig. 8, top). However, when the mt+ cpDNA was hypomethylated or damaged, mt− cpDNA would gain preferential access to the cpDNA replication machinery (Fig. 8, bottom). Because cpDNA is not remethylated during vegetative growth (Sano et al. 1984), the effect would be transient and self-limiting, occurring only during zygote germination and the first few mitotic divisions thereafter.
Figure 8.
Models for cpDNA replication during germination. (Top) Schematic of cpDNA methylation and replication during germination of a normal cross. Mature zygotes contain ∼100 times more mt+ than mt− cpDNA. During germination, the heavily methylated mt+ cpDNA replicates better than the mt− cpDNA, generating a final difference of ∼1000-fold in mt+ versus mt− cpDNA levels. (Bottom) Schematic of cpDNA methylation and replication during germination when the mt+ parental cpDNA has been hypomethylated by 5adc. Mature zygotes contain a similar low level of mt− cpDNA as the normal cross (∼1%), but the residual mt− cpDNA outcompetes the hypomethylated mt+ cpDNA for replication during germination, increasing its representation by 10-fold in the resulting progeny.
The differential replication model is appealing given the established links between DNA methylation and DNA replication. In prokaryotes, methylation of DNA replication origins controls the timing of initiation (Zyskind and Smith 1992), and in eukaryotes, DNA methylation may also be required for proper initiation (Knox et al. 2000). Moreover, a replication-based mechanism appears to be involved in the unusual uniparental mitochondrial inheritance in males of the mussel, Mytilus edulis. Whereas male somatic tissue contains a mixture of maternally and paternally derived mitochondria, the gonad and sperm cells are greatly enriched for paternal mitochondria, and it is thought that this enrichment is due to enhanced replication of paternal versus maternal mitochondria in the germ tissue (Skibinski et al. 1994a,b; Zouros et al. 1994a,b). It would be of interest to learn whether DNA methylation operates in this system.
Possible roles of cpDNA methylation
The extensive cpDNA methylation that takes place during gamete and zygote differentiation in Chlamydomonas is one of the most dramatic examples known of developmentally programmed DNA methylation. If this methylation is not necessary for protection of cpDNA from methylation-sensitive nucleases during early zygote development as we have demonstrated, what is its role? Our work indicates that one role is to enhance mt+ cpDNA replication during zygote germination, as discussed above. A potentially related function could be that of packaging cpDNA nucleoids in a protective conformation during zygote dormancy. DNA packaging by a set of acid soluble nucleoid-binding proteins in Bacillus subtilis is known to be important for spore resistance to DNA damage and other environmental stresses (Setlow 1988). In Chlamydomonas, methylcytosine residues in cpDNA might serve as binding sites for such proteins or play a role similar to that of nuclear methylcytosine, which is thought to help stabilize heterochromatin in a tight or inaccessible conformation (Lewis and Bird 1991). Methylation-mediated cpDNA packaging might even play a dual role, both protecting cpDNA during zygote dormancy and facilitating cpDNA replication during zygote germination.
Materials and methods
Strains
All strains used in this work besides CJU10 were obtained from the Chlamydomonas stock center at Duke University. Wild-type, high efficiency-mating strains CC620 (mt+) and CC621 (mt−) are also referred to as R3 and NO, respectively. CJU10 was constructed by use of biolistic transformation (Boynton et al. 1988) of strain NO with vector cg20-atpB-Int (Zerges and Rochaix 1994) and selection for spectinomycin resistance.
Growth of strains and mating
To obtain gametes, cells were spread on TAP plates (Harris 1989) and allowed to grow in the light for 5–7 d. During vegetative growth on the plates, nitrogen is depleted and cells become gametic (Martin and Goodenough 1975). Gametes were scraped off plates and resuspended in nitrogen-free HSM (Harris 1989) at a concentration of 2 × 107/mL, and the gamete suspensions were kept in the light for 1–2 h prior to mating.
5adc (Sigma) and L-ethionine (Sigma) were dissolved in water at concentrations of 50 mM and 100 mM, respectively, divided into single-use aliquots, and stored at −70°C. The drugs were spread on plates at the appropriate concentration just prior to plating cells for gametogenesis (see above). Antibiotics used for screening colonies were prepared as described (Harris 1989) and used at a concentration of 100 μg/mL in TAP medium.
For mating, equal volumes of gamete suspensions were mixed together at t = 0 h. For genetic analysis, 200-μL aliquots were plated on HSM agar after 1–2 h of mating. To obtain single zygote colonies, the aliquot was diluted prior to plating. Zygotes were incubated in the light for 20–24 h and then placed in the dark for 5 d. After 5 d, the plates were chloroform-treated for 45 sec to kill unmated gametes and then placed in the light for germination. After germination, cells were grown for an additional 5–7 d. For random progeny analysis, the cells were scraped from plates, resuspended in TAP, diluted, and replated on TAP to obtain single colonies. Antibiotic resistance was scored by replica plating to the appropriate media.
Mutagenesis
For EMS treatment, 4 × 108 R3 gametes were washed and resuspended in 10 mL of 50 mM KPO4 (pH 7.0), 200 μL of methanesulfonic acid ethyl ester (EMS; Sigma) was added, and the cells were incubated for 30 min to 2 h with agitation. After mutagenesis, the cells were washed 3 times with 10 mL of 50 mM KPO4 (pH 7.0), and resuspended in nitrogen-free HSM. After an overnight recovery period in the light, the cells were mated, and their progeny were analyzed as described above.
For 5adc mutagenesis, gamete suspensions in nitrogen-free HSM were prepared in triplicate after growth with the indicated amount of drug in the media (as described for mating experiments).
For determining mutagenic rates, 108 cells (EMS or 5adc treated) were plated on the appropriate antibiotic media or diluted and plated to obtain single colonies on TAP to determine viability. Mutagenic rates (number of resistant colonies/number of viable cells) on the three plates were averaged. A mock-mutagenized control was always included to determine the background mutation rate.
Vegetative diploid selection
Vegetative diploids were selected by preparing gametes from CC425 and CC2663, mating them for 1–2 h, and plating zygotes on TAP medium containing 22 μg/mL 3-acetyl-pyridine (Sigma), which facilitates negative selection against nicotinamide auxotrophs. Plates were kept in continuous light and Nic+ Arg+ diploid colonies were collected and analyzed for chloroplast markers as described above.
DNA preparation
For each time point to be analyzed, 3-mL aliquots of mating mixtures were removed for DNA isolation. For early zygotes or gametes, aliquots were spun down and resuspended in 0.4 mL of TEN buffer (10 mM Tris at pH 8.0, 10 mM EDTA, 150 mM NaCl). Small-scale DNA preps were then made as described below, but without the sonication, washes, or zirconium beads. For later zygote DNA preps (6 h+), aliquots were resuspended in 0.5 mL of TEN buffer with 0.2% NP-40 in 1.5-mL tubes and sonicated (Fisher Sonic Dismembrator Model 300) on full power for 30 sec to lyse unmated gametes. Zygotes were repelleted and washed 2–3 times more in 1 mL of TEN with 0.2% NP-40. After a final suspension in 0.4 mL of TEN buffer, 40 μL of 20% SDS, and 40 μL of 20% sarkosyl, 0.4 mL of 1.0-mm zirconium beads (Biospec Products, Inc.) were added. The zygotes were vortexed at full power for 5 min resulting in >99% breakage (determined microscopically). Then, 20 μL of 10 mg/mL pronase was added, and samples were incubated at 37°C for 30 min. Next, 500 μL of 1:1 phenol/chloroform was added and the samples vortexed for 3 min. After a brief centrifugation, the aqueous phase was removed and precipitated with 2 volumes of ethanol. Pellets were washed with 70% ethanol, vacuum dried, and resuspended in 50 μL of TE.
The 5-day-old zygote DNA was prepared as follows: 3 mL of zygotes was spun down 2 h after mating and spread on a cellulose filter (gift of David Cove, University of Leeds, UK) on a HSM plate. After 24 h in the light and 5 d in the dark, cells were scraped into 0.4 mL of TEN buffer and DNA prepared as described above.
Germinated progeny DNA was prepared by plating zygotes at 2 h and allowing them to mature by the normal procedure (described above). After chloroform treatment to kill unmated gametes, the plates were placed in the light for 5–7 d until a lawn of germinated cells formed. Approximately 5 × 107 progeny cells were then scraped off the plate and processed as described for gamete and early zygote DNA preparations.
Southern blots
Each DNA (5 μL) was digested overnight at 37°C with 5 units of MboI (Promega), Sau3AI (NEB), or EcoRV (NEB) in 20 μL using the recommended buffers, 100 μg/mL BSA, 25 μg/mL RNase, and 2 mM spermidine (Sigma). Southern blots were prepared from samples fractionated on 0.75% agarose gels (Sambrook et al. 1989) and hybridized as described (Church and Gilbert 1984). Bands were quantitated by use of a phosphorImager (Molecular Dynamics or BioRad).
Radiolabeled probes were made by random primer labeling (Sambrook et al. 1989) of PCR products. The atpB probe was an ∼1-kb PCR fragment amplified from plasmid cg20-atpB-Int (Zerges and Rochaix 1994) with primers atpB1, 5′-CCT TGGGGCATATTAATTCCACTT-3′, and atpB2, 5′-TTT TAATGAAGCAGCTTTACTAAT-3′. The aadA probe was an ∼0.8-kb PCR fragment amplified from the same plasmid with primers AAD5, 5′-GTGAAGCGGTTATCGCCGAAG-3′, and AAD3, 5′-TTGCCAACTACCTTAGTGATC-3′. A cDNA probe for the nuclear gene ezy1 (Armbrust et al. 1995) was used to normalize samples for quantitation of cpDNA.
Quantitative PCR
Mature, 5-day-old zygotes were prepared as described above, with 3 × 107 zygotes spread on each of 6 nitrogen-free HSM plates containing 4% agar. Mating efficiencies for control and 5adc-treated crosses exceeded 95%. For germination, zygotes were chloroform treated, scraped off the plate with a razor blade, and suspended in TEN buffer with 0.2% NP-40. After sonication and washing to remove unmated gametic DNA (see above), the zygotes were washed three times with TAP and resuspended in 250 mL of TAP in a 1-liter flask. For DNA preparation, 30 mL was removed (0 h time point), and the flask was shaken under the light to begin germination. An aliquot was also removed to measure cell concentration and for plating to determine germination efficiency. For preparation of DNA, 30 mL of the culture was used at each time point thereafter. DNA preparation for each sample was as described for mature zygotes (see above).
To ensure that our PCR assay was quantitative, we serially diluted DNA from each time point in TE buffer with 0.01% NP-40. Then, we performed PCR with each primer set to determine the dilution range that resulted in a linear amplification signal. For fus1, 5′-ATGCCTATCTTTCTCATTCT-3′, and fus2, 5′-GCAAAATACACGTCTGGAAG-3′, (nuclear DNA), this dilution was 10−2; for aadA3, 5′-CCGAAGTATCGACT CAACTATC-3′, and aadA4, 5′-GGCGAGTTCCATAGCGT TAAGG-3′ (mt− cpDNA), it was 10−3; and for atpB13, 5′-CGTC CACTAATATTTATATTCCC-3′, and atpB,14 5′-TGAAAC TATTGAAGGTTTTGG-3′ (mt+ cpDNA), it was 10–5. Each reaction contained 5 μL of diluted DNA in a total volume of 25 μL. Klentaq polymerase (Sigma) was used at a 500-fold dilution in 1× buffer (supplied by the manufacturer) containing 200 μM concentrations of each dNTP, and 1 μM concentrations of each primer. Thermal cycling was performed in a Omn-E machine (Hybaid) with the following program: 94°C for 4 min followed by 30 cycles of (47°C for 30 sec; 72°C for 30 sec; 94°C for 30 sec) and concluding with one cycle of (47°C for 30 sec; 72°C for 5 min). Of each reaction, 10 μL was loaded per lane on a 1% agarose TBE gel with ethidium bromide (0.5 μg/mL) and electrophoresed. The amplification products from the primer pairs were ∼500–600 bp long. Ethidium-stained bands were quantitated by use of a Bio-Rad Fluor-S scanner and Quantity One software. Each dilution and PCR experiment was repeated three times, yielding similar results each time.
Acknowledgments
We thank David Kirk, Craig Pikaard, and Eric Richards for comments on the manuscript. This work was supported by National Institutes of Health fellowship F32 GM18502 (J.U.) and United States Department of Agriculture grant 1999–01364 (U.W.G. and J.U.)
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ursula@biosgi.wustl.edu; FAX (314) 935-5125.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.906701.
References
- Ahmad I, Rao DN. Chemistry and biology of DNA methyltransferases. Crit Rev Biochem Mol Biol. 1996;31:361–380. doi: 10.3109/10409239609108722. [DOI] [PubMed] [Google Scholar]
- Armbrust EV. (eds. J.-D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant). Kluwer Academic Pub, Dordrecht, The Netherlands. 1998. Uniparental inheritance of chloroplast genomes. In The molecular biology of chloroplasts and mitochondria in Chlamydomonas. [Google Scholar]
- Armbrust EV, Ibrahim A, Goodenough UW. A mating type-linked mutation that disrupts the uniparental inheritance of chloroplast DNA also disrupts cell-size control in Chlamydomonas. Mol Biol Cell. 1995;6:1807–1818. doi: 10.1091/mbc.6.12.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky CW. Relaxed and stringent genomes: Why cytoplasmic genes don't obey Mendel's laws. J Hered. 1994;85:355–365. [Google Scholar]
- ————— Uniparental inheritance of mitochondrial and chloroplast genes: Mechanisms and evolution. Proc Natl Acad Sci. 1995;92:11331–11338. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolen PL, Grant DM, Swinton D, Boynton JE, Gillham NW. Extensive methylation of chloroplast DNA by a nuclear gene mutation does not affect chloroplast gene transmission in chlamydomonas. Cell. 1982;28:335–343. doi: 10.1016/0092-8674(82)90351-8. [DOI] [PubMed] [Google Scholar]
- Boynton JE, Gillham NW, Harris EH, Hosler JP, Johnson AM, Jones AR, Randolph-Anderson BL, Robertson D, Klein TM, Shark KB, et al. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988;240:1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- Burton WG, Grabowy CT, Sager R. Role of methylation in the modification and restriction of chloroplast DNA in Chlamydomonas. Proc Natl Acad Sci. 1979;76:1390–1394. doi: 10.1073/pnas.76.3.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang K-S, Sueoka N. Replication of chromosomal and cytoplasmic DNA during mitosis and meiosis in the eucaryote Chlamydomonas reinhardi. J Cell Physiol. 1967;70:89–112. doi: 10.1002/jcp.1040700408. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman AW. The fate of chloroplast DNA during cell fusion, zygote maturation and zygote germination in Chlamydomonas reinhardi as revealed by DAPI staining. Exp Cell Res. 1984;152:528–540. doi: 10.1016/0014-4827(84)90655-4. [DOI] [PubMed] [Google Scholar]
- Corriveau JL, Coleman AW. Monitoring by epifluorescence microscopy of organelle DNA fate during pollen development in five angiosperm species. Dev Biol. 1991;147:271–280. doi: 10.1016/s0012-1606(05)80024-7. [DOI] [PubMed] [Google Scholar]
- Cox R. Studies on DNA methyltransferase and alteration of the enzyme activity by chemical carcinogens. Toxicol Pathol. 1986;14:477–482. doi: 10.1177/019262338601400416. [DOI] [PubMed] [Google Scholar]
- Ebersold WT. Heterozygous diploid strains of Chlamydomonas reinhardi. Genetics. 1963;48:888. doi: 10.1126/science.157.3787.447. [DOI] [PubMed] [Google Scholar]
- ————— Chlamydomonas reinhardi: Heterozygous diploid strains. Science. 1967;157:447–449. doi: 10.1126/science.157.3787.447. [DOI] [PubMed] [Google Scholar]
- Feng TY, Chiang KS. The persistence of maternal inheritance in Chlamydomonas despite hypomethylation of chloroplast DNA induced by inhibitors. Proc Natl Acad Sci. 1984;81:3438–3442. doi: 10.1073/pnas.81.11.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Goodenough UW. Mating type in Chlamydomonas is specified by mid, the minus-dominance gene. Genetics. 1997;146:859–869. doi: 10.1093/genetics/146.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbara S, Bhagwat AS. The mechanism of inhibition of DNA (cytosine-5-)-methyltranferases by 5-azacytosine is likely to involve methyl transfer to the inhibitor. Biochem J. 1995;307:87–92. doi: 10.1042/bj3070087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillham NW, Boynton JE, Harris EH. Transmission of plastid genes. In: Bogorad L, Vasil IK, editors. Cell culture and somatic cell genetics. New York, NY: Academic Press; 1991. pp. 55–92. [Google Scholar]
- Goodenough UW. Chlamydomonas mating interactions. In: Dworkin M, editor. Microbial cell–cell interactions. Washington, DC.: American Society for Microbiology; 1991. pp. 71–112. [Google Scholar]
- Harris EH. The Chlamydomonas sourcebook: A comprehensive guide to biology and laboratory use. San Diego, CA: Academic Press; 1989. [DOI] [PubMed] [Google Scholar]
- Harris EH, Boynton JE, Gillham NW. Interaction of nuclear and chloroplast mutations in biogenesis of chloroplast ribosomes in Chlamydomonas. In: Wiessner W, Robinson DG, editors. Molecular and cellular aspects of algal development. Berlin, Germany: Springer-Verlag; 1987. pp. 142–149. [Google Scholar]
- Jackson-Grusby L, Laird PW, Magge SN, Moeller BJ, Jaenisch R. Mutagenicity of 5-aza-2‘-deoxycytidine is mediated by the mammalian DNA methyltransferase. Proc Natl Acad Sci. 1997;94:4681–4685. doi: 10.1073/pnas.94.9.4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JD, Araujo FD, Bigey P, Slack AD, Price GB, Zannis-Hadjopoulos M, Szyf M. Inhibition of DNA methyltransferase inhibits DNA replication. J Biol Chem. 2000;275:17986–17990. doi: 10.1074/jbc.C900894199. [DOI] [PubMed] [Google Scholar]
- Kuroiwa T, Kawano S, Nishibayashi S, Sato C. Epifluorescent microscopic evidence for maternal inheritance of chloroplast DNA. Nature. 1982;298:481–483. doi: 10.1038/298481a0. [DOI] [PubMed] [Google Scholar]
- Lewis J, Bird A. DNA methylation and chromatin structure. FEBS Lett. 1991;285:155–159. doi: 10.1016/0014-5793(91)80795-5. [DOI] [PubMed] [Google Scholar]
- Martin NC, Goodenough UW. Gametic differentiation in Chlamydomonas reinhardtii. I. Production of gametes and their fine structure. J Cell Biol. 1975;67:587–605. doi: 10.1083/jcb.67.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matagne RF. Transmission of chloroplast alleles in somatic fusion products obtained from vegetative cells and/or ‘gametes’ of Chlamydomonas reinhardtii. Curr Genet. 1981;3:31–36. doi: 10.1007/BF00419578. [DOI] [PubMed] [Google Scholar]
- Matagne RF, Beckers M-C. Perturbation of chloroplast gene transmission in diploid and triploid zygotes of Chlamydomonas reinhardi by 5-fluorodeoxyuridine. Curr Genet. 1983;7:335–338. doi: 10.1007/BF00445872. [DOI] [PubMed] [Google Scholar]
- Matagne RF, Deltour R, Ledoux L. Somatic fusion between cell wall mutants of Chlamydomonas reinhardi. Nature. 1979;278:344–346. [Google Scholar]
- Munaut C, Dombrowicz D, Matagne RF. Detection of chloroplast DNA by using fluorescent monoclonal anti-bromodeoxyuridine antibody and analysis of its fate during zygote formation in Chlamydomonas reinhardtii. Curr Genet. 1990;18:259–263. doi: 10.1007/BF00318390. [DOI] [PubMed] [Google Scholar]
- Rosen H, Newman SM, Boynton JE, Gillham NW. A nuclear mutant of Chlamydomonas that exhibits increased sensitivity to UV irradiation, reduced recombination of nuclear genes, and altered transmission of chloroplast genes. Curr Genet. 1991;19:35–41. doi: 10.1007/BF00362085. [DOI] [PubMed] [Google Scholar]
- Sager R, Lane D. Molecular basis of maternal inheritance. Proc Natl Acad Sci. 1972;69:2410–2413. doi: 10.1073/pnas.69.9.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R, Grabowy C. Differential methylation of chloroplast DNA regulates maternal inheritance in a methylated mutant of Chlamydomonas. Proc Natl Acad Sci. 1983;80:3025–3029. doi: 10.1073/pnas.80.10.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R, Grabowy C, Sano H. The mat-1 gene in Chlamydomonas regulates DNA methylation during gametogenesis. Cell. 1981;24:41–47. doi: 10.1016/0092-8674(81)90499-2. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sano H, Grabowy C, Sager R. Loss of chloroplast DNA methylation during dedifferentiation of Chlamydomonas reinhardi gametes. Mol Cell Biol. 1984;4:2103–2108. doi: 10.1128/mcb.4.10.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA–cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci. 1984;81:6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow P. Small, acid-soluble spore proteins of Bacillus species: Structure, synthesis, genetics, function, and degradation. Annu Rev Microbiol. 1988;42:319–338. doi: 10.1146/annurev.mi.42.100188.001535. [DOI] [PubMed] [Google Scholar]
- Skibinski DOF, Gallagher C, Beynon CM. Mitochondrial DNA inheritance. Nature. 1994a;368:817–818. doi: 10.1038/368817b0. [DOI] [PubMed] [Google Scholar]
- ————— Sex-limited mitochondrial DNA transmission in the marine mussel, Mytilus edulis. Genetics. 1994b;138:801–809. doi: 10.1093/genetics/138.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Lemieux C, Lee RW. Net synthesis of chloroplast DNA throughout the synchronized vegetative cell cycle of Chlamydomonas. Curr Genet. 1980;2:229–232. doi: 10.1007/BF00435691. [DOI] [PubMed] [Google Scholar]
- Uchida H, Kawano S, Nakamura S, Kuroiwa T. A study on the effects of UV on preferential digestion of chloroplast nuclei in young zygote of Chlamydomonas reinhardtii. Cytologia. 1992;57:395–399. [Google Scholar]
- Whatley JM. Ultrastructure of plastid inheritance: Green algae to angiosperms. Biol Rev. 1982;57:527–569. [Google Scholar]
- Wurtz EA, Boynton JE, Gillham NW. Perturbation of chloroplast DNA amounts and chloroplast DNA gene transmission in Chlamydomonas reinhardtii by 5-fluorodeoxyuridine. Proc Natl Acad Sci. 1977;74:4552–4556. doi: 10.1073/pnas.74.10.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerges W, Rochaix JD. Mol. Cell. Biol. 14: 5268–5277. 1994. The 5′ leader of a chloroplast mRNA mediates the translational requirements for two nucleus-encoded functions in Chlamydomonas reinhardtii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouros E, Ball AO, Saavedra C, Freeman KR. Mitochondrial DNA inheritance. Nature. 1994a;368:818. doi: 10.1038/368818a0. [DOI] [PubMed] [Google Scholar]
- ————— An unusual type of mitochondrial DNA inheritance in the blue mussel Mytilus. Proc Natl Acad Sci. 1994b;91:7463–7467. doi: 10.1073/pnas.91.16.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyskind JW, Smith DW. DNA replication, the bacterial cell cycle, and cell growth. Cell. 1992;69:5–8. doi: 10.1016/0092-8674(92)90112-p. [DOI] [PubMed] [Google Scholar]