Abstract
From the characterization of enzyme activities and the analysis of genomic sequences, the complement of DNA methyltransferases (MTases) possessed by the cyanobacterium Anabaena PCC 7120 has been deduced. Anabaena has nine DNA MTases. Four are associated with Type II restriction enzymes (AvaI, AvaII, AvaIII and the newly recognized inactive AvaIV), and five are not. Of the latter, four may be classified as solitary MTases, those whose function lies outside of a restriction/modification system. The group is defined here based on biochemical and genetic characteristics. The four solitary MTases, DmtA/M.AvaVI, DmtB/M.AvaVII, DmtC/M.AvaVIII and DmtD/M.AvaIX, methylate at GATC, GGCC, CGATCG and rCCGGy, respectively. DmtB methylates cytosines at the N4 position, but its sequence is more similar to N6-adenine MTases than to cytosine-specific enzymes, indicating that it may have evolved from the former. The solitary MTases, appear to be of ancient origin within cyanobacteria, while the restriction MTases appear to have arrived by recent horizontal transfer as did five now inactive Type I restriction systems. One Mtase, M.AvaV, cannot reliably be classified as either a solitary or restriction MTase. It is structurally unusual and along with a few proteins of prokaryotic and eukaryotic origin defines a structural class of MTases distinct from all previously described.
INTRODUCTION
DNA methyltransferases (MTases) in bacteria are most often associated with cognate restriction endonucleases (1), serving to protect the host organism from self-digestion. However, some MTases modify DNA sequences for purposes distinct from restriction. These enzymes, sometimes called orphans, will be termed solitary MTases to emphasize their functional role rather than their possible origins. The best studied enzyme in this group is Dam of Escherichia coli (2). Methylation of DNA by Dam at GATC permits the cell to distinguish recently synthesized (hemimethylated) DNA from older (fully methylated) DNA. The cell uses this information to regulate the timing of DNA synthesis and to choose the proper strand as a template in order to correct mismatched bases. MTases nearly identical to Dam in sequence and function appear to be widespread amongst enteric bacteria (see Discussion), and MTases of similar function have been observed in the α division proteobacteria (3,4). Little is known, however, about a second MTase in E.coli, the CCwGG-specific enzyme Dcm (2), nor about the range of functions served in bacteria by solitary MTases.
Modification of GATC sequences is also prevalent amongst the cyanobacteria (5), an ancient class of photosynthetic eubacteria. Characterization of GATC modification in cyanobacteria has extended little past demonstrating the general inability of adenine-specific, GATC-recognizing restriction endonucleases to cut genomic DNA (5,6). One gene, mbpA, in the fully sequenced genome of the cyanobacterium Synechocystis PCC 6803 has been identified whose putative product bears a striking resemblance to Dam of E.coli and to other MTases that modify the adenine within GATC (http://www.kazusa.or.jp/cyano/). The insensitivity of genomes of the filamentous cyanobacteria within the genera Anabaena and Nostoc to digestion by the GGCC-specific endonuclease HaeIII (6,7) coupled with the apparent absence of a corresponding restriction enzyme (8), led us to speculate that these cyanobacteria carry at least two solitary MTases.
Type II DNA MTases may be grouped according to the positions of the modifications they catalyze. Enzymes methylating cytosine at the 5-carbon of the pyrimidine ring form a coherent group, according to a common ordering of 10 well conserved blocks within their amino acid sequences (9). Enzymes that methylate the exocyclic N4 amine of cytosine or N6 amine of adenine form a looser group whose members share some sequence similarities. The latter group has been subdivided into three types, α, β and γ, according to their different orderings of common motifs and sequence similarities within these motifs (10). Crystal structures have recently been deduced for two C5-methylcytosine (5mC) MTases (11,12) and for an N4-methylcytosine (N4mC) or N6-methyladenine (N6mA) MTase from each of the three types (13–15). These have led to the view that all MTases share a common architecture (16).
Type I DNA MTases differ markedly from Type II in both structure and mechanism of action (17). Most notably, the former consist of two distinct subunits, a catalytic subunit (M, encoded by hsdM) and a subunit (S, encoded by hsdS) responsible for specific binding to DNA. The addition of a third subunit (R, encoded by hsdR) confers upon the complex the additional ability to cleave DNA unmethylated at the target recognition site. Type I DNA MTases have been divided into four families, IA–ID, originally on the basis of antigenic crossreactivity.
The possibility that DNA replication, modulated by a solitary MTase, might regulate differentiation within filaments of Anabaena PCC 7120 (henceforth referred to as Anabaena) motivated us to screen the strain for genes encoding such enzymes. Unusual features in the deduced amino acid sequences of protein encoded by two of the genes found led us to examine them more closely within the context of current understanding of structure–function relationships.
MATERIALS AND METHODS
DNA isolation and cloning
DNA was isolated from Anabaena grown on BG11, modified as previously described (18), by vortexing with glass beads in phenol (19). Plasmids bearing expressed DNA MTase genes avaMV, dmtB/avaMVII and dmtD/avaMIX were isolated essentially according to the protocol of Kiss et al. (20). In brief, Anabaena DNA partially digested with Sau3AI to an apparent average size of 2–3 kb, was ligated with pBluescript II KS+ (Stratagene). The ligated DNA was electroporated into E.coli strain GM4715 (dam– mcrB–) (2), for cloning of avaMV, or K803 (hsdS3 mcrB) (21), for cloning of dmtB and dmtD. Approximately 400 000 colony forming units were incubated in 4 ml LB for 1 h, then 100 ml LB + 50 µg/ml ampicillin for 6 h. Plasmid DNA was isolated through a Qiaprep spin column. Approximately 1 µg plasmid DNA was digested for at least 6 h with excess restriction enzyme (DpnII for avaMV, HaeIII for dmtB and BsrFI for dmtD). The DNA was precipitated, and 20% of it was used to electroporate the same strain of E.coli to ampicillin resistance. For dmtB and dmtD, this was sufficient to isolate several colonies carrying the gene. For avaMV, it was necessary to go through a second round of amplification, starting with several hundred colonies scraped off the transformation plate.
The gene encoding DmtA was isolated by PCR amplification using the primers 5′-ATATCATCAGGTGATCGCGC-3′ and 5′-TTTGGCGCTGGGATAGTACC-3′. The gene encoding M.AvaIII and its downstream open reading frame (ORF) were isolated on the same PCR-amplified fragment, using the primers 5′-CAGTATGCTTCAGGGGGAAA-3′ and 5′-GTTGTTGATGCTTTGAGCGA-3′. Both sets of primers were derived from available genomic sequence (http://www.kazusa.or.jp/cyano/) of the regions identified as described below and in the Results.
Routine plasmid isolation and manipulations followed standard procedures. The presence of small DNA digestion fragments was assessed by electrophoresis on 3% gels made with Metaphor (FMC). DNA was prepared for pulsed-field gel electrophoresis as previously described (22).
Analysis of DNA and protein sequences
Plasmid DNA purified through a Qiaprep minispin column was sequenced by the University of Chicago DNA Sequencing Facility. Most of the DNA, and all regions where there were any ambiguities, were sequenced from both strands. ORFs were identified and translated using EditBase, provided by Niels Nielsen (Purdue University, IN) and ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Protein sequences were aligned by ClustalX (23), which uses distance matrices and nearest neighbor joining, and displayed by ClustalInColor, a locally written program that facilitates the coloring of amino acids to accentuate relationships between groups of sequences. Tree structures were drawn using TreeView (24) and colored by hand.
In sequence comparisons, an amino acid was judged to be similar to the consensus amino acid if the entry of the pair in BLOSUM62, a matrix of amino acid similarities (25), gives a positive value. Groups of amino acids were judged to be similar to each other if they fell into one of the following sets defined by positive values in BLOSUM62 with respect to the first listed amino acid: DEN, EDKQ, HNY, LIMV, SANT, QEKR, YFHW. Since BLOSUM62 groups amino acids by the frequency with which they occur at the same position in aligned positions, the sets do not necessarily correspond to obvious biochemical characteristics.
DNA MTases and restriction endonucleases were found by searching GenBank with Gapped BLAST (26) and searching other databases listed below (last searched June 2000). DNA sequences from Synechocystis PCC 6803 and preliminary DNA sequences from Anabaena were obtained from CyanoBase (www.kazusa.or.jp/cyano/ ). Preliminary DNA sequences from Nostoc punctiforme ATCC 29133 (henceforth called N.punctiforme), and Enterococcus faecalis V583 were obtained from the DOE Joint Genome Institute (http://spider.jgi-psf.org/JGI_microbial/html/nostoc_homepage.html) and from The Institute for Genomic Research (http://www.tigr.org), respectively. DNA MTase sequences were identified in genomic sequences by three means: annotation of the sequence, annotation in REBASE (27; http://rebase.neb.com), a depository of information regarding restriction endonucleases and DNA MTases, and BLAST searches using the following archetypical sequences, with each followed by its class (10,17) and accession/enzyme number: M.EcoBI (IA; rebase:3381), M.EcoEI (IB; rebase:3386), M.EcoR124I (IC; rebase:3392), M.StySBLI (ID; rebase:3571), M.HaeIII (II-5mC; sp:P20589), M.DpnIIA (II-N6mAα; sp:P04043), M.NlaIII (II-N4mCα; pir:XYNHAL), M.HinFI (II-N6mAβ; rebase:3429), M.PvuII (II-N4mCβ; gb:AAA96336), M.EcoPI (III-N6mAβ; sp:P08763), M.PstI (II-N6mAγ; rebase:3483). An ORF was judged to encode a DNA MTase if it matched a positively identified DNA MTase with a BLAST score better than 10–4. Unidentified ORFs were searched for known motifs using EMOTIF (28; http://motif.stanford.edu/emotif/). Possible membrane-spanning regions were found in protein sequences by the Dense Alignment Surface method (DAS) (29; http://www.sbc.su.se/~miklos/DAS/maindas.html).
Chemical analysis of base composition
Plasmid DNA isolated from the dam– dcm– strain E.coli GM48 (2) was hydrolyzed by a procedure modified from that described by Gehrke et al. (30). About 50 µg DNA in 100 µl deionized water was denatured by boiling for 5 min and plunging the microfuge tube into ice water. 200 µl 30 mM sodium acetate pH 5.2 and 10 µl 20 mM zinc sulfate was added, followed by 6 U of nuclease P1 (Roche). After 6 h of incubation at 37°C, 40 µl of 0.5 M Tris (not adjusted for pH) and 5 U shrimp alkaline phosphatase (Roche) was added prior to an overnight incubation at 37°C. The resulting nucleosides were purified through a microcon YM-3 filter (Millipore). The purified nucleosides were analyzed on a 3.9 × 150 mm Nova-Pak reverse phase C18 column (Waters) using a previously described step gradient (30).
RESULTS
Cloning of three genes from Anabaena encoding DNA modification enzymes
As part of a general screen, DNA from Anabaena was digested with a number of restriction enzymes and run out on conventional and sometimes pulsed-field gels. We confirmed previously published results (6,31) that DNA from Anabaena could not be cut by HaeIII, which recognizes GGCC, nor by DpnII, which recognizes GATC. It is, however, cut by DpnI, which cuts only at methylated GATC. In addition, Anabaena DNA could not be cut by BsrFI, which recognizes the degenerate sequence rCCGGy (data not shown). The three genes responsible for the protection of Anabaena from digestion by DpnII, HaeIII and BsrFI were cloned by their ability to extend protection in E.coli to plasmid DNA bearing the genes and characterized as described below.
Analysis of avaMV, encoding a GATC-specific DNA MTase
A plasmid isolated as resistant to cutting by DpnII (which recognizes GATC) proved to be sensitive to digestion by DpnI (which recognizes GmeATC) and Bsp143I (which recognizes GATC regardless of adenine methylation). A 1.3 kb fragment subcloned from that plasmid (forming pUR147) retained these characteristics. Analysis of the original plasmid from dam– E.coli by HPLC showed a minor base with the same retention time as N6mA. These results are consistent with pUR147 encoding an enzyme that modifies the adenine residue within GATC.
The sequence of the insert within pUR147 revealed a single large ORF (Fig. 1), named avaMV (see Discussion), potentially encoding a protein of 210 amino acids. The region of the Anabaena genome surrounding avaMV was carefully examined to determine whether there may be any genes nearby that could conceivably encode a restriction endonuclease (Fig. 1). Such genes are often not recognizable by sequence, but of 227 sequenced Type II restriction endonucleases found in GenBank, only eight have fewer than 200 amino acids, and the smallest, R.PvuII, has 157 amino acids. Thus it is reasonable to expect a gene of at least that size. Furthermore, of 110 cases where the relative positions of the genes encoding the restriction enzyme and corresponding MTase(s) were readily available, only one was found where the gap between the two genes was >1000 bp (1451 bp for Eco47I), and the average gap size was only 82 bp. Furthermore, only one pair of genes (R.CviAI and M.CviAI from a chlorella virus) was found where a gene not part of the restriction/modification (RM) system intervened between the two. Thus we deemed it unnecessary to look beyond 2 kb and more than two large ORFs from a gene encoding a MTase to seek a gene encoding a corresponding endonuclease.
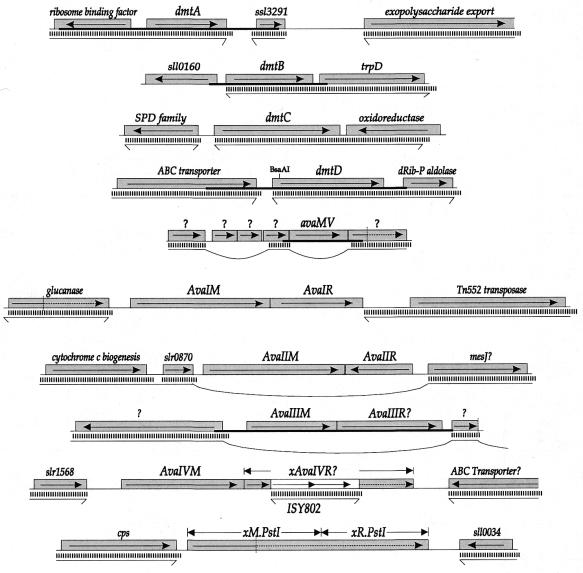
Figure 1.
Genetic context surrounding genes encoding or potentially encoding DNA MTases of Anabaena. Gray boxes represent ORFs, with the direction of translation shown. The white box indicates an insertion sequence. Vertical dotted lines indicate the point at which a frameshift occurs. Thick horizontal lines indicate the region that was cloned. Thin horizontal lines separated from the Anabaena genes by vertical bars indicate that a contiguous DNA sequence from N.punctiforme exhibits sequence similarity in that region. Horizontal lines with bent ends represent DNA sequences that continue without similarity to the Anabaena sequence. Loops indicate interpolations of sequences without similarity to the Anabaena sequence. Genes with names preceded by ‘x’ are defective by reason of a frameshift, nonsense codon or insertion sequence.
The available 1968 base sequence 5′ to the avaMV gene lacks any ORF capable of encoding a protein >119 amino acids. Immediately 3′ to avaMV lies an ORF capable of encoding a protein of 200 amino acids similar through most of its length to hypothetical proteins from the plant Arabidopsis thaliana (AAD23616) and from Streptomyces coelicolor (gb:CAB93740), but it is disrupted by a frameshift in the 5′-end of the gene. Other ORFs within the 2500 bp region 3′ to avaMV are either very small (the largest is 106 amino acids) or show great similarity to proteins of known function.
It was of interest to see whether avaMV is found generally in cyanobacteria. The sequence surrounding avaMV is found in the closely related strain N.punctiforme, but avaMV itself is not (Fig. 1). Instead it is replaced almost precisely by a different ORF that is not significantly similar to any in GenBank. No protein in the region shows any significant similarity to proteins of the distantly related cyanobacterium Synechocystis PCC 6803.
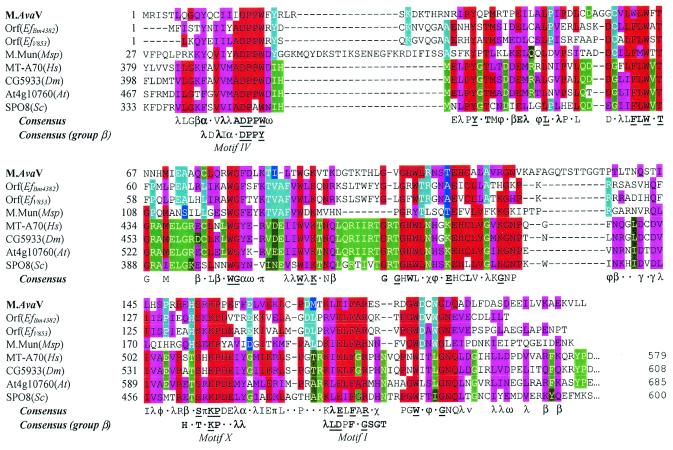
All previously characterized MTases with GATC specificity and one cyanobacterial ORF (mbpA) highly similar to genes encoding such enzymes fall into either the α class (nine genes) or β class (two genes) of MTases, and we anticipated that M.AvaV would do the same. In fact, a search through known protein sequences brought up three bacterial and several eukaryotic proteins (Table 1 and Fig. 2) that do not readily fit into any described class. One, M.MunI, is the MTase of a RM system, and another, MT-A70, is the S-adenosylmethionine (AdoMet)-binding subunit of a human RNA MTase that acts on adenine residues (32). The functions of the other proteins are unknown.
Table 1. Proteins similar to putative solitary Type II DNA MTases of Anabaena.
| Proteina |
Recognition sequenceb |
Source |
Scorec |
Accession no.d |
| Protein sequences similar to M.AvaV | ||||
| 1. Ef-Orf18 | ? | Enterococcus | 4 × 10–21 | gb:AAF72345.1 |
| 2. Ef10277.13628 | ? | Enterococcus | 1 × 10–20 | Genome sitee |
| 3. DmCG5933 | ? | Drosophila | 6 × 10–17 | gb:AAF56221.1 |
| 4. MT-A70 | ? | Homo | 2 × 10–16 | gb:AAB71850.1 |
| 5. AT4g10760 | ? | Arabidopsis | 2 × 10–16 | gb:CAB81177.1 |
| 6. Mouse ORF | ? | Mus | 2 × 10–16 | gb:AAD33673.1 |
| 7. Spo8 | ? | Saccharomyces | 3 × 10–15 | ref:NP_011323.1 |
| 8. M.MunI | CAATTG | Mycoplasma | 4 × 10–12 | sp:P43641 |
| Protein sequences similar to DmtA/M.AvaVI | ||||
| 1. DmtA(Np) | GATC? | Nostoc | 1 × 10–18 | Genome sitef |
| 2. M.PgiI | GA6TC | Porphyromonas | 3 × 10–66 | pir:S34414 |
| 3. Ph-orf | GATC? | Pyrococcus | 8 × 10–60 | pir:C71096 |
| 4. St8-orf3 | GATC? | Streptococcus | 3 × 10–59 | gb:CAB46541.1 |
| 5. M.DpnIIA | GA6TC | Diplococcus | 9 × 10–59 | sp:P04043 |
| 6. M.LlaDCHIA | GA6TC | Lactococcus | 8 × 10–58 | sp:P50179 |
| 7. M.MjaIII | GATC | Methanococcus | 7 × 10–53 | sp:Q58015 |
| 8. MbpA | GATC? | Synechocystis | 6 × 10–52 | pir:S77170 |
| 22. DmtB | GGCC | Anabaena | 7 × 10–17 | gb:AAF75229 |
| Protein sequences similar to DmtB/M.AvaVII | ||||
| 1. DmtB(Np) | GGCC? | Nostoc | 1 × 10–123 | Genome sitef |
| 2. Sll0729 | GGCC? | Synechocystis | 1 × 10–88 | pir:76841 |
| 3. St8-orf3 | GATC? | Streptococcus | 6 × 10–20 | gb:CAB46541.1 |
| 4. M.PgiI | GA6TC | Porphyromonas | 4 × 10–18 | pir:S34414 |
| 5. M.MjaIII | GATC | Methanococcus | 2 × 10–16 | sp:Q58015 |
| 6. M.LlaDCHIA | GA6TC | Lactococcus | 5 × 10–14 | sp:P50179 |
| 7. M.CviAI | GA6TC | Chlorella virus | 5 × 10–14 | pir:T18083 |
| 8. M.DpnIIA | GA6TC | Diplococcus | 2 × 10–13 | sp:P04043 |
| 9. M.CviQVI | GA6nTC | Chlorella virus | 5 × 10–13 | gb:AAC03126.1 |
| 10. MbpA | GATC? | Synechocystis | 9 × 10–13 | pir:S77170 |
| 11. M.StsI | GGA6TC | Streptococus | 1 × 10–11 | sp:P29347 |
| 16. M.BalI | TGGCCA | Brevibacterium | 8 × 10–7 | pir:S71506 |
| Protein sequences similar to DmtC/M.AvaVIII | ||||
| 1. DmtC(Np) | CGATCG? | Nostoc | 1 × 10–188 | Genome sitef |
| 2. M.XorII | CGATCG | Xanthomonas | 1 × 10–128 | sp:P52311 |
| 3. SynMI | CGATCG | Synechocystis | 1 × 10–97 | pir:S76359 |
| 4. M.NspHI | rC5ATGy | Nostoc | 6 × 10–43 | gb:AAC97192.1 |
| 5. M.NspI | rC5ATGy | Nostoc | 1 × 10–42 | gb:AAC97190.1 |
| 6. M.DdeI | C5TnAG | Desulfovibrio | 5 × 10–42 | sp:P05302 |
| Protein sequences similar to DmtD/M.AvaIX | ||||
| 1. DmtD(Np) | rCCGGy? | Nostoc | 1 × 10–176 | Genome sitef |
| 2. M.NmeDI | rCCGGy? | Neisseria | 1 × 10–119 | gb:CAB59897.1 |
| 3. M.Cfr10I | rC5CGGy | Citrobacter | 1 × 10–106 | Personal communicationg |
| 4. M.Bse634I | rCCGGy | Bacillus | 4 × 10–58 | Personal communicationg |
| 5. M.HaeIII | GGC5C | Haemophilus | 9 × 10–24 | sp:P20589 |
| 6. M.MthTI | GGCC | Methanobacter | 3 × 10–23 | sp:P29567 |
| 7. M.Phi3TII | TC5GA | Bacillus phage | 6 × 10–23 | pir:S47248 |
| 25. SynMI | CGATCG | Synechocystis | 1 × 10–14 | pir:S76359 |
aProtein sequences obtained by BLAST search (26) of the combined GenBank, PDB, SwissProt, PIR and PRF databases (unless otherwise noted), last accessed June 2000, ranked by score. Cyanobacterial sequences are shown in bold.
bRecognition sequences of MTases are given along with the position, if known, where they methylate: A6, N6-methyladenine; C5, C5-methylcytosine.
cScore represents expected number of proteins at least as similar to target within the combined databases.
dgb, GenBank; sp, SwissProt; pir, Protein Information Resource; ref, NCBI Reference Sequence project.
eSequence obtained from the E.faecalis web site (http://www.tigr.org), last accessed June 2000.
fSequences obtained from the N.punctiforme web site (http://spider.jgi-psf.org/JGI_microbial/html/nostoc_homepage.html), last accessed June 2000.
gSequences graciously provided by Virgis Siksnys (Fermentas).
Figure 2.
Alignment of M.AvaV with known and putative DNA and RNA MTases. Sequences listed in Table 1 (except the mouse sequence, which is substantially the same as the human sequence) were aligned as described in Materials and Methods. For each position, amino acids substantially conserved over the entire group are colored red and similar amino acids colored magenta. Identical amino acids within the bacterial subgroup (first four sequences) are colored cyan and similar amino acids dark blue. Identical amino acids within the eukaryotic subgroup (last four sequences) are colored bright green and similar amino acids dark green. A consensus sequence over the entire group is shown below the alignment and for the corresponding region of Group β N6mA DNA MTases (10). A letter is shown if >50% of the sequences have the same amino acid residue or class of amino acids at the given position. The letter is in bold type if the frequency is >75% and underscored if the position is invariant. α, aromatic (FHWY); β, big hydrophilic (EKQR); λ, leucine family (ILMV); ν, negative (DE); π, positive (KR); γ, glutamate family (EQDK); χ, (NHY); φ, (ANST); ω, aspartate family (END).
These proteins all have the region, Motif IV, most highly conserved in MTases and regions that could possibly correspond to Motifs X and I, as previously noted for M.MunI (33). All three regions participate in the binding of AdoMet (10). The Motif I assignment is particularly dissatisfying because the aligned sequences all lack a hydrophobic residue four amino acids prior to F and a G residue two amino acids after F, both found in almost all known adenine MTase sequences (34).
Analysis of dmtB, encoding a GGCC-specific DNA MTase
A second plasmid, pUR109, was isolated repeatedly from the library of Anabaena inserts, based on its ability to withstand digestion by HaeIII, which recognizes and cuts at GGCC. We expected that the enzyme encoded by pUR109 would determine a GGCC-specific MTase, which, like all others known, methylated at 5mC. To test this, pUR109 DNA was isolated from the dam– dcm– strain Gm48 and hydrolyzed. Surprisingly, gas chromatography showed a minor base that eluted with the same retention time as N4mC, not 5mC. HaeIII is sensitive to N4 methylation at the outer cytosine (35), but nothing has been reported regarding its sensitivity to N4 methylation at the inner cytosine.
To identify the precise site of N4mC methylation, pUR109 was digested with MspI. pUR109 contains one site (GGCCCGG) where a GGCC sequence overlaps by one base with the recognition site (CCGG) of MspI. A derivative of pUR109 was made that contains an additional site (GGCCGG) that overlaps by two bases. The ability of MspI to cut at the first site but not the second is consistent with known sensitivity of MspI to N4mC methylation at the outer cytosine of CCGG (36) only if the enzyme encoded by pUR109 methylates at the inner cytosine of GGCC.
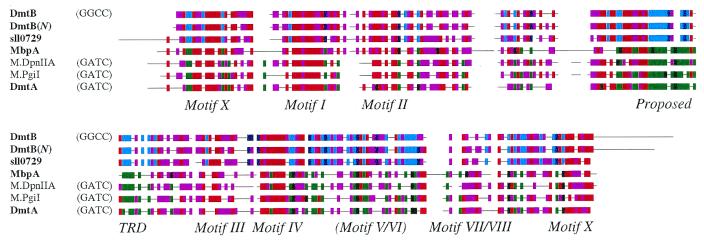
The sequence of the 1211 bp insert within pUR109 contains one large ORF (Fig. 1), denoted dmtB (for DNA MTase; see Discussion), capable of encoding a protein of 293 amino acids. There are at least 11 distinct GGCC-specific MTases from diverse sources that have been characterized sufficiently to distinguish the MTase type, and all methylate at the C5 position (seven of the MTases are from a Bacillus or its phage and the remaining are from Fusobacterium, Haemophilus, Methanobacterium and Neisseria) (27). The deduced protein sequence of DmtB does not fall into this class but instead shows significant similarity to N6mA-specific MTases of the α class, particularly those that recognize GATC (Table 1). The first similar sequence with known specificity that does not methylate at GATC is that of M.BalI, an N4mC MTase. DmtB is most similar to two predicted proteins from cyanobacteria: Orf352.2360R from N.punctiforme and Sll0729 from Synechocystis PCC 6803. They share 73 and 55% amino acid identity, respectively, over the full length of the protein.
In order to assess the relationship of a GGCC-modifying enzyme to those that modify at GATC, eight such MTases of the α class were compared to the predicted amino acid sequence of DmtB, and a representative part of the comparison is shown in Figure 3. The regions of similarity to these MTases are focused on conserved motifs spread throughout the protein. The target recognition domain (TRD), the region through which MTases make contact with DNA, is highly conserved amongst the adenine-modifying GATC-specific MTases but in this region there is little similarity with DmtB. The corresponding region in DmtB is highly similar to that in Sll0729 but bears no resemblance to the TRDs of known GGCC-specific MTases. The motifs that are particularly well conserved (Motifs X, I–IV) are those that participate in the binding of AdoMet and in the case of Motif IV serves as the catalytic site. Those motifs that are least conserved (Motifs V–IX) are required for the structural integrity of the protein (15).
Figure 3.
Alignment of DmtB and DmtA with representative Group α Nm6A/Nm4C DNA MTases. Names of cyanobacterial sequences are shown in bold. Coloring conventions are as described in the legend to Figure 2, with the cyan/dark blue and green/dark green groups representing DNA MTases of proven and putative GGCC-specificity and GATC-specificity, respectively. The motifs (including the TRD, target recognition domain) are those described by Tran et al. (15).
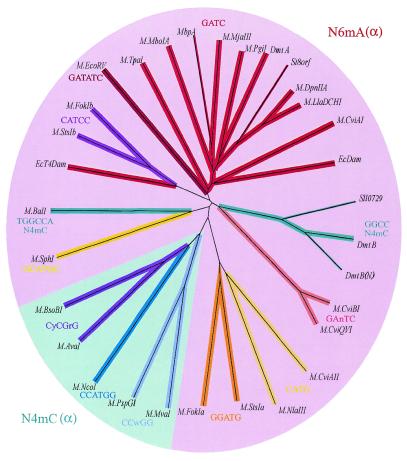
It seemed peculiar that a MTase that presumably methylates a cytosine within GGCC should appear so similar to MTases that methylate an adenine within GATC, so a dendrogram was constructed from all known MTase sequences of the α class, including those that methylate cytosine at the N4 position (Fig. 4). The tree uses sequences in which the TRD region has been removed, so as not to bias the relationships by regions that are known to be constrained by target-specificity, but the resulting tree was not appreciably different from that constructed with full sequences (not shown). In both cases, the sequences of the N6-adenine-methylating enzymes clustered together, apart from sequences of the N4-cytosine-methylating enzymes. DmtB fell into the N6mA group, as did a second N4mC MTase, M.BalI.
Figure 4.
Dendrogram of all available sequences of N4 and N6 MTases of the α group. The sequences compared consisted of the N- and C-termini, with the central TRD removed. The TRD was liberally defined as the block corresponding to residues 77 through 172 according to M.DpnII coordinates (15). The branches of all N6mA MTases are some hue of red and of all N4mC MTase some hue of blue. Thin lines indicate that the target sequence of the enzyme has not been established by biochemical criteria. Besides those sequences listed in Table 1, the following were found through the indicated accession nos: M.AvaI (CAA66984), M.BsoBI (CAA66933), M.CviAII (S27901), M.CviBI (Q01511), M.EcDam (P00475), M.EcoRV (P04393), M.EcoT4Dam (X17641), M.FokI (P14871), M.MboIA (P34720), M.MvaI (P14244), M.NcoI (AAC23514), M.NlaIII, M.PspGI (AAC67523), M.SphI (AAB40377) and M.TpaI (AAB82782). M.FokI and M.StsI consist of two separable MTases that are fused as a single polypeptide. The N-terminal portions (M.FokIa and M.StsIa) recognize one asymmetric target and the C-terminal portions (M.FokIb and M.StsIb) recognize the other.
If DmtB serves to protect GGCC sequences from a corresponding restriction enzyme of Anabaena, then one would expect the genes encoding the RM system to lie nearby one another. Immediately downstream lies a gene very similar to that encoding anthranilate synthetase (TrpD) from diverse bacteria. All other ORFs capable of determining a protein >100 amino acids and lying within 2 kb of either side of dmtB encode membrane-spanning proteins (data not shown) or a protein that exists in Nostoc in four copies. These were judged unlikely candidates to encode a restriction endonuclease.
Analysis of dmtD, encoding an rCCGGy-specific DNA MTase
One element (pUR115) of a library of Anabaena DNA was consistently isolated after selection for resistance to BsrFI. Plasmid DNA was fully protected only when isolated from cells in the stationary phase of growth. The rCCGGy sites within pUR115 were resistant not only to BsrFI but also to MspI, which cuts at the internal CCGG tetranucleotide. MspI is sensitive to methylation at the C5 position of the outer cytosine but not to methylation at the inner cytosine. The resistance of pUR115 to MspI, coupled with the sequence analysis later in this section, suggests that the enzyme encoded by the plasmid modifies at least the 5′ cytosine of rCCGGy at the C5 position.
From the inability of BsrFI to cut genomic DNA from Anabaena and its inability to cut pUR115 (which contains GCCGGC, GCCGGT and ACCGGC sites), it follows that the modification enzyme protects all sequences within rCCGGy. However, it is still conceivable that the enzyme modifies other sequences as well. To assess this possibility, the MspI digest of pUR115 was examined more closely. pUR115 contains 11 of the 12 possible nCCGGn sequences not described by rCCGGy (only TCCGGA is lacking). Since MspI cuts at all of these sites (data not shown), the sequence recognized by the encoded enzyme cannot be more redundant at its outer bases than rCCGGy.
Only one intact ORF was found within the sequenced 2071 bp fragment (Fig. 1). When that fragment was interrupted at the BsaAI site by insertion of a kanamycin resistance cassette, protection against BsrFI was lost. The intact ORF evidently encodes a DNA-modifying enzyme and was named dmtD. It is surrounded by two genes of readily identifiable function. Further downstream from dmtD lies a gene encoding a protein of unknown function with strong similarity to a protein in Synechocystis, which does not possess a DmtD-like MTase. No other significant ORFs lie within 1700 bp of dmtD.
The 385 amino acid protein sequence of DmtD, deduced using the first potential start codon of the ORF, was compared to known protein sequences, and >100 sequences with e-values of <10–5 were found. Most of these had been identified as 5mC DNA MTases. Particularly prominent were the DNA MTases M.Cfr10I and M.Bse634I, both of which recognize and modify rCCGGy, and many DNA MTases recognizing the sequence GGCC (Table 1).
The sequence of DmtD contains all six highly conserved motifs common to 5mC MTases and matches all 21 of the most conserved residues (data not shown), those residues common to almost all 5mC MTases. Alignments with 20 5mC MTases revealed lesser but significant similarity between DmtD and the less well conserved motifs II, III, V and VII. DmtD shows no significant similarity to most 5mC MTases in the TRD region, where the enzyme makes contact with its DNA substrate. However, it shares in this region (residues 257–313) 43 of 67 amino acid residues with M.Cfr10I and 35 of 67 with M.Bse634I.
Analysis of dmtA and dmtC, encoding GATC- and CGATCG-specific DNA MTases
The release of the partial genomic sequence of Anabaena made it possible to screen the strain for genes encoding MTases on the basis of sequence similarity rather than function. The proteins predicted from the available sequence were compared to several representative 5mC, N4mC and N6mA MTases covering all established groups, and significant hits were further analyzed.
One predicted protein showed considerable sequence similarity to class α N6mA MTases, particularly those that recognize GATC. Since the GATC-recognition ability of M.AvaV had been established, we were keen on determining whether Anabaena truly possesses a second such enzyme. The gene was cloned by PCR on a fragment containing no other significant ORFs and placed parallel to the lac promoter of pBluescript. The resulting plasmid was partially digested by both DpnI (cutting at GmeATC) and DpnII (cutting at GATC). Activity, while evident, was not sufficient either to protect the plasmid from digestion by DpnII or to permit full digestion by DpnI. On the basis of this activity, the cloned gene was named dmtA. The limited protection afforded by dmtA in E.coli may explain why the gene was not picked up by the strategy that led to the cloning of avaMV. dmtA is flanked by two large ORFs of recognizable function. Besides these, there are no ORFs capable of encoding protein greater than 95 amino acids within 2000 bp of dmtA.
In addition, a predicted 5mC MTase was found that is very similar to M.XorII, which methylates CGATCG, and to SynMI, a cyanobacterial enzyme (37) with the same specificity. The similarity is particularly striking in the variable TRD region: 56 and 46% amino acid identity, respectively. Since PvuI, a CGATCG-specific endonuclease sensitive to cytosine methylation, fails to cut Anabaena DNA (38), even though recognition sites can be found in the genome with a frequency of one in about 1100 bp (J.Elhai, unpublished data), we concluded that the predicted protein actually functions in the cyanobacterium and on this basis named the gene that encodes it dmtC.
The gene is preceded by an ORF capable of encoding a 188 amino acid protein. This upstream gene, and close variants of it, occurs at least 11 times in the Anabaena genome, seven times in the genome of N.punctiforme and six times in the genome of Synechocystis PCC 6803 (data not shown). Its putative product shows no obvious similarity to noncyanobacterial protein. The gene family has been termed SPD, after a three amino acid sequence found in almost all variants. Apart from this ORF, there is no possible gene 1117 bp upstream or 2000 bp downstream of dmtC capable of encoding a protein greater than 95 amino acids that does not have a function readily recognizable from the sequence.
Analysis of genes capable of encoding Type II RM systems
The presence of three restriction activities in Anabaena (8) implies the existence of corresponding MTases, and three likely genes were found. One gene shows 100% nucleotide identity with the previously described gene encoding M.AvaI (accession no. X98339) cloned from Anabaena PCC 7118, and the adjacent gene is identical to R.AvaI (which also recognizes CyCGrG). Likewise, a second gene pair initially identified for the similarity of their predicted products to the 5mC MTase M.SinI (P09795) and its corresponding restriction enzyme R.SinI (an isoschizomer of R.AvaII) have 100% amino acid identity to the proteins predicted from the cloned M.AvaII and R.AvaII genes from Anabaena PCC 7118 (G.G.Wilson and K.D.Lunnen, personal communication). The enzymes from Anabaena PCC 7120 were originally given names [e.g. M.Asp(7120)I] to distinguish them from the corresponding enzymes from Anabaena PCC 7118 (39,40). Since the proteins are evidently identical, those names may now be abandoned. In fact, the near identity between the two strains over all known nucleotide sequences (four differences in 4783 bases; data not shown) compels us to accept the suggestion of Rippka (41) that the two strains are derived from the same natural isolate.
Detection of the gene encoding M.AvaIII (which recognizes ATGCAT) was more problematic, since no sequence has been reported of a MTase with the same target specificity. It seemed likely that the MTase was encoded by Orf337.35147, evidently an N6mA or N4mC MTase (Table 2), since that is the only ORF in the available Anabaena genome sequence predicted to encode a MTase that remained unaccounted for. This ORF and the one downstream from it was cloned (Fig. 1), and the resulting plasmid gained resistance to digestion by NsiI, which shares the same recognition sequence as AvaIII. On this basis, we judged that Orf337.35147 encodes M.AvaIII.
Table 2. Proteins similar to proven and putative Type II DNA RM MTases of Anabaena.
| Target |
Proteina |
Recognition sequenceb |
Source |
Scorec |
Accession no.d |
| Protein sequences similar to AvaI RM proteins | |||||
| M.AvaI | 1. M.NspIII | CyCGrG | Nostoc PCC 7524 | 4 × 10–141 | gb:AAC97192 |
| 2. M.BsoBI | CyCGrG | Bacillus | 6 × 10–83 | gb:CAA66933 | |
| R.AvaI | 1. R.BsoBI | CyCGrG | Bacillus | 1 × 10–104 | gb:CAA66932 |
| 2. R.NspIII | CyCGrG | Nostoc PCC 7524 | 1 × 10–103 | gb:AAC97193 | |
| Next match | >5 | ||||
| Protein sequences similar to AvaII RM proteins | |||||
| M.AvaII | 1. M.SinI | GGwC5C | Salmonella | 1 × 10–149 | sp:P09795 |
| 2. M.EcoO109I | rGGnCCy | Escherichia | 4 × 10–64 | gb:AAF06965 | |
| R.AvaII | 1. M.SinI | GGwCC | Salmonella | 4 × 10–66 | sp:P09796 |
| 2. M.Eco47I | GGwCC | Escherichia | 7 × 10–66 | sp:P50194 | |
| Next match | >0.2 | ||||
| Protein sequences similar to putative AvaIII RM proteins | |||||
| M.AvaIII | 1. ? | ? | Escherichia | 1 × 10–76 | sp:P28638 |
| 2. M.HpaI | GTTAA6C | Haemophilus | 3 × 10–31 | sp:P29538 | |
| 3. M.BglII | AGATC4T | Bacillus | 1 × 10–24 | gb:AAC45061 | |
| R.AvaIII? | No match | >3 | |||
| Protein sequences similar to AvaIV RM proteins | |||||
| M.AvaIV | 1. M.BlpI | GCTnAGC | Bacillus | 1 × 10–102 | NEB databasee |
| 2. M.DdeI | C5TnAG | Desulfovibrio | 1 × 10–84 | sp:P05302 | |
| 3. M.Bpu10IC1 | GC5TnAGG | Bacillus | 1 × 10–79 | gb:CAA74996 | |
| R.AvaIV | 1. R.BlpI | GCTnAGC | Bacillus | 2 × 10–98 | NEB databasee |
| (reconstructed)f | 2. R.Bpu10Iβ | GCTnAGG | Bacillus | 6 × 10–19 | gb:CAA74999 |
| 3. R.Bpu10Iα | GCTnAGG | Bacillus | 9 × 10–7 | gb:CAA74998 | |
| Next match | >1 | ||||
| Protein sequences similar to PstI RM-like proteins | |||||
| Orf362.31394R | 1. M.PstI | CTGCA6G | Providencia | 5 × 10–53 | sp:P00474 |
| (M-portion) | 2. Orf-F60 | ? | Aeromonas | 2 × 10–52 | gb:AAF45040 |
| 3. M.BsuBI | CTGCA6G | Bacillus | 7 × 10–52 | sp:P33563 | |
| Orf362.31394R | 1. R.BsuBI | CTGCAG | Bacillus | 6 × 10–89 | sp:P33562 |
| (M-portion) | 2. R.PstI | CTGCAG | Providencia | 2 × 10–72 | sp:P00640 |
| 3. R.Rle39BI | CTGCAG | Rhizobium | 9 × 10–20 | gb:CAA67875 | |
| 4. R.XphI | CTGCAG | Xanthomonsa | 5 × 10–14 | gb:AAF22367 | |
| Next match | >2 | ||||
aProtein sequences obtained by BLAST search (26) of the combined GenBank, PDB, SwissProt, PIR and PRF databases (unless otherwise noted), last accessed June 2000, ranked by score. Cyanobacterial sequences are shown in bold.
bRecognition sequences of MTases are given along with the position, if known, where they methylate: A6, N6-methyladenine; C5, C5-methylcytosine.
cScore represents expected number of proteins at least as similar to target within the combined databases.
dgb, GenBank; sp, SwissProt; pir, Protein Information Resource.
eSequence graciously provided by Geoffrey Wilson (New England Biolabs).
fReconstructed by removing insertion sequence from Orf323B.1084′R and Orf339B.′706 and combining the results.
The adjacent ORF, Orf337.36048, most likely encodes R.AvaIII. The failure to find a match between that ORF and known restriction enzymes, while not very informative, is expected, since restriction enzymes seldom show similarity to proteins that recognize different sequences (1; also compare with R.AvaI and R.AvaII in Table 2).
Two other gene pairs are similar to those encoding Type II RM enzymes. Orf323B.3213 is capable of encoding a 5mC DNA MTase highly similar to M.BlpI, while the adjacent gene is highly similar to R.BlpI (Table 2), except for an intervening insertion sequence near the 5′-end of the gene (Fig. 1). Since neither BlpI (data not shown) nor its isoschizomer EspI (38) cut Anabaena DNA, despite the presence of at least 16 sites (J.Elhai, unpublished results), it is likely that such sites are methylated owing to the action of the M.BlpI-like gene product. On that basis, the putative MTase has been named M.AvaIV, though the cognate restriction enzyme is surely inactive.
Orf362.31394R shows strong similarity (Table 2) to M.PstI and R.PstI fused together (also noted independently by Bill Buikema, University of Chicago, IL). The gene is interrupted by a stop codon appearing between a region encoding Motif VIII of the MTase and its TRD, so it is not surprising that Anabaena does not methylate at PstI sites (38).
None of the five predicted RM proteins show significant sequence similarity to proteins potentially encoded by N.punctiforme genes, beyond the general similarity shown by all members of their particular classes; nor do the genes that encode the proteins show similarity to N.punctiforme DNA (Fig. 1). In two cases, M/R.AvaII and M/R.AvaIII, the genes encoding the RM system replace a significant amount of N.punctiforme DNA: ∼740 and 5720 bp, respectively. The novel Anabaena sequences are not flanked by discernible direct or indirect repeats that might be indicative of the ends of insertion sequences.
Analysis of genes capable of encoding Type I MTases
Five regions of the Anabaena genome were found containing genes similar to those encoding elements of Type I RM systems (Table 3). In all cases, one or more genes were defective, as judged by frameshifts or insertion sequences within the gene. Two regions bear genes related to Type IB RM systems. The two sets of genes are nearly identical to each other, except for the specificity subunit (HsdS) and in both cases defects in that subunit should render restriction and modification inactive. The two modification (HsdM) and restriction (HsdR) proteins show striking resemblance to those of the EcoA and EcoE systems of E.coli strains T15 and A58. In fact, these E.coli systems are more similar to the Type IB systems from Anabaena than to those found thus far in any other organism, including the closely related enteric bacterium Salmonella typhimurium (data not shown).
Table 3. Proteins similar to those of putative Type I RM systems of Anabaena.
| Targeta |
Proteinb |
Source |
Scorec |
Accession no.d |
| Sequences similar to orf308A Type IB RM proteins | ||||
| HsdM | -. Orf311A | Anabaena | 1 × 10–153 | Genome sitee |
| (271 amino acid C-terminus) | 1. EcoE HsdM | Escherichia | 2 × 10–94 | pir:I41293 |
| 2. EcoA HsdM | Escherichia | 2 × 10–94 | pir:A47200 | |
| 3. StySKI HsdM | Salmonella | 9 × 10–93 | gb:CAA71895 | |
| -. Orf658 | Nostoc | 6 × 10–23 | Genome sitef | |
| -. Orf588 | Nostoc | 1 × 10–4 | Genome sitef | |
| HsdS | -. Orf311A | Anabaena | 3 × 10–90 | Genome sitee |
| (550 amino acid) | 1. CfrA | Citrobacter | 3 × 10–58 | pir:S06097 |
| 2. EcoE | Escherichia | 2 × 10–47 | pir:P19705 | |
| 3. EcoA | Escherichia | 2 × 10–46 | pir:P19704 | |
| -. Orf658 | Nostoc | 4 × 10–8 | Genome sitef | |
| HsdR | -. Orf308B | Anabaena | <10–200 | Genome sitee |
| (430 amino acid C-terminus) | 1. EcoA | Escherichia | 1 × 10–141 | pir:I41291 |
| 2. EcoE | Escherichia | 1 × 10–140 | pir:I41292 | |
| 3. StySKI | Salmonella | 2 × 10–53 | gb:CAA71894 | |
| -. Orf658 | Nostoc | 7 × 10–8 | Genome sitef | |
| Sequences similar to Orf308B/311A Type IB RM proteins | ||||
| HsdM | 1. EcoE HsdM | Escherichia | 1 × 10–101 | pir:I41293 |
| (296 amino acid C-terminus) | 2. EcoA HsdM | Escherichia | 1 × 10–101 | pir:A47200 |
| 3. StySKI HsdM | Salmonella | 1 × 10–100 | gb:CAA71895 | |
| HsdS | 1. EcoA | Escherichia | 4 × 10–81 | sp:P19704 |
| (542 amino acid) | 2. StySKI | Salmonella | 1 × 10–40 | gb:CAA71896 |
| 3. CfrA | Citrobacter | 7 × 10–35 | gb:CAA35604 | |
| 4. EcoE | Escherichia | 8 × 10–30 | sp:P19705 | |
| HsdR | 1. EcoA | Escherichia | <10–200 | pir:I41291 |
| (776 amino acid) | 2. EcoE | Escherichia | <10–200 | pir:I41292 |
| 3. StySKI | Salmonella | 3 × 10–53 | gb:CAA71894 | |
| Protein sequences similar to Orf260 Type IC RM protein | ||||
| HsdM | 1. LldI | Lactococcus | 1 × 10–120 | pir:T09460 |
| (537 amino acid) | 7. HsdM | Helicobacter | 1 × 10–93 | pir:E71886 |
| -. Orf339A | Anabaena | 1 × 10–67 | Genome sitee | |
| -. Orf376B | Anabaena | 4 × 10–47 | Genome sitee | |
| -. Orf641 | Nostoc | 4 × 10–21 | Genome sitef | |
| HsdR | 1. HsdR | Helicobacter | 1 × 10–127 | pir:B71890 |
| (933 amino acid) | -. Orf376B | Anabaena | 1 × 10–25 | Genome sitee |
| -. Orf641 | Nostoc | 3 × 10–10 | Genome sitef | |
| Sequences similar to Orf339A Type IC RM protein | ||||
| HsdM | 1. HsdM | Helicobacter | 1 × 10–180 | pir:C71810 |
| (803 amino acid) | -. Orf376B | Anabaena | 6 × 10–43 | Genome sitee |
| -. Orf641 | Nostoc | 8 × 10–19 | Genome sitef | |
| HsdS | 1. HsdS | Methanococcus | 8 × 10–22 | pir:B64316 |
| (425 amino acid) | -. Orf641 | Nostoc | 3 × 10–3 | Genome sitef |
| HsdR | 1. HsdR(179) | Klebsiella | 6 × 10–65 | pir:T30818 |
| (179 amino acid N-terminus) | -. Orf376B | Anabaena | 2 × 10–4 | Genome sitee |
| Sequences similar to Orf376B Type ID RM protein | ||||
| HsdM | 1. Cj1553c | Campylobacter | 1 × 10–171 | gb:CAB73544 |
| (527 amino acid) | 2. HsdM | Pasteurella | 1 × 10–135 | gb:AAC44666 |
| -. Orf641 | Nostoc | 4 × 10–20 | Genome sitef | |
| HsdS | 1. Cj1551c | Campylobacter | 6 × 10–29 | gb:CAB73967 |
| (390 amino acid) | 2. MJECL41 | Methanobacter | 5 × 10–15 | pir:H64514 |
| HsdR | 1. Cj1549c | Campylobacter | <10–200 | gb:CAB73965 |
| (1011 amino acid) | 2. HI1285 | Haemophilus | <10–200 | pir:F64114 |
| 3. HsdR | Pasteurella | 1 × 10–198 | pir:JC5216 | |
| -. Orf641 | Nostoc | 6 × 10–8 | Genome sitef | |
aProtein sequence from Anabaena used in BLAST search (26) of the combined GenBank, PDB, SwissProt, PIR and PRF databases, followed by length of sequence in amino acid residues. Partial sequences were used when full sequences were unavailable. Cyanobacterial sequences are shown in bold.
bProteins found by BLAST search, ranked by score. Scores of proteins not found in combined databases were obtained by a pairwise BLAST comparison.
cScore represents expected number of proteins at least as similar to target within the combined databases.
dgb, GenBank; sp, SwissProt; pir, Protein Information Resource.
eSequence obtained from the Anabaena web site (http://www.kazusa.or.jp/cyano/anabaena/), last accessed June 2000.
fSequences obtained from the N.punctiforme web site (http://spider.jgi-psf.org/JGI_microbial/html/nostoc_homepage.html), last accessed June 2000.
The three other Type I systems of Anabaena, two of Type IC and one of Type ID, follow the same pattern: the HsdM and HsdR proteins are very similar to known Type I enzymes, while the HsdS protein (when present) is less similar. In contrast, the Anabaena proteins show much less resemblance to Type I RM proteins from the cyanobacterium N.punctiforme (Table 3). None of the systems have restriction capability, by reason of a defective HsdR subunit, defective HsdS subunit or both. There are no obvious defects in the genes encoding the Type ID MTase and specificity subunits, so that system may be functional.
DISCUSSION
Nine genes that encode MTases with demonstrated or deduced activity have been identified from the cyanobacterium Anabaena (Table 4). Of these, four have characteristics associated with solitary MTases, and a fifth may also fall into that category. While this number is considerably higher than that established thus far for any other bacterium, solitary MTases may be common amongst bacteria.
Table 4. DNA MTases in Anabaena.
| MTase |
Locusa |
Specificityb |
Typec |
Found ind |
Restriction enzymee |
Comments |
||||||
| DNA MTases of demonstrated functionality | ||||||||||||
| M.AvaI | Orf308a.5063 | CyCGrG (AvaI) | N4mC(α) | — | Orf308A.6511 | = RM.Ava I | ||||||
| M.AvaII | Orf333.17488 | GGwCC (AvaII) | 5mC | — | Orf333.19647R | = RM.Ava II | ||||||
| M.AvaIII | Orf337.35114 | ATGCAT (NsiIII) | N6mA(β) | — | Orf337.36048? | = RM.Ava III? | ||||||
| M.AvaIV | Orf323b.2328R | GCTnAGC (BlpI)? | 5mC | — | (Orf323B.1084′R/339B.′706) | ∼ M.BlpI R.Ava IV interrupted by IS | ||||||
| M.AvaV | AF220506 | GATC (Dam) | N6mA(β2) | — | None | Solitary? | ||||||
| DmtA/M.AvaVI | Orf304.22874 | GATC (Dam) | N6mA(α) | NS | None | Solitary | ||||||
| DmtB/M.AvaVII | AF220507 | GGCC (HaeIII) | N4mC(α) | NS | None | Solitary | ||||||
| DmtC/M.AvaVIII | Orf350.1117 | CGATCG (PvuI) | 5mC | NS | None | Solitary | ||||||
| DmtD/M.AvaIX | AF220508 | rCCGGy (Cfr10I) | 5mC | N | None | Solitary | ||||||
| Putative or defective DNA MTases | ||||||||||||
| Defective? | Orf362.31394R | CTGCAG (PstI)? | N6mA(γ) | – | Orf362.31394R | Stop before TRD; fused to R.PstI | ||||||
| Defective? | Orf275.9656R | ? | N4mC(β) | None | Frame shift in TRD region | |||||||
| Defective? | Orf335.3576 | ? | 5mC | – | None | Motifs I, IV, VI and VIII only | ||||||
| Defective? | Orf351a.9898 | ? | 5mC | – | None | Motifs I, IV, VI and VIII (≈Orf335.3576) | ||||||
| Defective? | Orf308a.17412R | ? | Type IB | – | Orf308A.15244R (HsdS) Orf308A.′19733R (HsdR) | IS in hsdS | ||||||
| Defective? | Orf308b.11683′/311a.′30178 | ? | Type IB | – | Orf311A.29298 (HsdS) Orf308B.8327 (HsdR) | Stop at 5′ end of hsdM; frameshift in hsdS | ||||||
| Defective? | Orf260.11347R | ? | Type IC | – | Orf260.4629R (HsdR) | Frameshift, no start in hsdR; no nearby hsdS | ||||||
| Defective? | Orf339a.24489R | ? | Type IC | – | Orf339A.22077R (HsdS) Orf339A.20008R’ (HsdR) | Frameshift in hsdM;IS in hsdR | ||||||
| | ||||||||||||
| M.Ava V? | Orf376b.55687 | ? | Type ID | – | Orf376B.58994 (HsdS) Orf376B.60879 (HsdR) | Frameshift in hsdR at C-terminus | ||||||
aLocus given is either the accession number or the contig deposited in the Anabaena web site (http://www.kazusa.or.jp/cyano/anabaena/). The number after ‘Orf’ refers to the number of the contig and the number after the period refers to the position within the contig of the start codon. Numbers followed by ‘R’ indicate that the ORF lies on the strand opposite to that given in the contig. Numbers followed by an apostrophe indicate that the contig stops in the middle of the gene. Numbers preceded by an apostrophe indicate that the contig begins in the middle of the gene. Italicized names refer to ORFs with obvious defects (stop codons, frameshifts or insertion sequences within the gene).
bSpecificities were determined either as described in the text (M.AvaV, DmtB and DmtD) or by comparison of the TRD region with MTases of known specificity. A well-known RM system or solitary MTase with the same specificity is given in parentheses.
cThe subtype of MTase is according to Malone et al. (10) for Type II MTases (except for M.AvaV) or Murray et al. (17) for Type I MTases.
d‘N’ or ‘S’ is given if the MTase is substantially similar to one found in N.punctiforme or Synechocystis PCC 6803, respectively. A MTase is judged to be substantially similar if it exhibits greater similarity than most MTases of the same type or subtype.
eNearby gene encoding putative restriction enzyme or (in the case of Type I enzymes) specificity subunit (HsdR) or restriction subunit (HsdR). Naming conventions are the same as described abovea.
Definition of solitary MTases
Solitary MTases are defined primarily by a cellular role distinct from RM. The demonstration of such a function is no trivial task, but in the absence of proven function, it is possible to recognize solitary MTases by other characteristics. The failure to detect a corresponding restriction endonuclease is supportive evidence but by no means definitive, only in part because of the impossibility of proving that an activity is not present in a strain. It is more telling that a gene encoding a MTase has no nearby gene that can conceivably encode an active endonuclease. Here too, however, caution must be exercised. A MTase not long ago associated with a restriction endonuclease may momentarily exist alone, before selective pressures swallow it as well.
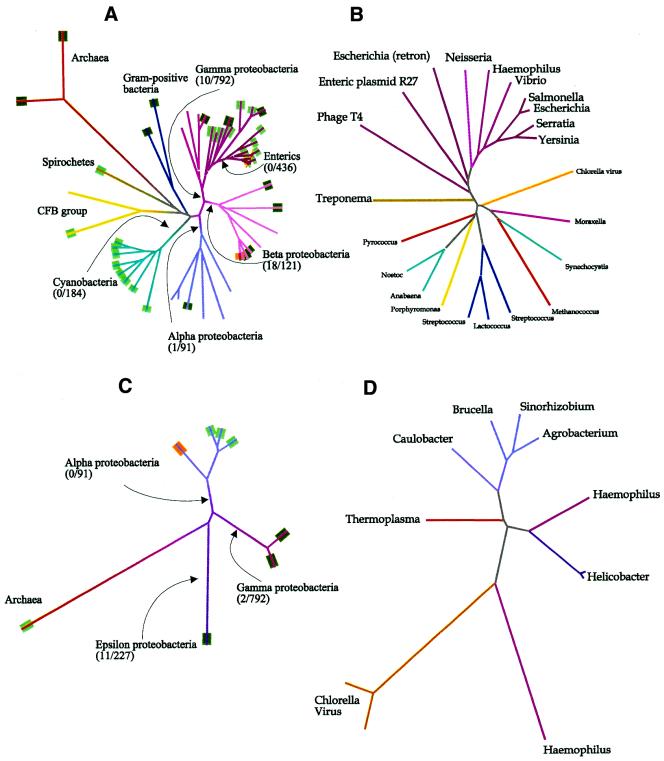
What sets solitary MTases apart from transiently orphaned MTases is stability over evolutionary time, conferred by the selective advantage its function confers on their hosts. This is seen in the two cases, Dam and CcrM, where independent function of a MTase has been demonstrated (2,3,42–44). Modification of GATC sites is found generally in enteric bacteria and closely related γ proteobacteria (Fig. 5A), and the lineage of the responsible MTase closely follows that of 16S rRNA (Fig. 5B). In contrast, the ability to restrict GATC sites is sporadic, and the lineage of restriction-associated GATC MTases shows a heavy influence of lateral DNA transfer (45). The same phenomenon is seen with CcrM and other GAnTC-specific MTases (Figs. 5C and D). Just as the lineage of GATC-specific MTases is coherent within a subset of the γ proteobacteria, so is the lineage of GAnTC-specific MTases within the α proteobacteria.
Figure 5.
Relationship amongst enzymes functionally similar to known solitary MTases, Dam and CcrM. Aligned 16S rRNA sequences were obtained from the Ribosomal Database Project (http://www.cme.msu.edu/RDP/html/index.html), and only the part of the alignment common to all was used in constructing the tree. (A) Dendrogram of 16S rRNA sequences from organisms that have been tested for GATC-specific adenine methylation (5) or that possess sequenced GATC-specific MTases of type N6mA(α). The number of restriction endonucleases known to recognize GATC is given followed by the total number of known endonucleases, for all bacteria beyond the indicated branch point. Lines are colored according to taxonomic groups. Tags at the end of branches are green if GATC-specific methylation has been demonstrated, dark green if the strain possesses a GATC-specific MTase and orange if the strain possesses a proven solitary GATC-specific MTase. (B) Dendrogram of all GATC-specific MTases of type N6mA(α) for which sequences are available. The lines are colored as in (A). The source of the sequences are given in the legend to Figure 4, except for Dam from Yersinia pseudotuberculosis (AAG23175), Dam from Serratia marcescens (P45454), Dam from S.typhimurium (P55893), gene from V.cholerae O395 (AAG23174), gene from H.influenzae Rf (P44431), Dam from Neisseria meningitidis BF13 (AAD34292), gene from E.coli retron Ec67 (P21311) and gene from enteric plasmid R27 (AAF69879). (C) Dendrogram of 16S rRNA sequences from organisms that possess sequenced GAnTC-specific MTases. The conventions are analogous to those described in (A). (D) Dendrogram of all GAnTC-specific MTases for which sequences are available. The lines are colored as in (C). The source of the sequences are (reading counterclockwise from Agrobacterium): gene from Agrobacterium tumefaciens C58 (Brad Goodner, personal communication), gene from Sinorhizobium meliloti 1021 (AAB71350), M.BabI (O30570), CcrM (Q45971), gene from Thermoplasma acidophilum DSM1728 (CAC12293), M.CviBI (Q01511), M.CviQVI (AAC03126), M.HhaII (P00473), M.HpyAIV (H64688), M.Hpy99IX (F71827) and M.HinfI (P20590).
One might have expected that the presence of a MTase would make it easier for the host to acquire a RM system that recognizes the same target sequence, but just the opposite is the case with Dam and CcrM. It is striking that while methylation at GATC is perhaps universal amongst enteric bacteria, there is no known instance of a restriction enzyme carried by a member of this group that cuts at GATC (Fig. 5A). Similarly, GAnTC-specific restriction enzymes are absent from the α proteobacteria (Fig. 5C), though the rarity of such enzymes in general makes the observation not particularly informative. Perhaps the functional constraints placed on Dam and CcrM methylation do not permit the total modification of DNA required by the presence of a restriction system. It should be noted that restriction enzymes that target the recognition sequence (CCwGG) of Dcm are very common in enteric bacteria.
In recognition of the needs of two scientific communities, those that study the structures of MTases and restriction enzymes and those that study bacterial physiology, we have given two names to solitary MTases. Like other MTases, they have been given names of the form M.AvaX (encoded by avaMX). In addition, to emphasize their cellular roles distinct from RM, they have been given names of the form DmtX (encoded by dmtX). A new name was chosen for its generality. It seemed inappropriate to use CcrM for any enzyme except that from Caulobacter crescentus, and Dam fits only deoxyadenine-specific MTases.
Multiple solitary MTases in Anabaena
On the basis of the criteria set forth above, Anabaena appears to have four solitary MTases, denoted as DmtA–D. None of the genes encoding these MTases lies near any plausible candidate to encode a corresponding restriction endonuclease. Restriction activities have not been found in Anabaena that recognize any of the four target sequences (8,46). Indeed, in two cases, dmtB and dmtD, null mutants are viable (A.V.Matveyev, K.T.Young and J.Elhai, unpublished observations), indicating that their activities are not required to protect the host against self-digestion by a cognate restriction enzyme. Supportive of the absence of such restriction enzymes in Anabaena is the observation that the efficiency of transfer from E.coli to Anabaena of plasmids bearing recognition sites for unmethylated DmtB, DmtC and DmtD is high. In contrast, the presence of unmethylated sites recognized by R.AvaI, R.AvaII or R.AvaIII drastically reduces the efficiency of conjugal transfer (8).
Another important criterion is evolutionary stability. GATC methylation (as specified by DmtA) is widely distributed and perhaps universal amongst cyanobacteria (Fig. 5A) (5,6) and yet none of the 162 characterized cyanobacterial restriction enzymes (27) have GATC specificity. The other cases are less clear. GGCC methylation (as specified by DmtB) is not universal (6,7), though it is widespread, and GGCC-specific restriction enzymes have been found in six cyanobacterial genera, including Anabaena (27). Little is known about the prevalence of CGATCG (DmtC) or rCCGGY (DmtD) methylation amongst cyanobacteria. Restriction enzymes recognizing the former sequence have been described in two cyanobacterial genera (27), but those recognizing the latter have not yet been found.
The GATC-specific M.AvaV poses an unusual case in that it, like RM systems (see below), appears to be a relatively recent arrival to Anabaena. On the other hand, there is no plausible ORF in the vicinity that might encode a corresponding endonuclease. Conceivably, an intact RM system was recently transferred to Anabaena, and the gene encoding the restriction enzyme has deteriorated. However, we have found that mutation of avaMV appears to alter the frequency at which cells differentiate (A.V.Matveyev and J.Elhai, unpublished results), so the MTase has evidently acquired a physiological role during its brief stay in Anabaena, even though the cyanobacterium already possesses DmtA, another GATC-specific MTase.
Unusual structure of Type II MTase M.AvaV
The 5mC MTases of Anabaena (DmtD, DmtC, M.AvaII and M.AvaIV) fall readily within the definitions of established groups, as do four of the N6mA/N4mC MTases (DmtB, DmtA, M.AvaI and M.AvaIII). One MTase, M.AvaV, does not and deserves special mention.
From the order of the few motifs that can be identified and the limited sequence similarity (Fig. 2), M.AvaV appears to be a distant relative of group β N6mA MTases. Presuming the identifications of Motif X (and possibly I) to be valid, the order of motifs corresponds to that found in group β MTases (IV-V-VI-VII-VIII-TRD-X-I-II-III; 10). The consensus sequences for the three identified regions are closer to the corresponding sequences for motifs from group β than to those of any other group. However, the differences between M.AvaV (and similar proteins) and group β MTases are also striking. The two groups show no significant similarity, according to BLAST comparisons, and M.AvaV and two of the three similar bacterial proteins are smaller than any of the 43 DNA MTases used to establish the conserved motifs amongst N4mC/N6mA MTases (10). We propose the creation of a new subgroup, β2, to accommodate M.AvaV-like MTases, but only a three-dimensional structure determination can resolve whether these proteins share the common architecture of previously characterized MTases despite unusual primary sequences or instead represent a second mechanism found by nature to methylate DNA.
There are other MTases besides M.AvaV that fit poorly into the conventional classification scheme. GATC-specific adenine MTases from Vibrio cholerae, E.coli phage T1 and Haemophilus influenzae phage P1 exhibit no obvious similarity to other MTases motif IV (47,48).
Evolution
We have attempted to discern the evolutionary history of MTases of Anabaena by comparing their sequences to those of other bacteria and in particular to those of the closely related cyanobacterium N.punctiforme and the distantly related cyanobacterium Synechocystis PCC 6803 (49). From this analysis, the generality emerges that solitary MTases have ancient antecedents in the cyanobacterial lineage. DmtD is common to both Anabaena and N.punctiforme. DmtB, DmtA and DmtC homologs are found in Synechocystis as well, and probably throughout the cyanobacterial radiation [the DmtB-like protein sll0729 of Synechocystis is no doubt the GGCC-specific MTase postulated by Scharnagl et al. (37)].
In contrast, RM systems are transient visitors to Anabaena. The active RM systems of Anabaena are absent even in N.punctiforme, and most of the genes show striking resemblances to those that exist in organisms outside the cyanobacteria (Tables 2 and 3). However, the systems are not long lived: the Anabaena genome is a graveyard of RM genes in various states of decomposition, as is the genome of Helicobacter pylori J99 (50). A survey of such enzymes must therefore represent a snapshot that would differ markedly from one taken at a different moment in evolutionary time.
These results are consistent with a genome that is showered with DNA derived from diverse organisms. Most foreign genes presumably disappear without a trace, but there remain a few well documented examples of horizontal transfer into the cyanobacterial lineage (51–53). The great divergence between cyanobacteria and other eubacteria (54) makes it unlikely that genes whose products interact with host machinery would function in their new host. RM systems, which can operate independently, seem well suited to proliferate by horizontal gene transfer (55,56) and either confer on their new hosts a selective advantage, e.g. defense against phage attack (57), or act as molecular parasites (56).
The alternative hypothesis that inactive RM systems represent an internal pool to be drawn upon for defense against foreign DNA (50) is less likely. None of the genes encoding the four active RM systems in H.pylori J99 are found in the closely related strain H.pylori 26695, although the two strains share several inactive systems (http://www.tigr.org/ and unpublished observations). The life history for RM systems seems more consistent with a one way path from acquisition to decay than with internal genetic recycling.
The commonality of certain RM systems in cyanobacteria related to Anabaena supports the idea that DNA exchange occurs more frequently among themselves than with the bacterial world at large. Of the 40 strains of Anabaena and other heterocystous cyanobacteria [Sections IV and V as per Rippka et al. (49)], 11 have AvaI activity, 14 have AvaII activity and six have both (27). In contrast, AvaI and AvaII are relatively rare amongst other cyanobacteria, and the one characterized AvaI-like system, AquI (58), found in a distantly related cyanobacterium is part of a 5mC system, with no similarity to the enzymes of Anabaena. There are several other restriction enzymes that recur amongst heterocystous cyanobacteria. A self-consistent phylogeny accounting for these occurrences without recourse to multiple horizontal transfers requires an ancestor with more than six RM systems and bears little resemblance to the currently accepted taxonomy (J.Elhai, unpublished observations). We conclude, then, that horizontal transfer amongst heterocystous cyanobacteria has been responsible on several occasions for the spread of RM systems, perhaps by transduction by phage with a broad host range amongst heterocystous cyanobacteria (59) or by conjugation (60). A perusal of known RM systems organized by their hosts indicates that the spread of certain systems within taxonomic groups is common. One may view this either as infections of taxa by selfish DNA or as sharing of valuable resources amongst relatives.
The transient presence of RM systems commonly exchanged among related cyanobacteria explains the observation that Anabaena strains (61; J.Elhai, unpublished observations) and E.coli K-12 (62) are deficient in sites recognized by restriction enzymes of related strains. This would be the expected result if organisms selected against recognition sites for their own restriction enzymes and if there were rampant genetic exchange of RM systems amongst similar strains (62,63,64).
The evidence suggests an unusual origin for DmtB (recognizing GGCC). In general, N4mC MTases form a coherent phylogenetic group, distinct from N6mA MTases (65). DmtB, however, lies within the class α N6mA radiance, particularly close to MTases with GATC-specificity. In general, DNA MTases that modify the same target sequence at the same site share broad sequence similarity (66), despite the taxonomic diversity of their hosts (e.g. the archaeal M.MjaIII and eukaryotic virus-derived M.CviAI mixed amongst the eubacterial GATC-specific MTases in Fig. 4). The phylogenetic position of DmtB suggests that it evolved from the line leading to DmtA and other cyanobacterial GATC-dependent N6mA MTases. DmtB adds credence to the hypothesis of Bujnicki and Radlinska (65) that N4mC MTases may have occasionally originated from N6mA precursors.
Function
The present work was inspired by a desire to understand the function of solitary MTases in Anabaena. Pertinent results along those lines will be published elsewhere, but in brief, DmtA appears to be essential for the viability of Anabaena under standard laboratory conditions, DmtB lies in a region intimately involved in the cellular differentiation evoked by nitrogen starvation, and loss of DmtD causes a slight defect in growth under laboratory conditions (A.V.Matveyev, J.Rumble and J.Elhai, unpublished results). The functional importance of the GATC sequences modified by M.AvaV and DmtA MTases is further indicated by the absence of such sites in some phages that infect Anabaena (38).
The target sequence (CGATCG) of DmtC is provocative, because it is contained within a highly iterated sequence (GCGATCG; HIP1) found in many cyanobacteria (67) and also envelops the target sequence (GATC) of DmtA and M.AvaV. Loss of the DmtC homolog SynMI of Synechocystis leads to poor growth, especially under conditions most favorable to rapid doubling (37).
It is interesting that two DNA MTases, M.AvaV and DmtA, recognize the same sequence. There are several instances known of restriction enzymes accompanied by two MTases with the same target sequence. In one such case, Streptococcus pneumoniae possesses two GATC-specific MTases, whose genes are adjacent to that encoding the GATC-specific restriction endonuclease DpnII (68). As with M.AvaV and DmtA, the two MTases of Streptococcus are of different groups (N6mAα and N6mA). The presence of the two MTases may be explained by their different biochemical properties. Furthermore, a strain of E.coli has been described that carries a second functional GATC-specific MTase besides Dam (69), and V.cholerae may also carry an unusual GATC-specific enzyme (48) in addition to the conventional Dam-like MTase.
In short, very little is yet known about the function of cyanobacterial solitary MTases. In pursuing their functions, we hope to extend our understanding of the roles played by solitary MTases beyond what is known from Dam and CcrM.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Elaine Bucheimer for help in the initial stages of cloning dmtD, Jeff Elbich for assembling the sequence of dmtB, Rafael de Sá for advice on phylogenetic analysis, Suzanne O’Handley and Chris Stevenson for help with the HPLC analysis of the modified bases, Virgis Siksnys and Brad Goodner for providing sequences prior to publication, M.G.Marinus for providing strains, Bill Buikema for sharing his analysis of Anabaena genes potentially encoding restriction endonucleases, Geoffrey Wilson for many useful discussions and use of the New England Biolabs database and Richard Roberts and the two anonymous reviewers for helpful comments. Part of the work was supported by grant 9975002 from the National Science Foundation (to J.E.). K.T.Y. was supported with an Undergraduate Arts and Sciences Summer Fellowship from the University of Richmond and a National Science Foundation Research Experiences for Undergraduates award. Sequencing of E.faecalis V583 was accomplished with support given to The Institute of Genomic Research from the National Institute of Allergy and Infectious Disease.
References
- 1.Wilson G.G. and Murray,N.E. (1991) Restriction and modification systems. Annu. Rev. Genet., 25, 585–627. [DOI] [PubMed] [Google Scholar]
- 2.Palmer B.R. and Marinus,M.G. (1994) The dam and dcm strains of Escherichia coli – a review. Gene, 143, 1–12. [DOI] [PubMed] [Google Scholar]
- 3.Zweiger G., Marcynski,G. and Shapiro,L. (1994) A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J. Mol. Biol., 235, 472–485. [DOI] [PubMed] [Google Scholar]
- 4.Wright R., Stephens,C. and Shapiro,L. (1997) The CcrM DNA methyltransferase is widespread in the α subdivision of proteobacteria, and its essential functions are conserved in Rhizobium meliloti and Caulobacter crescentus. J. Bacteriol., 179, 5869–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbeyron T., Kean,K. and Forterre,P. (1984) DNA adenine methylation of GATC sequences appeared recently in the Escherichia coli lineage. J. Bacteriol., 160, 586–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Padhy R.N., Hottat,F.G., Coene,M.M. and Hoet,P.P. (1988) Restriction analysis and quantitative estimation of methylated bases of filamentous and unicellular cyanobacterial DNAs. J. Bacteriol., 170, 1934–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmerman W.J. and Culley,D.E. (1991) Genetic variation at the apcAB, cpcAB, gvpA1, and nifH loci and in DNA methylation among N2-fixing cyanobacteria designated Nostoc punctiforme. Microbiol. Ecol., 21, 199–209. [DOI] [PubMed] [Google Scholar]
- 8.Elhai J., Vepritskiy,A., Muro-Pastor,A.M., Flores,E. and Wolk,C.P. (1997) Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol., 179, 1998–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar S., Cheng,X., Klimasauskas,S., Mi,S., Posfai,J., Roberts,R.J. and Wilson,G.G. (1994) The DNA (cytosine-5) methyltransferases. Nucleic Acids Res., 22, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malone T., Blumenthal,R.M. and Cheng,X. (1995) Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol., 253, 618–632. [DOI] [PubMed] [Google Scholar]
- 11.Klimasauskas S., Kumar,S., Roberts,R.J. and Cheng,X. (1994) HhaI methyltransferase flips its target base out of the DNA helix. Cell, 76, 357–369. [DOI] [PubMed] [Google Scholar]
- 12.Reinisch K.M., Chen,L., Verdine,G.L. and Lipscomb,W.N. (1995) The crystal structure of HaeIII methyltransferase convalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell, 82, 143–153. [DOI] [PubMed] [Google Scholar]
- 13.Labahn J., Granzin,J., Schluckebier,G., Robinson,D.P., Jack,W.E., Schildkraut,I. and Saenger,W. (1994) Three-dimensional structure of the adenine-specific DNA methyltransferase M.TaqI in complex with the cofactor S-adenosylmethionine. Proc. Natl Acad. Sci. USA, 91, 10957–10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong W., O’Gara,M., Blumenthal,R.M. and Cheng,X. (1997) Structure of PvuII DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucleic Acids Res., 25, 2702–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran P.H., Korszun,Z.R., Cerritelli,S., Springhorn,S.S. and Lacks,S.A. (1998) Crystal structure of the DpnM DNA adenine methyltransferase from the DpnII restriction system of Streptococcus pneumoniae bound to S-adenosylmethionine. Structure, 6, 1563–1575. [DOI] [PubMed] [Google Scholar]
- 16.Schluckebier G., O’Gara,M., Saenger,W. and Cheng,X. (1995) Universal catalytic domain structure of AdoMet-dependent methyltransferases. J. Mol. Biol., 247, 16–20. [DOI] [PubMed] [Google Scholar]
- 17.Murray N.E. (2000) Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol. Mol. Biol. Rev., 64, 412–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elhai J. and Wolk,C.P. (1990) Developmental regulation and spatial pattern of expression of the structural genes for nitrogenase in the cyanobacterium Anabaena. EMBO J., 9, 3379–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai Y. and Wolk,C.P. (1990) Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol., 172, 3138–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiss A., Posfai,G., Keller,C.C., Venetianer,P. and Roberts,R.J. (1985) Nucleotide sequence of the BsuRI restriction-modification system. Nucleic Acids Res., 13, 6403–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood W.B. (1966) Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J. Mol. Biol., 16, 118–133. [DOI] [PubMed] [Google Scholar]
- 22.Matveyev A.V., Rutgers,E., Soderback,E. and Bergman,B. (1994) A novel genome rearrangement involved in heterocyst differentiation of the cyanobacterium Anabaena sp PCC 7120. FEMS Microbiol. Lett., 116, 201–207. [Google Scholar]
- 23.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL:X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Page R.D. (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci., 12, 357–358. [DOI] [PubMed] [Google Scholar]
- 25.Henikoff S. and Henikoff,J.G. (1992) Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA, 89, 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts R.J. and Macelis,D. (2000) REBASE – restriction enzymes and methylases. Nucleic Acids Res., 28, 306–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nevill-Manning C.G., Wu,T.D. and Brutlag,D.L. (1998) Highly specific protein sequence motifs for genome analysis. Proc. Natl Acad. Sci. USA, 95, 5865–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cserzo M., Wallin,E., Simon,I., von Heijne,G. and Elofsson,A. (1997) Prediction of transmembrane α-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng., 10, 673–676. [DOI] [PubMed] [Google Scholar]
- 30.Gehrke C.W., McCune,R.A., Gama-Sosa,M.A., Ehrlich,M. and Kuo,K.C. (1984) Quantitative reversed-phase high-performance liquid chromatography of major and modified nucleosides in DNA. J. Chromatogr., 301, 199–219. [DOI] [PubMed] [Google Scholar]
- 31.Bancroft I. and Smith,R.J. (1988) An analysis of restriction endonuclease sites in cyanophages infecting the heterocystous cyanobacteria Anabaena and Nostoc. J. Gen. Virol., 69, 739–743. [DOI] [PubMed] [Google Scholar]
- 32.Bokar J.A., Shambaugh,M.E., Polayes,D., Matera,A.G. and Rottman,F.M. (1997) Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA, 3, 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 33.Siksnys V., Zareckaja,N., Vaisvila,R., Timinskas,A., Stakenas,P., Butkus,V. and Janulaitis,A. (1994) CAATTG-specific restriction-modification MunI genes from Mycoplasma: sequence similarities between R.MunI and R.EcoRI. Gene, 142, 1–8. [DOI] [PubMed] [Google Scholar]
- 34.Timinskas A., Butkus,V. and Janulaitis,A. (1995) Sequence motifs characteristic for DNA [cytosine-N4] and DNA [adenine-N6] methyltransferases. Classification of all DNA methyltransferases. Gene, 157, 3–11. [DOI] [PubMed] [Google Scholar]
- 35.Piekarowicz A. and Radlinska,M. (1998) Sensitivity of the restriction endonucleases HaeIII, BsrI, EaeI and CfrI to cytosine N4-methylation. Acta Microbiol. Polonica, 47, 405–407. [PubMed] [Google Scholar]
- 36.Nelson M., Christ,C. and Schildkraut,I. (1984) Alteration of apparent restriction endonuclease recognition specificities by DNA methylases. Nucleic Acids Res., 12, 5165–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scharnagl M., Richter,S. and Hagemann,M. (1998) The cyanobacterium Synechocystis sp. strain PCC 6803 expresses a DNA methyltransferase specific for the recognition sequence of the restriction endonuclease PvuI. J. Bacteriol ., 180, 4116–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bancroft I., Wolk,C.P. and Oren,E.V. (1989) Physical and genetic maps of the genome of the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol ., 171, 5940–5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murray K., Hughes,S.G., Brown,J.S. and Bruce,S.A. (1976) Isolation and characterization of two sequence-specific endonucleases from Anabaena variabilis. Biochem. J., 159, 317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roizes G., Nardeux,P.C. and Monier,R. (1979) A new specific endonuclease from Anabaena variabilis. FEBS Lett., 104, 39–44. [DOI] [PubMed] [Google Scholar]
- 41.Rippka R. (1988) Recognition and identification of cyanobacteria. Methods Enzymol., 167, 28–67. [DOI] [PubMed] [Google Scholar]
- 42.Torreblanca J. and Casadesus,J. (1996) DNA adenine methylase mutants of Salmonella typhimurium and a novel dam-regulated locus. Genetics, 144, 15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia-Del Portillo F., Pucciarelli,M.G. and Casadesus,J. (1999) DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc. Natl Acad. Sci. USA, 96, 11578–11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bucci C., Lavitola,A., Salvatore,P., Del Giudice,L., Massardo,D.R., Bruni,C.B. and Alifano,P. (1999) Hypermutation in pathogenic bacteria: frequent phase variation in meningococci is a phenotypic trait of a specialized mutator biotype. Mol. Cell, 3, 435–445. [DOI] [PubMed] [Google Scholar]
- 45.Raleigh E.A. and Brooks,J.E. (1997) Restriction modification systems: where they are and what they do. In de Bruijn,F.J., Lupski,J.R. and Weinstock,G.M. (eds), Bacterial Genomes: Physical Structure and Analysis. Chapman & Hall, New York, NY, pp. 78–92.
- 46.Duyvesteyn M.G.C., Korsuize,J., de Waard,A., Vonshak,A. and Wolk,C.P. (1983) Sequence-specific endonucleases in strains of Anabaena and Nostoc. Arch. Microbiol., 134, 276–281. [Google Scholar]
- 47.Schneider-Scherzer E., Auer,B., de Groot,E.J. and Schweiger,M. (1990) Primary structure of a DNA (N6-adenine)-methyltransferase from Escherichia coli virus T1. DNA sequence, genomic organization, and comparative analysis. J. Biol. Chem., 265, 6086–6091. [PubMed] [Google Scholar]
- 48.Bandyopadhyay R. and Das,J. (1994) The DNA adenine methyltransferase-encoding gene (dam) of Vibrio cholerae. Gene, 140, 67–71. [DOI] [PubMed] [Google Scholar]
- 49.Rippka R., Deruelles,J., Waterbury,J.B., Herdman,M. and Stanier,R.Y. (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol., 111, 1–61. [Google Scholar]
- 50.Kong H., Lin,L.-F., Porter,N., Stickel,S., Byrd,D., Posfai,J. and Roberts,R.J. (2000) Functional analysis of putative restriction-modification system genes in the Helicobacter pylori J99 genome. Nucleic Acids Res., 28, 3216–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moens L., Vanfleteren,J., Vandepeer,Y., Peeters,K., Kapp,O., Czeluzniak,J., Goodman,M., Blaxter,M. and Vinogradov,S. (1996) Globins in nonvertebrate species dispersal by horizontal gene-transfer and evolution of the structure–function-relationships. Mol. Biol. Evol., 13, 324–333. [DOI] [PubMed] [Google Scholar]
- 52.Delwiche C.F. and Palmer,J.D. (1996) Rampant horizontal transfer and duplication of rubisco genes in eubacteria and plastids. Mol. Biol. Evol., 13, 873–882. [DOI] [PubMed] [Google Scholar]
- 53.Rudi K. and Jakobsen,K.S. (1997) Cyanobacterial tRNA-Leu(UAA) group I introns have polyphyletic origin. FEMS Microbiol. Lett., 156, 293–298. [DOI] [PubMed] [Google Scholar]
- 54.Feng D.F., Cho,G. and Doolittle,R.F. (1997) Determining divergence times with a protein clock: update and reevaluation. Proc. Natl Acad. Sci. USA, 94, 13028–13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jain R., Rivera,M.C. and Lake,J.A. (1999) Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl Acad. Sci. USA, 96, 3801–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kobayashi I., Nobusato,A., Kobayashi-Takahashi,N. and Uchiyama,I. (1999) Shaping the genome–restriction-modification systems as mobile genetic elements. Curr. Opin. Genet. Dev., 9, 649–656. [DOI] [PubMed] [Google Scholar]
- 57.Bickle T.A. and Krüger,D.H. (1993) Biology of DNA restriction. Microbiol. Rev., 57, 434–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karreman C. and De Waard,A. (1990) Agmenellum quadruplicatum M.AquI, a novel modification methylase. J. Bacteriol., 172, 266–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Currier T.C. and Wolk,C.P. (1979) Characteristics of Anabaena variabilis influencing plaque formation by cyanophage N-1. J. Bacteriol., 139, 88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muro-Pastor A.M., Kuritz,T., Flores,E., Herrero,A. and Wolk,C.P. (1994) Transfer of a genetic marker from a megaplasmid of Anabaena sp strain PCC 7120 to a megaplasmid of a different Anabaena strain. J. Bacteriol., 176, 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Herrero A., Elhai,J., Hohn,B. and Wolk,C.P. (1984) Infrequent cleavage of cloned Anabaena variabilis DNA by restriction endonucleases from A. variabilis. J. Bacteriol., 160, 781–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elhai ,J. (2001) Determination of bias in the relative abundance of oligonucleoticles in DNA sequences. J.Comput. Biol., in press. [DOI] [PubMed] [Google Scholar]
- 63.Gelfand M.S. and Koonin,E.V. (1997) Avoidance of palindromic words in bacterial and archaeal genomes: a close connection with restriction enzymes. Nucleic Acids Res., 25, 2430–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rocha E.P.C., Viari,A. and Danchin,A. (1998) Oligonucleotide bias in Bacillus subtilis: general trends and taxonomic comparisons. Nucleic Acids Res., 26, 2971–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bujnicki J.M. and Radlinska,M. (1999) Molecular evolution of DNA-(cytosine-N4) methyltransferases: evidence for their polyphyletic origin. Nucleic Acids Res., 27, 4501–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Noyer-Weidner M. and Trautner,T.A. (1993) Methylation of DNA in prokaryotes. In Jost,J.P. and Saluz,H.P. (eds), DNA Methylation: Molecular Biology and Biological Significance. Birkhäuser Verlag, Basel, Switzerland, pp. 39–108.
- 67.Robinson N.J., Robinson,P.J., Gupta,A., Bleasby,A.J., Whitton,B.A. and Morby,A.P. (1995) Singular over-representation of an octameric palindrome, HIP1, in DNA from many cyanobacteria. Nucleic Acids Res., 23, 729–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cerritelli S., Springhorn,S.S. and Lacks,S.A. (1989) DpnA, a methylase for single-strand DNA in the DpnII restriction system, and its biological function. Proc. Natl Acad. Sci. USA, 86, 9223–9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hsu M.-Y., Inouye,M. and Inouye,S. (1990) Retron for the 67-base multicopy single-stranded DNA from Escherichia coli: a potential transposable element encoding both reverse transcriptase and Dam methylase functions. Proc. Natl Acad. Sci. USA, 87, 9454–9458. [DOI] [PMC free article] [PubMed] [Google Scholar]