Abstract
The human ISWI-containing factor RSF (remodeling and spacing factor) was found to mediate nucleosome deposition and, in the presence of ATP, generate regularly spaced nucleosome arrays. Using this system, recombinant chromatin was reconstituted with bacterially produced histones. Acetylation of the histone tails was found to play an important role in establishing regularly spaced nucleosome arrays. Recombinant chromatin lacking histone acetylation was impaired in directing transcription. Histone-tail modifications were found to regulate transcription from the recombinant chromatin. Acetylation of the histone tails by p300 was found to increase transcription. Methylation of the histone H3 tail by Suv39H1 was found to repress transcription in an HP1-dependent manner. The effects of histone-tail modifications were observed in nuclear extracts. A highly reconstituted RNA polymerase II transcription system was refractory to the effect imposed by acetylation and methylation.
Keywords: RSF, chromatin assembly, histones tails, chromatin transcription, acetylation, methylation
In eukaryotic cells, genes are complexed with core histones and other chromosomal proteins in the form of chromatin. The basic repeating unit of chromatin, the nucleosome, includes two copies of each of the four core histones—H2A, H2B, H3, and H4—wrapped by 146 base pairs of DNA (Luger et al. 1997a). With the aid of additional proteins, including histone H1, nucleosomes are further packaged into 30-nm fibers with six nucleosomes per turn in a spiral or solenoid arrangement (for reviews, see Kornberg and Lorch 1999; Hayes and Hansen 2001). The 30-nm fiber unfolds to generate a template for transcription, an 11-nm fiber or “beads on a string,” by a mechanism that is not entirely clear (for review, see Orphanides and Reinberg 2000). However, it is thought that unfolding involves posttranslational modifications, particularly acetylation, of the core histone N-terminal tails. The 11-nm fiber is also repressive to processes requiring access of proteins to DNA.
Over the past decade, studies have revealed that there are different types of protein complexes that can alter the repressive nature of chromatin (for reviews, see Vignali et al. 2000; Wolffe and Guschin 2000). These factors apparently can maintain chromatin in a dynamic state. One family of factors function by covalently modifying the histone polypeptides, through phosphorylation, acetylation, and/or methylation, primarily within the histone tails (for reviews, see Cheung et al. 2000a; Roth et al. 2001; Zhang and Reinberg 2001). A second class of factors involves multiprotein complexes that mobilize and/or alter the structure of nucleosomes (Kingston and Narlikar 1999; Orphanides and Reinberg 2000; Vignali et al. 2000). A third family of factors function to re-establish the structure of chromatin (for reviews, see Verreault 2000; Mello and Almouzni 2001). Importantly, the chromatin-related factors do not function in isolation; factors that alter the structure of nucleosomes function in concert with histone acetyltransferases (Krebs et al. 1999; Agalioti et al. 2000; Hassan et al. 2001), and factors that re-establish chromatin structure require specific patterns of histone acetylation (Verreault et al. 1996; Tyler et al. 1999). Most of the histone acetylation occurs within the tails, which protrude out of the nucleosome (Luger et al. 1997a). Therefore, acetylation does not directly alter the structure of the nucleosome but appears to affect the ability of nucleosomes to organize into high-order structures, by affecting internucleosomal interactions (Luger et al. 1997a) and/or interactions of the histone tail with linker DNA (Angelov et al. 2001). However, acetylation of the histone tails affects the mobility of the nucleosomes, and therefore it facilitates the access of sequence specific DNA-binding proteins to nucleosomal DNA (Wolffe and Hayes 1999; Brown et al. 2000). Moreover, recent studies have shown that acetylation at some residues within the histone tails serves as marks or recognition signals for factors to bind to acetylated tails (for reviews, see Strahl and Allis 2000; Turner 2000; Roth et al. 2001).
Recent studies have also shown that methylation of the histone tails affects transcription and chromatin structure (for reviews, see Rice and Allis 2001; Zhang and Reinberg 2001). Moreover, increasing evidence suggests that the different modifications of the histone tails are functionally interconnected (for reviews, see Jenuwein and Allis 2001; Zhang and Reinberg 2001). There are different examples showing interplay between phosphorylation, acetylation, and methylation of histones (Cheung et al. 2000b; Jenuwein and Allis 2001; Zhang and Reinberg 2001). For example, phosphorylation of histone H3–Ser 10 promotes acetylation of histone H3–Lys 14; moreover, these two modifications prevent methylation at Lys 9 (Berger 2001). PRMT1 (protein arginine methyltransferase-1), which stimulates transcription, specifically methylates histone H4–arginine-3, resulting in the enhancement of H4-tail acetylation by p300 (Wang et al. 2001). On the other hand, Suv39H1, which functions as a repressor, methylates histone H3–Lys 9 and acts as a signal for the recruitment of HP1, inducing the formation of heterochromatin (Bannister et al. 2001; Lachner et al. 2001). Histone H3–Lys 9 is also a substrate for acetylation, and methylation by Suv39H1 requires the action of histone deacetylases (for review, see Berger 2001; Jenuwein and Allis 2001; Nakayama et al. 2001; Zhang and Reinberg 2001).
A different class of factors that manipulate chromatin structure is the chromatin remodeling complexes. These factors use the energy derived from ATP hydrolysis to mobilize and/or disrupt the nucleosome structure, making DNA accessible to DNA-binding proteins (Kingston and Narlikar 1999; Kornberg and Lorch 1999; Wolffe and Hayes 1999; Vignali et al. 2000). ATP-dependent chromatin remodeling factors are highly conserved between species. These factors contain a related subunit that belongs to the SNF2 family and can be divided into three classes: SWI2/SNF2, ISWI, and CHD (Vignali et al. 2000). Although these complexes contain structurally related catalytic subunits, the factors differ in the mechanism by which they manipulate nucleosomes. Some of these complexes, represented by Swi/Snf, can mobilize nucleosomes and alter the structure of a nucleosome to increase access to DNA (Vignali et al. 2000). These factors can also transfer an entire octamer of histone proteins from one DNA site to another (Lorch et al. 1999; Phelan et al. 2000). Also, the ISWI-containing factor, NURF (nucleosome remodeling factor), can mobilize a nucleosome and alter its structure; however, apparently NURF cannot transfer the nucleosome in trans (Hamiche et al. 1999). NURF is therefore functionally different from the Swi/Snf-containing factors and also different from the other ISWI-containing factors, ACF (ATP-utilizing chromatin assembly and remodeling factor), CHRAC (chromatin accessibility factor), and RSF (remodeling and spacing factor) (see below), which can remodel chromatin to allow access of transcription factors to DNA. Apparently, the remodeling reaction does not result in the alteration of nucleosome structure but is a consequence of the ability of these factors to mobilize or slide nucleosomes (for reviews, see Kingston and Narlikar 1999; Vignali et al. 2000).
With the existence of different factors that can mobilize and/or alter the structure of the nucleosomes, it is not surprising to find factors that have the ability to deposit nucleosome arrays to re-establish the structure of chromatin. In general, there are two classes of chromatin assembly factors: DNA replication coupled factors and DNA replication independent factors. The bulk of chromatin assembly occurs during S-phase, when the DNA is being replicated, and it is achieved by CAF-1 (chromatin assembly factor-1; for reviews, see Verreault 2000; Mello and Almouzni 2001) and RCAF (replication-coupling assembly factor; Tyler et al. 1999). In this process, newly synthesized histones carrying a specific pattern of acetylation, specifically at lysines 5 and 12 on the N-terminal tail of histone H4, are recognized by CAF-1, as well as by RCAF, and are deposited onto newly replicated DNA (Verrault et al. 1996; Tyler et al. 1999). During interphase, however, a fraction of nucleosomal histones is lost as a consequence of histone turnover and/or because of different processes in which DNA is used as template. Chromatin can be re-established through the action of specific factors. Histones chaperones, such as NAP-1 (nucleosome assembly factor-1), together with factors that use the energy derived from ATP-hydrolysis, mobilize nucleosomes and mediate chromatin assembly uncoupled to DNA replication (for review, see Ito et al. 1997b). Two factors have been identified that are able to assemble regularly spaced chromatin in the presence of NAP-1: ACF (Ito et al. 1997a) and CHRAC (Varga-Weisz et al. 1997). These factors were first identified in Drosophila and subsequently in Xenopus laevis and human cells (Bochar et al. 2000; Guschin et al. 2000; LeRoy et al. 2000; Poot et al. 2000). ACF is composed of two subunits, Acf-1 and ISWI (ATPase), whereas CHRAC appears to be composed by Acf-1, ISWI, and two small subunits (CHRAC-16 and CHRAC-14). These factors are able to mobilize nucleosomes in cis (Langst et al. 1999).
The formation of regularly spaced nucleosomes by ACF and CHRAC is likely the result of two independent activities that function in concert. NAP-1 can mediate nucleosome deposition, whereas the ISWI subunit of ACF and CHRAC can space nucleosomes in an ATP-dependent manner on its own (Ito et al. 1999). The efficiency of nucleosome array formation is enhanced by the common larger subunit composing ACF and CHRAC (Acf-1, BAF-1A, WCRF-180). Nucleosome formation is likely to occur in three steps. The first step, which appears to be rate limiting, is the deposition of the H3/H4 tetramers onto DNA, followed by the fast deposition of the H2A/H2B dimers. The last step is the wrapping of the DNA around the histone octamer. Once the nucleosomes are deposited on the DNA, ACF and/or CHRAC (in a reaction requiring ATP hydrolysis) mobilize nucleosomes to create regularly spaced nucleosome arrays (Ladoux et al. 2000; Nakagawa et al. 2001).
In the present study, we found that RSF, which is an ISWI-containing factor (LeRoy et al. 1998), is able to deposit regularly spaced nucleosomes in the absence of chaperones. We use the RSF-mediated nucleosome array reaction and fully reconstitute chromatin with recombinant bacteria-expressed histones. This allowed us to directly study the role of different histone modifications in the chromatin assembly reaction and in transcription.
Results
RSF mediates the assembly of nucleosome arrays
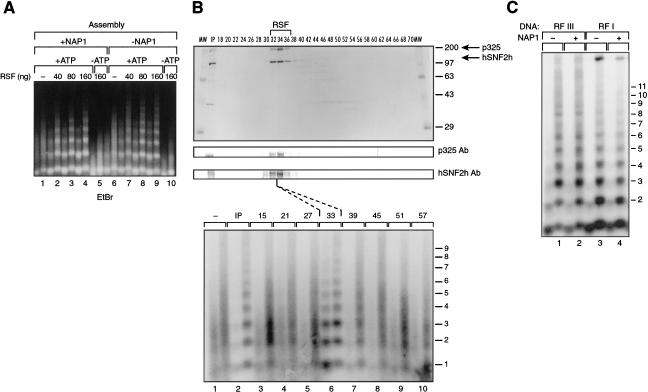
We previously reported the ability of RSF to convert irregularly spaced nucleosomes into regularly spaced nucleosome arrays (LeRoy et al. 1998). This activity is also present in the Drosophila and human ACF and CHRAC, which can also assemble regularly spaced nucleosome arrays in the presence of histone chaperone NAP-1. These findings prompted us to analyze whether RSF has the ability to assemble chromatin. Chromatin assembly reactions were performed using a 3-kb supercoiled DNA, HeLa-purified core histones, and the histone chaperone NAP-1. Partial micrococcal nuclease digestion shows that RSF assembles chromatin, as detected by the generation of regularly spaced micrococcal nuclease DNA ladders (Fig. 1A). The reaction is ATP dependent (Fig. 1A, lane 5) and requires RSF (Fig. 1A, cf. lane 1 with 2–4). Fractionation of RSF on a gel filtration column in the presence of high salt (0.5 M KCl) shows that the activity generating micrococcal nuclease ladders coeluted with the p325 and hSNF2h subunits of RSF as shown by SDS-PAGE and Western blot analyses (Fig. 1B).
Figure 1.
RSF mediates the assembly of nucleosome arrays. (A) Ethidium bromide staining of micrococcal nuclease digestions of chromatin assembled by RSF. Chromatin assembly was performed in the presence (left) and absence (right) of NAP-1, with increasing amounts of RSF. The highest amount of RSF was also assayed in the absence of ATP, as indicated on the panel. Products of the assembly reaction were partially digested with two different concentrations of micrococcal nuclease. Each lane number, at the bottom of the panel, denotes two different concentrations of micrococcal nuclease used in the assays. (B) Fractionation of RSF by gel filtration. Fractions of the Superose 6 column were assayed by Coomassie blue staining (top), Western blot (middle), and chromatin assembly (bottom). Each lane number on the micrococcal nuclease assay is between lanes and denotes two different concentrations of micrococcal nuclease used in the assay. RSF fractionated between fractions 32 and 36, which corresponds to a complex of ∼450 kD. (C) Southern blot of micrococcal nuclease digestions of RSF-assembled chromatin. Supercoiled (pG5MLP) DNA was linearized with EcoRI and used as a substrate in the RSF-assembly reaction (RFIII). As a control, supercoiled DNA was assayed in parallel (RFI). Lane numbers, at the bottom of the panel, denote two different concentrations of micrococcal nuclease used in the assays.
Characterization of the chromatin assembly reaction establishes that RSF functions stoichiometrically with DNA, as approximately one molecule of RSF was found to be required to completely assemble one molecule of DNA (data not shown; see below). According to the length of the DNA, RSF allowed the deposition of 17 nucleosomes under the conditions used (see Fig. 2). The RSF-mediated reaction is mechanistically different from the reaction catalyzed by ACF. Previous studies have established that ACF functions catalytically and that one molecule of ACF can catalyze nucleosome deposition on 90 molecules of DNA (Ito et al. 1997a). The differences observed between RSF and ACF are likely explained by the finding that RSF not only spaces chromatin, but RSF is directly involved in the deposition of histones onto the DNA (see below).
Figure 2.
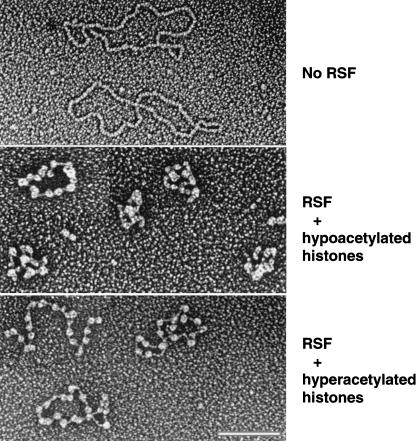
Electron microscopic analysis of RSF-assembled chromatin. (Top) An electron microscopic visualization of the RSF-assembled chromatin performed in the absence of RSF. DNA size is 3 kb. (Middle) The RSF-assembled chromatin performed with hypoacetylated histones purified from HeLa cells. (Bottom) The reaction with RSF and hyperacetylated histones purified in the presence of the histone deacetylase inhibitors TSA and Na-butyrate. The pictures are representatives of the majority of the molecules visualized on the grid. Bar, 116 nm.
Micrococcal nuclease digestion (Fig. 1A) and supercoiling experiments (data not shown; see below) show that RSF is able to assemble regularly spaced chromatin in a histone chaperone NAP-1-independent manner with linear or supercoiled DNA (Fig. 1C). To directly visualize RSF-assembled chromatin, we characterized the reaction products by electron microscopy using hypo- and hyperacetylated histones. In the absence of RSF, the majority of the DNA was naked or contained one or two nucleosomes (Fig. 2, top panel; data not shown). In the presence of RSF, the DNA was fully packaged into chromatin (Fig. 2). In general, 79% of the DNA contained nucleosomes, and the fully assembled DNA was in an array of correctly spaced nucleosomes (Fig. 2, middle and bottom panels). In reactions reconstituted with hypoacetylated histones, the nucleosomes interacted more readily (Fig. 2, middle panel), in contrast to hyperacetylated chromatin, where chromatin was found to be less compacted (Fig. 2, bottom panel), in agreement with previous observations (Garcia-Ramirez et al. 1995). In general, the levels of N-tail acetylation have been associated with a specific type of chromatin; hypoacetylated histones are associated with the formation of higher-order repressed chromatin structure. On the other hand, hyperacetylated histones are mostly associated with an open, transcriptionally active, chromatin structure. These previous observations correlate with the behavior of the RSF-assembled arrays in transcription (see below).
Mechanism of RSF-mediated chromatin assembly
To gain insight into the mechanism by which RSF mediates chromatin assembly, we investigated the possible role of RSF as a histone chaperone. Histone chaperones can be divided into two families: One class, represented by NAP-1, interacts strongly with histones H2A/H2B. The other class, represented by CAF-1, interacts strongly with histones H3/H4 (for review, see Ito et al. 1997b). The function of these chaperones is likely to facilitate association between histones and DNA by reducing electrostatic associations between positively charged histones and negatively charged DNA.
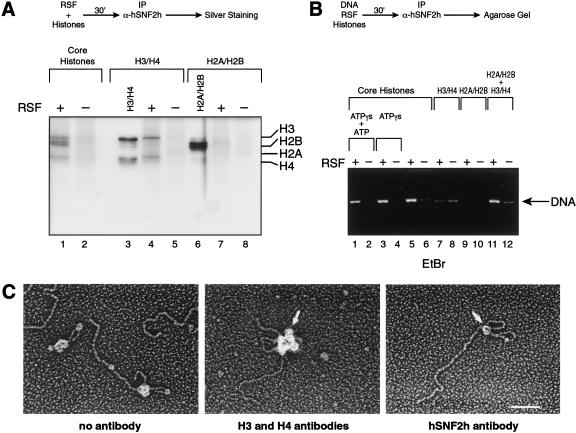
To address whether RSF interacts with histone polypeptides, immunoprecipitation assays were performed using antibodies against hSNF2h, the small subunit of RSF. These antibodies were able to coimmunoprecipitate HeLa core histones in the presence of RSF (Fig. 3A). The coimmunoprecipitate observed was specific for RSF; reactions performed in the absence of RSF did not coimmunoprecipitate histones (Fig. 3A, lane 2). Importantly, when the H3/H4 tetramers and the H2A/H2B dimers were used instead of the histone octamer, the antibodies were able to coimmunoprecipitate the H3/H4 tetramers but not the H2A/H2B dimers (Fig. 3A, lanes 4 and 7). From these results, we conclude that RSF interacts with the histone octamer through an association with the H3/H4 tetramer.
Figure 3.
RSF interacts with the histone tetramer, and its association with DNA requires the octamer. (A) Silver staining of an immunoprecipitation of the RSF–histone complexes. (Top) A schematic representation of the assay used to analyze the interaction of RSF with histones is depicted. The reaction was performed in the presence and absence of RSF, with HeLa core histones, H3/H4 tetramers, or H2A/H2B dimers, as indicated above the figure. H3/H4 tetramers and H2A/H2B dimers were separated over a Mono S column as described (Chang and Luse 1997). Immunoprecipitation reactions were performed as described in Materials and Methods. The immunocomplexes were washed with a buffer containing 0.5 M KCl and 0.01% NP-40. Immunoprecipitates were separated by SDS-PAGE followed by silver staining. (Right) The migration of each of the histones. (B) Ethidium bromide staining of an agarose gel showing a requirement for core histones for the establishment of an RSF–DNA complex.(Top) Schematic representation of the steps used to analyze binding of RSF to DNA. EcoRI-linearized DNA, RSF and core histone octamers were incubated in the presence and absence of ATP (0.6 μM) and ATP-γS (6 mM ATP), as indicated. H3/H4 tetramers, H2A/H2B dimers, and H3/H4 tetramers plus H2A/H2B dimers were incubated with or without RSF for 30 min at 30°C. The complexes were immunoprecipitated, and the immunoprecipitates were washed with a buffer containing 0.5 M KCl and 0.01% NP40 as described in Materials and Methods. Products of the immunoprecipitation reaction were separated by electrophoresis on an agarose gel. The arrow indicates the migration of DNA. (C) Electron microscopic visualization of intermediates in the RSF-chromatin assembly reaction. RSF was incubated with EcoRI-linearized plasmid and core histones in the presence of 6 mM ATP-γS plus 0.6 μM ATP for 30 min at 30°C. (Left) The products of the reaction visualized by electron microscopy. (Middle, right) RSF was incubated with EcoRI-linearized DNA and histones as above. The products of the reactions were first incubated with antibodies against histones H3 (monoclonal) and H4 (polyclonal; middle) or against the RSF subunit hSNF2h (monoclonal; right). This was followed by the incubation with a secondary antibody conjugated with gold (10 nm for H3 and H4 antibodies, 5 nm for hSNF2h antibody), as described in Material and Methods. The samples were visualized by electron microscopy. The arrow shows the secondary antibody gold particle. The big protein complex that appears in the reaction with antibodies against H3 and H4 (middle) represents multiples secondary antibodies bound to the primary antibody. The pictures are representatives of the majority of the molecules visualized on the grid. Bar, 85 nm.
We next analyzed whether RSF interacts with DNA by immunoprecipitation, as described above. The results established that RSF does not directly bind to DNA (data not shown; see below), and although RSF interacts with the H3/H4 tetramer, this interaction is not sufficient for stable binding of RSF to DNA. The formation of an RSF–DNA complex requires the octamer (Fig. 3B). Moreover, although ATP and hydrolysis of the highly energetic β–γ bond are required for the establishment of arrays, ATP had no effect on octamer-dependent binding of RSF to DNA (Fig. 3B).
To gain further insights into the mechanism by which RSF mediates deposition of nucleosome arrays, we examined intermediates of the reaction by electron microscopy. Here, chromatin assembly reactions were performed in the presence of ATP-γS to trap reaction intermediates. Under limiting ATP concentrations, we observed that most DNA molecules contained a single large protein complex at the base of a DNA loop and nucleosomes at various positions along the DNA molecule (Fig. 3C, left panel). The large protein complexes observed on the DNA were composed of one RSF complex and one or more nucleosomes, as shown by probing with gold-conjugated antibodies to RSF and histones H3 and H4 (Fig. 3C, right and middle panel, respectively). Increasing the amount of RSF to fivefold relative to the number of DNA molecules resulted in no change in the number and size of the large complexes on DNA, which agrees with the reaction stoichiometry of one RSF to one DNA molecule. Further, the size of the DNA loop, possibly generated by the RSF/nucleosome complex, was variable, but in most cases, the DNA loop contained one or two nucleosomes. From the above study of the role of ATP hydrolysis in this reaction (Fig. 3B), we suggest that the looped structure is an intermediate of the nucleosome spacing reaction, not an intermediate of the deposition reaction. Taken together, we suggest that one of the first steps in the chromatin assembly reaction is the binding of RSF to the DNA. This binding is dependent on the core histones and does not require energy.
Reconstitution of RSF-mediated chromatin assembly
We next investigated whether RSF could mediate the formation of nucleosome arrays with bacterially produced core histones. This would permit the analysis of the different histone-tail modifications involved during the establishment of nucleosome arrays, as well as in transcription.
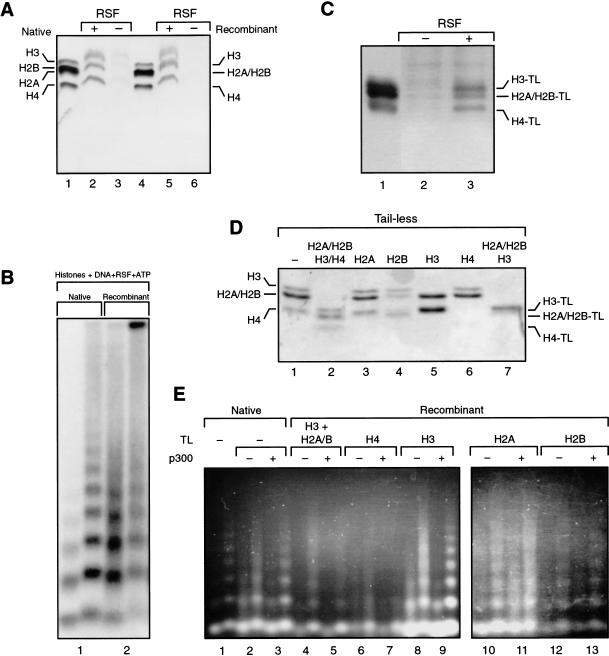
We first analyzed whether RSF is able to interact with the histone octamer reconstituted with bacterially expressed polypeptides. Immunoprecipitation studies, performed as described above, show that antibodies against hSNF2h coimmunoprecipitated the bacterially produced histones (Fig. 4A). This result established that RSF interacts with the histone octamer regardless of posttranslational histone modifications. We expanded this observation and analyzed whether RSF could generate nucleosome arrays using the bacterially produced histones. Micrococcal nuclease digestion shows the formation of regularly spaced nucleosomal arrays (Fig. 4B), although the reaction with bacterially expressed histones was less efficient than that observed with histones purified from HeLa cells (Fig. 4B, cf. lanes 1 and 2).
Figure 4.
Reconstitution of RSF-mediated chromatin assembly with recombinant histones. (A) RSF interacts with recombinant histones. Silver staining of coimmunoprecipitates obtained using antibodies against hSNF2h. (Lanes 4–6) Results with RSF and recombinant histones. (Lanes 1–3) As a control, HeLa purified core histones were used. The immunoprecipitates were washed with a buffer containing 0.5 M KCl and 0.01% NP-40. Indicated are the migration of recombinant histones (right) and the migration of native histones (left). (B) Southern blot analysis of partial micrococcal nuclease digestion of RSF assembly with native and recombinant histones, as indicated. Each lane number, at the bottom of the panel, denotes two different concentrations of micrococcal nuclease used in the assays. (C) Silver staining of RSF immunoprecipitates from reactions using recombinant histone octamer deleted of N-terminal tails. Immunoprecipitation was performed using antibodies against hSNF2h. The immunoprecipitates were washed with a buffer containing 0.5 M KCl and 0.01% NP-40. (Right) The migration of recombinant tailless histones. (D) Coomassie blue staining of a 15% SDS-PAGE containing different octamers formed with histone polypeptides deleted of the tails, as indicated at the top of the panel. The migration of wild-type histones (left) and tailless histones (right). (E) Ethidium bromide staining of partial micrococcal nuclease digestion of products obtained in the RSF-assembled chromatin using recombinant octamers with different combinations of histone tails (indicated at top). The octamers used were those analyzed in D. Reactions were performed in the presence and the absence of p300 plus 10 μM acetyl-CoA as indicated. As a control, the first lane represents chromatin assembled with native histones. Each lane number, at the bottom of the panel, denotes two different concentrations of micrococcal nuclease used in the assays.
Histone tails and their acetylation have been shown to affect chromatin assembly. DNA replication coupled chromatin assembly, catalyzed by CAF-1 and RCAF, requires a specific pattern of histone-tail acetylation (Verreault et al. 1996; Tyler et al. 1999). On the other hand, the tail of histone H4 has been shown to be essential to stimulate the ATPase activity of Drosophila ISWI (Georgel et al. 1997; Clapier et al. 2001) and to attain spaced nucleosomes (Clapier et al. 2001). In light of these previous observations, and the finding that nucleosome arrays generated with bacterially isolated histones are spaced less efficiently than those obtained with human histones (Fig. 4B), we investigated whether the RSF-chromatin assembly reaction requires histone tails and could be affected by acetylation.
We began the analysis by asking whether the tails of the core histones are important for RSF–octamer interaction. Octamers reconstituted with core histones with deleted tails (Fig. 4D, lane 2) were incubated with RSF, and the possible interaction was analyzed by immunoprecipitation as above. Antibodies against the small subunit of RSF immunoprecipitated the tailless octamer in an RSF-dependent manner (Fig. 4C); therefore, we conclude that the tails are not necessary for interaction with RSF. Although RSF was able to interact with the tailless octamer, we could not see RSF-dependent histone deposition with the tailless octamer (data not shown; see below). This prompted us to analyze the function of each of the core histone tails on RSF-mediated histone deposition onto DNA and on RSF-mediated nucleosome array formation and whether acetylation affects any of these reactions.
We found that p300-mediated acetylation of the histone polypeptides before assembly improved RSF-mediated nucleosome array formation (Fig. 4E, cf. lanes 2 and 3). This effect was caused by acetylation, as reactions performed in the absence of AcCoA failed to stimulate nucleosome array formation (data not shown). To investigate which of the histone tails are targeted by p300 acetylation in the assembly reaction, octamers were generated using different combinations of histone polypeptides with the tail deleted (Fig. 4D). These octamers were used in the chromatin assembly reaction in the presence and absence of p300 and analyzed for arrays using the micrococcal nuclease digestion assay. As expected (Clapier et al. 2001), the tail of histone H4 was necessary to attain correctly spaced nucleosome arrays, regardless of whether the tails of the other core histones were acetylated (Fig. 4E, lanes 6 and 7). From the micrococcal nuclease digestion pattern, which demonstrate the presence of mononucleosomes, it is possible to conclude that the histones were deposited onto the DNA, but the nucleosomes could not be spaced.
We investigated whether the tails of the core histones, other than that of H4, were required. Assembly reactions were analyzed with wild-type H4, but the tails of the histones H3, H2A, and H2B were deleted. Under these conditions, poor nucleosome array formation was observed, and p300-mediated acetylation (of histone H4) failed to enhance the efficiency of array formation (Fig. 4E, lanes 4 and 5). These results collectively establish that the histone tails and their acetylation are important for RSF-dependent nucleosome array formation.
We then assessed the role of the H3, H2A, and H2B tails, individually. Interestingly, the deletion of the histone H3 tail enhanced nucleosome deposition and, in a p300-acetylation–dependent manner, greatly enhanced the formation of nucleosome arrays (Fig. 4E, lanes 8 and 9). This result strongly suggests that acetylation of the H2A and/or H2B tail is important in establishing nucleosome arrays. This was analyzed using octamers missing the H2A or H2B tail. Interestingly, octamers deleted of the H2A tail were deposited, and p300 failed to stimulate array formation (Fig. 4E, lanes 10 and 11). Similarly, array formation using H2B tailless octamer was not efficient, and p300-mediated acetylation could not stimulate the reaction (Fig. 4E, lanes 12 and 13). From these results, we conclude that the tails of H2A and H2B are important for assembly and that p300-mediated stimulation of array formation requires these tails. Under the conditions of our assay, the histone H3 tail appears to interfere with p300-mediated stimulation of array formation (Fig. 4E, cf. lanes 3 and 9). It is important to consider that although octamers were provided in the chromatin assembly reaction, the conditions for optimal RSF activity included 0.05 M KCl, a salt concentration that is not permissive for octamers (Thomas and Kornberg 1975; Greyling et al. 1983). Therefore, the histones in the chromatin assembly reaction are likely to exist as dimers (H2A/H2B) and tetramers (H3/H4). Therefore, we suggest that first the H3/H4 tetramer–RSF complex forms and subsequently H2A/H2B contacts the H3/H4–RSF complex. Importantly, acetylation appears not to affect the binding of RSF to the histones, as native histones, bacterially produced histones, and tailless histones bind to RSF, with similar apparent affinities (data not shown).
Transcription on RSF-assembled chromatin templates
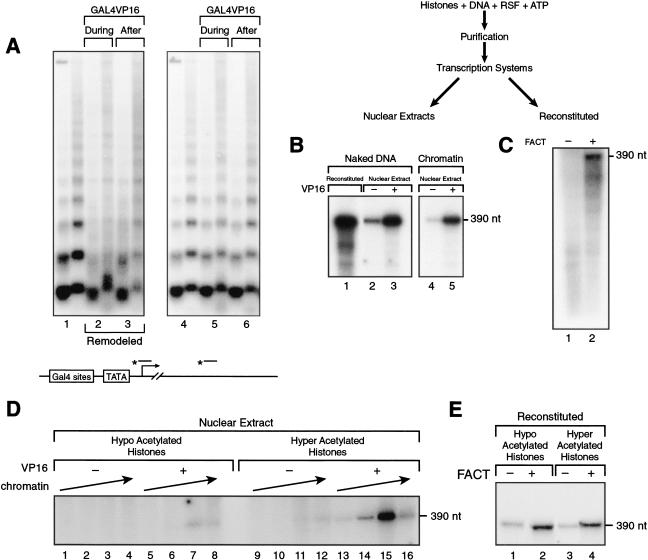
We next analyzed the RSF-assembled chromatin in transcription performed in vitro. In our initial studies, we used RSF-assembled chromatin with histones derived from HeLa cells. We began the analysis by testing whether the reconstituted assembled chromatin could be remodeled. The DNA used contains a 390-nt G-less cassette with five Gal4 DNA-binding sites upstream of the adenovirus major late promoter. The activator Gal4VP16 was added during or after chromatin assembly. As a result, chromatin was remodeled (Fig. 5A), as shown by the loss of the micrococcal nuclease digestion ladder and the appearance of smeared DNA. Remodeling was dependent on the presence of the activator (Fig. 5A, cf. lane 1 with 2 and 3) and occurred only within the vicinity of the activator binding sites (Fig. 5A, cf. left and right panels), as shown by the presence of micrococcal nuclease DNA ladder distal to the activator binding sites.
Figure 5.
Transcription on RSF-assembled chromatin. (A) Southern blot of micrococcal nuclease digestions of RSF-assembled and remodeled chromatin. Chromatin was assembled with RSF in the presence and absence of the activator Gal4VP16, added during the assembly or after the assembly as indicated. The Southern blot was probed with a promoter probe (left), followed by stripping off the probe and reprobing with a distal probe (right). The illustration (bottom) shows schematically the location of the probes (for details, see Orphanides et al. 1998). Each lane number, at the bottom of the panel, denotes two different concentrations of micrococcal nuclease used in the assays. (B) Transcription on RSF-assembled chromatin. (Top) A scheme of the procedure used to deposit chromatin and its analysis on transcription. Transcription was performed with nuclear extract in the presence and absence of the activator Gal4VP16. As a control, transcription from naked DNA is shown (lanes 1–3). The same amount of DNA was used in all lanes. Products of the transcription reactions were separated by electrophoresis on a denaturing polyacrylamide gel. (Right) Full-length transcripts (390 nt). (C) Reconstituted transcription on RSF-assembled chromatin. Transcription was performed using a reconstituted system, in the presence and absence of FACT. All reactions contained the activator Gal4VP16. Short products are observed as a result of transcription without FACT. Products of the transcription reactions were separated by electrophoresis on a denaturing polyacrylamide gel. (Right) Full-length transcripts obtained in the presence of FACT (390 nt). (D) Transcription on nuclear extract using chromatin templates reconstituted with hypo- and hyperacetylated histones isolated from HeLa cells. Increasing amounts of hypo- and hyperacetylated chromatin (equal molar amounts) were added to nuclear extract in the presence and absence of the activator Gal4VP16. Products of the transcription reactions were separated by electrophoresis on a denaturing polyacrylamide gel. (Right) Full-length transcripts (390 nt). (E) Analysis of the chromatin reconstituted with hypo- and hyperacetylated histones using a reconstituted transcription system. Transcription reactions were performed in the presence of Gal4–VP16, but with and without FACT, as described in Materials and Methods. Products of the transcription reactions were separated by electrophoresis on a denaturing polyacrylamide gel. (Right) Full-length transcripts (390 nt).
Having established that the RSF-assembled chromatin can be remodeled on activator binding, we analyzed the in vitro reconstituted chromatin templates in RNA polymerase II–dependent transcription reactions using nuclear extracts or a fully reconstituted transcription system. As expected, transcription directed by HeLa nuclear extract and using the reconstituted chromatin templates was repressed relatively to transcription directed from naked DNA (Fig. 5B, cf. lanes 2 and 4). However, in the presence of the activator Gal4VP16, transcription from the chromatin and naked DNA templates was stimulated (Fig. 5B, cf. lanes 3 and 5). We also tested RSF-assembled chromatin templates using a reconstituted transcription system. We reported previously that transcription from chromatin templates requires, in addition to an activator and the general transcription factors, two chromatin-related factors: RSF, which allows the formation of transcription initiation competent complexes, and FACT (facilitates chromatin transcription), which allows RNA polymerase II to traverse through nucleosomes (LeRoy et al. 1998). Consistent with our previous findings, the production of full-length transcripts on RSF-assembled chromatin templates was dependent on FACT; in its absence, short RNA molecules were detected (Fig. 5C). Transcription from the reconstituted system was also dependent on activator, as in its absence no remodeling was observed (Fig. 5A; for details, see LeRoy et al. 1998; data not shown). Therefore, these studies allow us to conclude that transcription from the reconstituted chromatin templates, assembled by RSF and human histones, behaved as previously established using chromatin assembled in extracts.
We expanded the studies and analyzed transcription from chromatin templates reconstituted with hypo- and hyperacetylated human histones using both nuclear extracts and the reconstituted transcription system. Using nuclear extracts, we found that the chromatin reconstituted with hyperacetylated histones was transcriptionally active, whereas the chromatin reconstituted with hypoacetylated histone was transcriptionally impaired (Fig. 5D). This effect was not caused by the use of different amounts of each chromatin, as approximately equimolar amounts of each template were used (Fig. 5D). Moreover, transcription from the hypo- and hyperacetylated chromatin responded similarly to the concentration of chromatin added to the reaction (Fig. 5D, cf. lanes 6–8 with 14–16); however, the template utilization was drastically different (Fig. 5D). Our findings are in agreement with previous studies showing that hypoacetylated chromatin correlates with the formation of higher-order chromatin structure and with transcriptional repression, whereas hyperacetylated chromatin correlates with active transcription (Garcia-Ramirez et al. 1995). Surprisingly, the reconstituted transcription system could not recapitulate the differences observed between hypo- and hyperacetylated chromatin in extracts (Fig. 5E; see below). Therefore, our studies also suggest that hypoacetylation is not sufficient for repression, but factors present in nuclear extract and missing in the reconstituted system use the hypoacetylated chromatin to establish repression (see below). Similarly, our studies suggest that acetylation per se is not sufficient to stimulate transcription, but factors present in nuclear extract and missing in the reconstituted system use the hyperacetylated chromatin to stimulate transcription.
Next we characterized the chromatin reconstituted with bacterially produced histones in transcription. We analyzed the recombinant chromatin for its response to two different histone-tail modifications, acetylation and methylation.
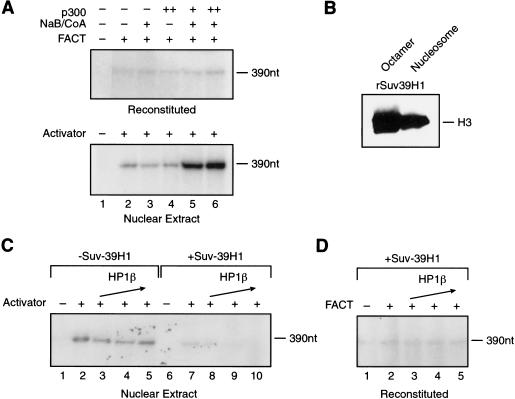
In agreement with the results presented above with hypoacetylated chromatin, the chromatin reconstituted with bacterially produced histones, which is devoid of any posttranslational modifications, was impaired. On remodeling and activator binding, transcription was observed, and the levels of transcription were similar to those observed with hypoacetylated chromatin (Fig. 6A). These low levels of transcription were observed in nuclear extract and in the reconstituted transcription system. We investigated whether acetylation of the chromatin might stimulate transcription. This was found to be the case in nuclear extracts, as the addition of p300 resulted in approximately fivefold stimulation of transcription, which was dependent on acetyl-CoA (Fig. 6A). Interestingly, similar treatment of the chromatin followed by transcription using the reconstituted system failed to stimulate transcription. The inability of the reconstituted system to respond to acetylation with recombinant chromatin appears consistent with the results presented above (Fig. 5E; see Discussion) and was owing to the assay conditions (see Materials and Methods). The reconstituted transcription system was healthy (Fig. 5B, lane 1) and capable of directing transcription from chromatin reconstituted with human histones (Fig. 5C; see Discussion). These results establish that the chromatin reconstituted with bacterially produced histone polypeptides is active but requires p300-mediated acetylation for optimal transcription activity. We next analyzed the response of the recombinant chromatin to methylation.
Figure 6.
Transcription using chromatin reconstituted with RSF and recombinant histones. (A) p300 stimulates transcription in nuclear extract but not in the reconstituted system. Chromatin templates were incubated (top, bottom) with recombinant p300 and 1 μM acetylCoA/2 mM Na-butyrate for 30 min before performing transcription. Assays performed in the reconstituted system (top) received Gal4VP16 and were performed in the presence or absence of FACT, as indicated. Transcriptions performed in extracts (bottom) received Gal4VP16 as indicated. Products of the transcription reactions were separated by electrophoresis on a denaturing polyacrylamide gel. (Right) Full-length transcripts (390 nt). (B) SUV39H1 methylates octamer and nucleosome. Baculovirus-expressed Suv39H1 was assayed for methylation on recombinant core histone octamers and recombinant nucleosomes. The substrates were incubated with 3H-SAM in the presence of Suv39H1 for 30 min at 30°C. Products were separated by SDS-PAGE, and labeled polypeptides were detected. (Right) Methylated histone H3. (C) Suv39H1 methylation inhibits transcription in an HP1β-dependent manner. Chromatin was incubated with recombinant Suv39H1 and 2.5 μM SAM for 30 min followed by 30-min incubation with HP1β. The reaction was further incubated under transcription conditions using nuclear extracts in the presence or absence of Gal4VP16, as indicated. Products of the transcription reactions were separated by electrophoresis on a denaturing polyacrylamide gel. (Right) Full-length transcripts (390 nt). (D) Suv39H1/HP1β-mediated repression is not observed in the reconstituted transcription system. Chromatin was incubated with recombinant Suv39H1 and 2.5 μM SAM for 30 min followed by 30-min incubation with HP1β followed by transcription in a reconstituted system. Reactions were performed in the presence of Gal4VP16 in the presence or absence of FACT, as indicated. Products of the transcription reactions were separated by electrophoresis on a denaturing polyacrylamide gel. (Right) Full-length transcripts (390 nt).
Methylation of histone tails plays an important role in transcription regulation. The identification of several proteins with histone methyl transferase (HMT) activity, together with the characterization of some of their biological functions has helped to elucidate the function of specific modifications occurring at the histone tails.
One of the HMT proteins is Suv39H1, which specifically methylates Lys 9 of histone H3 (Rea et al. 2000). This modification acts as a mark for the recruitment and binding of HP1 (Bannister et al. 2001; Lachner et al. 2001; Nakayama et al. 2001), which has been implicated in gene silencing and formation of higher-order, repressive chromatin structure (for review, see Eissenberg and Elgin 2000). Because Lys 9 of histone H3 can also be acetylated, and these modifications (acetylation and methylation) cannot coexist within the same residue, methylation of histone H3–Lys 9 requires the action of histone deacetylases (Nakayama et al. 2001). In our studies, described below, we bypassed the requirement for histone deacetylase by using recombinant chromatin, which is devoid of any histone-tail modifications. To directly analyze the effect of methylation by Suv39H1 on transcription, the recombinant chromatin was treated with the methylase and then added to transcription systems in the presence and absence of HP1. Baculovirus-expressed Suv39H1 was able to methylate octamers as well as nucleosomes (Fig. 6B). On treatment of the assembled chromatin with Suv39H1, the levels of transcription were decreased (Fig. 6C, cf. lanes 2 and 7), probably a reflection of HP1 present in the extract. However, the addition of HP1 further increases repression, in a concentration-dependent manner (Fig. 6C, lanes 8–10). Repression by HP1 was dependent on Suv39H1-mediated methylation; in the absence of SAM (data not shown) or in the absence of Suv39H1 (Fig. 6C, cf. lanes 3–5 with 8–10), HP1 was unable to repress transcription. HP1-mediated transcription repression was not only dependent on methylation by Suv39H1, but it also required chromatin; HP1 was without effect on transcription directed by naked DNA (data not shown). Importantly, chromatin, Suv39H1-mediated methylation, and HP1 are not sufficient to establish repression, as repression could not be recapitulated in a reconstituted chromatin transcription system (Fig. 6D). It is likely that other factors, present in extracts, are necessary to establish fully repressed transcription conditions.
The above observations suggest that acetylation and methylation apparently mark the chromatin for other factors that bind the acetylated or methylated mark to stimulate or repress transcription, respectively. These results are in agreement with the histone code hypothesis that predicts that the pattern of histone-tail modifications serves as a recognition code for the binding of proteins that modulate chromatin structure and function (for reviews, see Strahl and Allis 2000; Turner 2000).
Discussion
In these studies, we have established conditions to reconstitute regularly spaced nucleosome arrays using bacterially expressed histones. This system has allowed us to begin to analyze the role of the histone tails and their modifications in establishing regularly spaced nucleosome arrays and in transcription.
Reconstitution of recombinant chromatin was achieved with bacterially produced histones in a reaction catalyzed by RSF. Our studies established that RSF deposits nucleosomes in the absence of chaperones. Although we did not use recombinant RSF, and therefore cannot absolutely rule out the presence of catalytic amounts of contaminating histone chaperones in the RSF preparation, this seems unlikely. The RSF preparation used was highly purified and obtained after gel filtration chromatography in the presence of high salt. Also, antibodies against CAF-1 (p48) did not detect these polypeptides in fractions containing RSF. Moreover, the reaction catalyzed by RSF appears mechanistically different from the reaction catalyzed by ACF, which requires the histone chaperone NAP-1 (Ito et al. 1997b). RSF appears to function stoichiometrically with DNA, whereas ACF functions catalytically. The assembly of regularly spaced nucleosomes, catalyzed by RSF, appears to proceed by at least two steps. The first step is nucleosome deposition, and it is likely mediated by the large subunit of RSF. This subunit is encoded by a novel gene, for which we have recently obtained a cDNA (A. Loyola, G. LeRoy, and D. Reinberg, unpubl.). The amino acid sequence of p325 shows the presence of a large charged region, which we suggest might participate directly in the nucleosome assembly reaction. The small subunit of RSF, hSNF2h, does not interact directly with the histone octamer (Clapier et al. 2001); however, RSF, like histone chaperones, was found to interact with the H3/H4 tetramer and the octamer. Moreover, the binding of RSF to DNA is dependent on the histone octamer. The interaction of RSF with the octamer is independent of the histone tails and does not require posttranslational modifications, as RSF interacts with the histone octamer generated with bacterially produced histones and with recombinant octamers deleted of the histone tails. Although, the nucleosome deposition step catalyzed by RSF is independent of the histone tails, the second step of the reaction, the ATP-dependent nucleosome spacing, was found to be dependent on histone tails. Moreover, the efficiency of array formation was found to be stimulated by p300-mediated acetylation. In agreement with previous studies showing that the Drosophila ISWI polypeptide requires the H4 tails for ATP-dependent nucleosome mobilization (Clapier et al. 2001), we found that the histone H4 tail is necessary for array formation. However, the histone H4 tail was not sufficient for the formation of regularly spaced nucleosome arrays, as the tails of the other histones influenced array formation. p300-mediated stimulation of array formation was found to require acetylation of the tails of the H2A/H2B dimer. Surprisingly, however, we also found that the histone H3 tail negatively affects the p300-mediated stimulation of array formation. We speculate that the negative effect imposed by the histone H3 tail might be overcome by other modifications such as phosphorylation of histone H3 at Ser 10 or an alternative HAT mediates acetylation of this tail. This remains to be elucidated.
The establishment of conditions that permit the reconstitution of recombinant chromatin allows for the analysis of the effect of the different histone tail modifications in transcription. Toward this goal, we analyzed the ability of the recombinant chromatin to be used as template for transcription and specifically analyzed the effect of two histone-tail modifications: p300-mediated acetylation and Suv39H1-mediated methylation.
Although these two modifications can have opposite effects on transcription, these modifications were not recognized in a highly reconstituted transcription system; their effect was observed only in crude extracts. The inability of the reconstituted system to recognize these modifications was not owing to a poor transcription system. The reconstituted transcription system is healthy: In comparison to transcription directed by extracts, the reconstituted system is capable of directing basal transcription to levels higher than those observed in extracts, even in the presence of the activator and using approximately equimolar amounts of general transcription factors and RNA polymerase II (Fig. 5B). The reconstituted system under activation conditions was also unable to recognize alterations caused by acetylation or methylation. There are different explanations to these findings. The most logical explanation is that acetylation and/or methylation per se does not affect template utilization but affects the ability of the chromatin templates to be recognized by the transcription machinery. It is likely that these modifications provide marks on the histone tails that are recognized by factors present in extracts but missing in the reconstituted system that affects transcription. This hypothesis is supported by our findings with methylation and transcription. We found that HP1-mediated repression of transcription requires Suv39H1-mediated methylation of histone H3. This finding is in perfect agreement with recent results obtained in vivo showing that the binding of HP1 to chromatin requires methylation of histone H3–Lys 9 (Bannister at al. 2001; Lachner et al. 2001; Nakayama et al. 2001). Surprisingly, however, we found that chromatin, H3–Lys 9 methylation, and HP1 are not sufficient to establish repression, as this could not be reproduced in a reconstituted transcription system. It is likely that other factors are required to establish repression. The studies in yeast have shown that histone deacetylation is required to establish the appropriate substrate for methylation by Suv39H1 (Nakayama et al. 2001). Although the use of chromatin without pre-existing modification bypasses the requirement for the histone deacetylase enzymatic activity, it is possible that the histone deacetylases that targets histone H3–lysines 9 and 14 not only functions to generate the appropriate substrate but also might be active components of the Suv39H1-repressive complex.
With regards to acetylation, we observed that chromatin reconstituted with hypoacetylated human histone polypeptides was not optimal for transcription in crude extracts; however, the reconstituted system was indifferent to acetylation of the histone polypeptides. This finding is in agreement with the histone-code hypothesis (Strahl and Allis 2000; Turner 2000) and strongly suggests that factors in the extract, but lacking in the reconstituted system, might recognize the acetylated mark(s) to stimulate transcription. Using recombinant chromatin, we observed that acetylation of histone tails, specifically by p300, stimulates transcription in extracts. In agreement with the results obtained using chromatin reconstituted with hypo/hyperacetylated human histones, no effect was observed in a reconstituted transcription system. Although a possible explanation to this observation is the absence of a factor in the reconstituted system, the inability of the reconstituted transcription system to respond to acetylation of the recombinant chromatin might also be the result of the inability of p300 to acetylate specific residues on the histone tails. The recombinant chromatin is devoid of histone-tail modifications, and it is likely that p300-mediated acetylation of a specific residue might require other histone modifications. This possibility is supported by studies showing that phosphorylation of histone H3–Ser 10 modulates acetylation of histone H3–Lys 14 (Cheung et al. 2000a; Lo et al. 2000, 2001). The presence of a specific kinase in the extract might phosphorylate histone H3–Ser 10, resulting in efficient acetylation. Elucidation of the factors necessary for p300-mediated acetylation to result in optimal transcription and of the factor(s) required for Suv39–H1-mediated methylation to result in repression of transcription, and their exact mechanism of action, require further studies. The development of the system described in the present study, capable of generating recombinant chromatin will permit the setting of biochemical complementation assays to isolate the different factors involved in these processes as well as the elucidation of their mechanism of action.
Materials and methods
Purification of proteins
HeLa core histones were purified as described (Orphanides et al. 1998). RSF was purified from the HeLa nuclear pellet fraction as described (LeRoy et al. 1998). ACF was purified from HeLa nuclear extract as described (LeRoy et al. 2000). Bacterially expressed full-length and tailless Xenopus laevis histones were expressed and purified as described (Luger et al. 1997b). Recombinant histidine-tagged Drosophila NAP-1 was expressed from baculovirus in SF9 cells. The protein was purified by standard nickel affinity purification, followed by a purification on a HiTrap Q Sepharose HP eluted with a linear gradient from 0.2–0.5 M NaCl. NAP-1 eluted at 0.3 M NaCl. Recombinant Flag-tagged human HP1β, p300, and Suv39H1 were expressed from baculovirus in SF9 cells. The proteins were purified by standard M2 affinity purification.
Antibodies
Monoclonal antibodies against the RSF subunits p325 and hSNF2h were developed by Bios-Chile. Polyclonal antibodies against histones H4 and monoclonal antibodies against histone H3 were purchased from Upstate Biotechnology. Gold conjugated antibodies were purchased from Sigma.
Chromatin assembly reaction
Chromatin assembly reactions were performed as follows: 2 μg of a 3-kb supercoiled DNA (pG5MLP) was incubated with 1.8 μg of histones (native histones isolated from HeLa cells or bacterially produced wild-type or tailless histones), 0.3 μg of RSF, 150 μg of bovine serum albumin, 3 mM ATP, 30 mM phosphocreatine, 0.2 μg of phosphocreatine kinase, 5 mM MgCl2, 50 mM KCl, 10 mM HEPES (pH 7.6), 0.2 mM EDTA, and 5% glycerol. Reactions were incubated for 6 to 12 h at 30°C. When p300 was tested, histones were first incubated with p300 and 10 μM acetyl-CoA in the assembly buffer for 1 h at 30°C, followed by the chromatin assembly reaction.
Analysis of chromatin
Typically a standard chromatin assembly reaction was partially digested with two different concentrations of micrococcal nuclease. The DNA was purified and deproteinized, and the products were analyzed on an agarose gel. For supercoiling assay, the chromatin assembly reaction was performed as described above, using DNA relaxed with topoisomerase I (Promega). The reaction was performed in the presence of topoisomerase I for 5–6 h at 30°C, and the topoisomers were analyzed on an agarose gel.
Electron microscopy
Chromatin was assembled as described above. The products of the reaction were fixed with glutaraldehyde, and the DNA–protein complexes were purified by gel filtration (1 mL of A5M resin, Bio-Rad). The fractions containing chromatin were prepared for electron microscopy (Griffith and Christiansen 1978). For the characterization of the DNA loop-associated complexes, the reaction was incubated with antibodies against histones H3 (monoclonal) and H4 (polyclonal) and against the RSF subunit hSNF2h (monoclonal). After the first antibody incubation, the reaction was fixed with glutaraldehyde and purified by gel filtration. Then the reaction was incubated with rabbit antibodies conjugated with 5-nm gold particle and against mouse antibodies conjugated with 10-nm gold particles, then fixed with glutaraldehyde, purified, and prepared for electron microscopy (Griffith and Christiansen 1978). The protein–DNA complexes were mixed in a buffer containing 2 mM spermidine, adsorbed to glow-charged carbon-coated grids, washed with a water/graded ethanol series, and rotary shadow cast with tungsten. Samples were examined using a Philips 420 electron microscope. Micrographs are shown in reverse contrast. A Cohu CCD camera attached to a Macintosh computer programmed with National Institutes of Health (NIH) IMAGE software was used to form the images.
Immunoprecipitations
HeLa core histones (1.8 μg), RSF (0.3 μg) and BSA (150 μg) were mixed in the chromatin assembly reaction buffer (without ATP) for 20 min at 30°C. Antibody against hSNF2h was added and incubated with rotation for 1 h at 4°C in the presence of 0.01% NP-40 followed by the addition and incubation with Protein G-agarose for 1 h at 4°C. The beads were washed three times with 10× beads volume of BC500 plus 0.01% NP-40 and eluted with SDS-PAGE loading sample. The proteins were analyzed by 15% SDS-PAGE followed by silver staining.
Immunoprecipitation of the RSF–DNA complex was performed as follows: Linear pG5MLP template was incubated with RSF and histones in the chromatin assembly buffer for 30 min at 30°C. Immunoprecipitation was performed as described above, and the DNA products were analyzed on an agarose gel.
Chromatin transcription
Chromatin was purified by CL-4B gel filtration chromatography as previously described (Orphanides et al. 1998). Transcription was performed as previously described (Orphanides et al. 1998) using ∼40 ng of chromatin and 80 μg of HeLa nuclear extract in the presence and absence of the activator Gal4VP16. Reconstituted transcription was performed in the presence of the activator Gal4VP16. In its absence, no promoter remodeling occurred, and therefore it was required, regardless of whether the transcription was reconstituted with TBP, under basal transcription conditions, or with TFIID and the coactivators PC4 and Mediator. Basal transcription was reconstituted using recombinant TBP, TFIIB, TFIIE, TFIIF expressed in Escherichia coli, and highly purified RNA polymerase II and TFIIH, purified from HeLa cells as described (Maldonado et al. 1996). Transcription on chromatin templates was measured as a function of FACT. Transcription reconstituted under activation conditions contained TFIID and the coactivators PC4 and Mediator. Activator was added to all reactions, and transcription was analyzed as a function of FACT. No differences were observed using the different reconstituted transcription systems, the state of acetylation of the histone polypeptides was not apparent in reconstituted systems. RNA products were deproteinized, precipitated with ethanol, and analyzed on 6% acrylamide denaturing gels. For reactions performed with HATs, 40 ng of CL-4B purified chromatin was incubated for 30 min at 30°C with Gal4–VP16, 1 μM of acetyl CoA, 2 mM Na-butyrate, and p300 in transcription buffer. The templates were then used in transcription as indicated above. For reactions performed with Suv39H1, 40 ng of purified chromatin was incubated with 2.5 μM of S-adenosyl-L-methionine (SAM) and Suv39H1 in transcription buffer for 30 min at 30°C, followed by 30 min incubation with recombinant HP1β at 30°C. Transcription assays were performed as described above.
Acknowledgments
We thank Drs. Michael Hampsey, Brian Lewis, and Lynne Vales for helpful comments on the manuscript. We also thank Drs. Jim Kadonaga and Mark Levenstein for the gift of baculovirus-expressing Drosophila NAP-1 and purified NAP-1 protein; Dr. Karolyn Luger for the E. coli expression plasmid encoding Xenopus laevis full-length and tailless histones; Drs. Pat Nakatani, Hidesato Ogawa, and Hideaki Tagami for baculovirus and E. coli expression vector encoding human His-tagged Suv39H1 and for purified Suv39H1 complex isolated from SF9-infected cells; and Drs. Jacob Seeler, Alejandro Vaquero, and Howard J. Worman for cDNA of HP1β (Ye and Worman 1996). We also thanks members of the laboratory: Dr. Erika Friedl for recombinant p300, Dr. Kenishi Nishioka for Figure 6B, and Sergei Chuikov for the expression of recombinant histones. We also thank Dr. George Orphanides and members of the Reinberg laboratory for helpful suggestions. This work was supported by grants from the NIH (GM37120) and the Howard Hughes Medical Institute to D.R. and by a grant from the NIH (CA85826) to Y.W.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL reinbedf@umdnj.edu; FAX (732) 235-5294.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.937401.
References
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- Angelov D, Vitolo JM, Mutskov V, Dimitrov S, Hayes JJ. Preferential interaction of the core histone tail domain with linker DNA. Proc Natl Acad Sci. 2001;98:6599–6604. doi: 10.1073/pnas.121171498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Berger SL. Molecular biology: The histone modification circus. Science. 2001;292:64–65. [PubMed] [Google Scholar]
- Bochar DA, Savard J, Wang W, Lafleur DW, Moore P, Cote J, Shiekhattar R. A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc Natl Acad Sci. 2000;97:1038–1043. doi: 10.1073/pnas.97.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CE, Lechner T, Howe L, Workman JL. The many HATs of transcription coactivators. TIBS. 2000;25:15–18. doi: 10.1016/s0968-0004(99)01516-9. [DOI] [PubMed] [Google Scholar]
- Chang CH, Luse DS. The H3/H4 tetramer blocks transcript elongation by RNA polymerase II in vitro. J Biol Chem. 1997;272:23427–23434. doi: 10.1074/jbc.272.37.23427. [DOI] [PubMed] [Google Scholar]
- Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000a;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000b;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Längst G, Corona DFV, Becker PB, Nightingale KP. A critical role for the histone H4 N-terminus in nucleosome remodeling by ISWI. Mol Cell Biol. 2001;21:875–883. doi: 10.1128/MCB.21.3.875-883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Elgin SC. The HP1 protein family: Getting a grip on chromatin. Curr Opin Genet Dev. 2000;10:204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- Garcia-Ramirez M, Rocchini C, Ausio J. Modulation of chromatin folding by histone acetylation. J Biol Chem. 1995;270:17923–17928. doi: 10.1074/jbc.270.30.17923. [DOI] [PubMed] [Google Scholar]
- Georgel PT, Tsukiyama T, Wu C. Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J. 1997;16:4717–4726. doi: 10.1093/emboj/16.15.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greyling HJ, Schwager S, Sewell BT, von Holt C. The identity of conformational states of reconstituted and native histone octamers. Eur J Biochem. 1983;137:221–226. doi: 10.1111/j.1432-1033.1983.tb07818.x. [DOI] [PubMed] [Google Scholar]
- Griffith JD, Christiansen G. Electron microscopic visualization of chromatin and other DNA protein complexes. Annu Rev Biophys Bioeng. 1978;7:19–35. doi: 10.1146/annurev.bb.07.060178.000315. [DOI] [PubMed] [Google Scholar]
- Guschin D, Geiman TM, Kikyo N, Tremethick DJ, Wolffe AP, Wade PA. Multiple ISWI ATPase complexes from Xenopus laevis: Functional conservation of an ACF/CHRAC homolog. J Biol Chem. 2000;275:35248–35255. doi: 10.1074/jbc.M006041200. [DOI] [PubMed] [Google Scholar]
- Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- Hayes JJ, Hansen JC. Nucleosomes and the chromatin fiber. Curr Opin Genet Dev. 2001;11:124–129. doi: 10.1016/s0959-437x(00)00168-4. [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997a;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Ito T, Tyler JK, Kadonaga JT. Chromatin assembly factors: A dual function in nucleosome formation and mobilization? Genes Cells. 1997b;2:593–600. doi: 10.1046/j.1365-2443.1997.1500348.x. [DOI] [PubMed] [Google Scholar]
- Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes & Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis DC. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes & Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- Krebs JE, Kuo MH, Allis CD, Peterson CL. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes & Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Ladoux B, Quivy JP, Doyle P, du Roure O, Almouzni G, Viovy JL. Fast kinetics of chromatin assembly revealed by single-molecule videomicroscopy and scanning force microscopy. Proc Natl Acad Sci. 2000;97:14251–14256. doi: 10.1073/pnas.250471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langst G, Bonte EJ, Corona DF, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- LeRoy G, Orphanides G, Lane WS, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- LeRoy G, Loyola A, Lane WS, Reinberg D. Purification and characterization of a human factor that assembles and remodels chromatin. J Biol Chem. 2000;275:14787–14790. doi: 10.1074/jbc.C000093200. [DOI] [PubMed] [Google Scholar]
- Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- Lo WS, Duggan L, Lane W, Shiekhattar R, Berger SL. Snf1 is a histone kinase which works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science. 2001;293:1142–1146. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- Lorch Y, Zhang M, Kornberg R. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96:389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997a;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997b;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- Maldonado E, Drapkin R, Reinberg D. Purification of human RNA polymerase II and general transcription factors. Methods Enzymol. 1996;274:72–100. doi: 10.1016/s0076-6879(96)74009-0. [DOI] [PubMed] [Google Scholar]
- Mello JA, Almouzni G. The ins and outs of nucleosome assembly. Curr Opin Genet Dev. 2001;11:136–141. doi: 10.1016/s0959-437x(00)00170-2. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Bulger M, Muramatsu M, Ito T. Multistep chromatin assembly on supercoiled plasmid DNA by nucleosome assembly protein-1 and ATP-utilizing chromatin assembly and remodeling factor. J Biol Chem. 2001;276:27384–27391. doi: 10.1074/jbc.M101331200. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D. RNA polymerase II elongation through chromatin. Nature. 2000;407:471–475. doi: 10.1038/35035000. [DOI] [PubMed] [Google Scholar]
- Orphanides G, LeRoy G, Chang C, Luse D, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- Phelan ML, Schnitzler GR, Kingston RE. Octamer transfer and creation of stably remodeled nucleosomes by human SWI–SNF and its isolated ATPases. Mol Cell Biol. 2000;20:6380–6389. doi: 10.1128/mcb.20.17.6380-6389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot RA, Dellaire G, Hülsmann BB, Grimaldi MA, Corona DFV, Becker PB, Bickmore WA, Varga-Weisz PD. HuCHRAC, a human ISWI chromatin remodeling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 2000;19:3377–3387. doi: 10.1093/emboj/19.13.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Rice JC, Allis CD. Histone methylation versus histone acetylation: New insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–173. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis DC. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Thomas JO, Kornberg RD. An octamer of histones in chromatin and free in solution. Proc Nat Acad Sci. 1975;72:2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM. Histone acetylation and an epigenetic code. BioEssays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- Verreault A. De novo nucleosome assembly: New pieces in an old puzzle. Genes & Dev. 2000;14:1430–1438. [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, et al. Methylation of histone H4 at arginine 3 facilitates transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Hayes JJ. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP, Guschin D. Review: Chromatin structural features and targets that regulate transcription. J Struct Biol. 2000;129:102–122. doi: 10.1006/jsbi.2000.4217. [DOI] [PubMed] [Google Scholar]
- Ye Q, Worman HJ. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem. 1996;271:14653–14656. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes & Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]