Abstract
In Drosophila melanogaster, formation of the axes and the primordial germ cells is regulated by interactions between the germ line-derived oocyte and the surrounding somatic follicle cells. This reciprocal signaling results in the asymmetric localization of mRNAs and proteins critical for these oogenic processes. Mago Nashi protein interprets the posterior follicle cell-to-oocyte signal to establish the major axes and to determine the fate of the primordial germ cells. Using the yeast two-hybrid system we have identified an RNA-binding protein, Tsunagi, that interacts with Mago Nashi protein. The proteins coimmunoprecipitate and colocalize, indicating that they form a complex in vivo. Immunolocalization reveals that Tsunagi protein is localized within the posterior oocyte cytoplasm during stages 1–5 and 8–9, and that this localization is dependent on wild-type mago nashi function. When tsunagi function is removed from the germ line, egg chambers develop in which the oocyte nucleus fails to migrate, oskar mRNA is not localized within the posterior pole, and dorsal–ventral pattern abnormalities are observed. These results show that a Mago Nashi–Tsunagi protein complex is required for interpreting the posterior follicle cell-to-oocyte signal to define the major body axes and to localize components necessary for determination of the primordial germ cells.
Keywords: RNA localization, oogenesis, germ line
Cellular diversification during development often requires the subcellular localization of molecular cues that serve to instruct genetically identical cells to differentiate into distinct cell types. In the fruit fly Drosophila melanogaster, asymmetries generated during oogenesis provide information needed for the establishment of the anterior–posterior (AP) and dorsal–ventral (DV) axes of both the oocyte and embryo. Oogenesis in Drosophila is initiated when a germ-line stem cell divides asymmetrically to produce a daughter stem cell and a cystoblast. The cystoblast undergoes four rounds of mitotic division without complete cytokinesis to form a cyst of 16 germ line-derived cells interconnected by ring canals (Spradling 1993). One of the two cells possessing four ring canals develops into the oocyte. Somatically derived follicle cells surround the 16-cell cyst to form an egg chamber, consisting of an oocyte, 15 nurse cells, and associated follicle cells. Multiple, discrete steps during oogenesis are necessary to accurately establish the axes within the oocyte (van Eeden and St. Johnston 1999). Proper axis formation of the egg chamber results in the localization of protein and mRNA determinants within the oocyte that are used to generate the axes of the embryo (Bashirullah et al. 1998).
The mRNA for gurken (grk) is translated to produce a transforming growth factor-α-like (TGFα) protein that initiates two reciprocal signaling interactions between the oocyte and the surrounding follicle cells at distinct times during egg chamber development to define the AP and DV axes of the oocyte. During early oogenesis (prior to stage 1 and until stage 7), grk mRNA accumulates within the oocyte (Neuman-Silberberg and Schüpbach 1993). Before stage 7 of oogenesis, the first reciprocal signaling event is induced by Gurken protein present in the oocyte and activates the epidermal growth factor receptor (EGFr) in a subset of follicle cells that contact the oocyte to specify their fate as posterior follicle cells (González-Reyes et al. 1995; Roth et al. 1995; González-Reyes and St. Johnston 1998). Oocyte-to-follicle cell signaling by Gurken is followed by an unidentified return signal from the posterior follicle cells to the oocyte that results in microtubule reorganization within the oocyte during stages 6–7. As a consequence of the microtubule rearrangement, the oocyte nucleus migrates from a posterior to an anterior-cortical position. During migration, grk mRNA is associated with the oocyte nucleus, and this initially restricts Gurken protein to the anterior-cortical region (Neuman-Silberberg and Schüpbach 1993, 1996). Gurken then induces a second signaling event during stages 8–10 that activates the EGFr in nearby follicle cells to define the DV axis (Neuman-Silberberg and Schüpbach 1993).
The majority of proteins and mRNAs that are necessary within the oocyte to generate the AP and DV axes are synthesized by the nurse cells. These proteins and RNAs are transported through the ring canals into the oocyte and are likely to move in large transport complexes (Wilhelm et al. 2000). Inhibitor studies indicate that this intercellular transport requires an intact microtubule network (Pokrywka and Stephenson 1991, 1995; Theurkauf et al. 1993). A microtubule organizing center (MTOC) observed within the oocyte is directly connected to each of the nurse cells through the ring canals by a continuous microtubule network (Theurkauf et al. 1993). The follicle cell-to-oocyte signaling induces microtubule reorganization that is necessary for the localization of determinants for both AP and DV axis formation.
Subsequent to the microtubule reorganization, specific mRNAs that code for embryonic protein determinants become concentrated within the anterior or posterior end of the developing oocyte. The bicoid (bcd) mRNA, which codes for the anterior morphogen of the embryo, is localized to the anterior end of the oocyte (St Johnston et al. 1989). The osk mRNA is initially concentrated at the anterior end of the oocyte (stages 8–9) and then localizes within the posterior pole during stages 9–10 (Ephrussi et al. 1991; Kim-Ha et al. 1991). Proper localization of osk mRNA is necessary to establish the posterior pole plasm, which contains determinants that specify the primordial germ cells and abdomen of the developing embryo.
Mago Nashi (Mago) is the only protein known to function within the oocyte to interpret the follicle cell-to-oocyte signal that defines the AP and DV axes of the oocyte (Micklem et al. 1997; Newmark et al. 1997). Mutations in mago nashi (mago) result in a failure of the microtubule rearrangement as well as the subsequent anterior-directed movement of the oocyte nucleus (Micklem et al. 1997; Newmark et al. 1997), and consequently mago mutant oocytes exhibit alterations in both the AP and DV axes.
Mutational analysis has shown that the wild-type function of mago is also essential for the localization of osk mRNA within the posterior pole (Ephrussi and Lehmann 1992; Newmark and Boswell 1994). Molecular analysis reveals that Stau protein contains ds-RNA binding domains that are required for osk mRNA localization (Gatignol et al. 1993; Ramos et al. 2000). Experimental results suggest that Staufen is involved in transporting osk mRNA in a protein/mRNA complex (Ephrussi et al. 1991; Kim-Ha et al. 1991; St Johnston et al. 1991). Studies of Mago indicate that it is required for anchoring the Staufen/osk mRNA complex within the posterior pole of the oocyte (Newmark and Boswell 1994). Therefore, mutations in mago result in a defective germ plasm (Boswell et al. 1991). The molecular basis of Mago function is enigmatic. Mago does not exhibit significant sequence similarity with proteins of known biochemical function, and molecules that bind to Mago have not been identified.
To gain a deeper understanding of the function of Mago during oogenesis we set out to identify proteins that bind directly to Mago. The Mago protein is highly conserved throughout evolution (Newmark and Boswell 1994; Micklem et al. 1997; Newmark et al. 1997). Because Mago does not exhibit any similarity to proteins of known function, the identification of Mago-binding proteins might help to elucidate its role during Drosophila development and indicate a function for Mago homologs in other organisms. Here we report the identification and characterization of Tsunagi, an RNA-binding protein that forms an in vivo complex with Mago. Our data support a role for Tsunagi both in the localization of germ-cell determinants and axis formation during oogenesis.
Results
Identification of a Mago-binding protein, Tsunagi, and molecular organization of the gene
To identify Mago-binding proteins, a full-length Mago fusion protein was used as bait, and an ovarian cDNA library of prey was screened in a yeast two-hybrid assay (Gyuris et al. 1993; Lie and Macdonald 1999). We recovered 17 positive clones from a screen of ∼sixfold representation of the library. Sequencing revealed that all 17 positive clones were derived from the same gene. The gene was named tsunagi (tsu), which is Japanese for “connection” or “link.” Tsunagi binds Mago specifically in the yeast two-hybrid assay: it does not activate transcription on its own, nor does it interact with several unrelated prey proteins that were tested (see Materials and Methods).
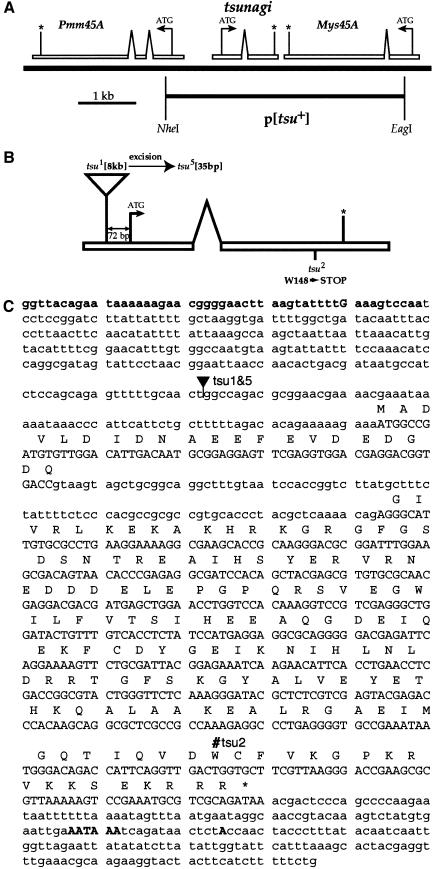
Hybridization of a probe from tsu cDNA to polytene chromosomes was used to ascertain the location of tsu in the genome. A single focus of hybridization on the right arm of chromosome 2, in polytene chromosome interval 45A4, was detected. Two genomic DNA contigs in the region, Dbp45A and hig (Berkeley Drosophila Genome Project; BDGP), were examined by PCR for the presence of tsu sequence. A 30–40-kb P1 phagemid clone, DS02099 (BDGP), that maps to the distal end of Dbp45A was found to contain tsu. The organization of tsu was determined by sequencing an 8295-bp NheI restriction endonuclease genomic fragment of DS02099 and comparing the sequence to that obtained from tsu cDNA (Fig. 1A).
Figure 1.
The genomic organization and sequence of tsu. (A) Genomic organization of tsu illustrating the relative position of the flanking genes Pmm45A and Mys45A, and the p[tsu+] transgene. For each gene the translation initiation (ATG), the translation termination (*), and the direction of transcription (arrows) are indicated. Proximal is to the left and distal to the right. (B) The relative positions of mutations within the tsu gene. The tsu gene with the point of insertion of the EP P element that produced tsu1, an allele (tsu5) induced by excision of the P element identified in tsu1, and the EMS-induced tsu2 allele. (C) Sequence of the Drosophila tsunagi gene. Lowercase bold letters in the 5′ end indicate the predicted promoter. An uppercase bold G designates the transcription start site. At the 3′ end, the asterisk defines the position of the translational stop site, and the bold uppercase letters show the position of the polyadenylation site. Intronic sequence is illustrated as lowercase letters nested between uppercase letters. Mutations tsu1 and tsu5 (filled triangle); tsu2 (#). (D) Amino acid sequence alignment of Drosophila Tsunagi with related proteins from yeast to humans. Amino acid residues are indicated as follows: identical amino acid residues are outlined in black, conserved amino acids are shaded, (∼) are amino acid residues that are absent, and dots represent gaps. The proteins are designated as follows: (RBM8) Homo sapiens; (MmTsu) Mus musculus; (DrTsu) Danio rerio; (Y14) Xenopus laevis; (DmTsu) Drosophila melanogaster; (CeTsu) Caenorhabditis elegans; (SpTsu) Schizosaccharomyces pombe.
A single tsu open reading frame (ORF) was identified and is present in its entirety in all 17 cDNA clones isolated by two-hybrid screening (Fig. 1B,C). RNA blot analysis of poly(A)+ RNA isolated from various stages throughout the life cycle, including embryogenesis, larval instars, pupariation, and adults identified a single ∼1.2-kb RNA when probed with a tsu cDNA (data not shown). The size of the RNA is in good agreement with the ∼1.1-kb size estimated from sequence data.
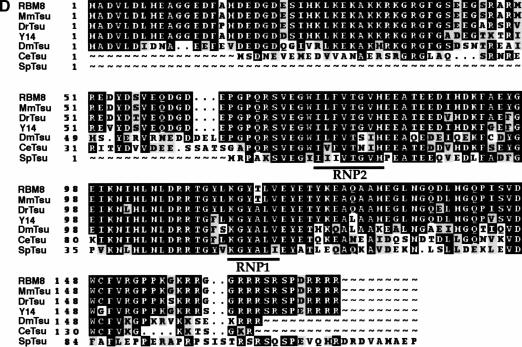
The predicted protein encoded by tsu is 165 amino acids in length with an estimated molecular weight of 19 kD (Fig. 1C). Immunoblot analysis of proteins extracted from adult females, adult males, all stages of embryogenesis, and larvae probed with an α-Tsunagi affinity-purified polyclonal antisera identified a single protein with an apparent molecular weight of 19 kD and revealed that Tsunagi protein is detected throughout the life cycle of the fly (Fig. 2A).
Figure 2.
Tsunagi and Mago form an in vivo complex. (A) Tsunagi protein is detected during all stages of Drosophila development. Affinity-purified anti-Tsunagi rabbit polyclonal antisera recognize a 19-kD protein that matches the size predicted from sequencing of the tsu gene. Protein extracts were resolved on a 10%–20% SDS-PAGE gradient gel. (B) Tsunagi coimmunoprecipitates with Myc-tagged Mago (Myc-Mago). Extracts from wild-type (OreR) or transgenic flies expressing Myc-Mago were immunoprecipitated with the anti-Myc Mab 9E10. Immunoprecipitated proteins were resolved on a 10%–20% SDS-PAGE gradient gel, and the presence of Myc-Mago (left blot) and Tsunagi (right blot) was determined by probing immunoblots with the indicated antisera.
Database searches reveal that Tsunagi is significantly similar to the ribonucleoprotein (RNP) family of RNA-binding proteins with a single RNP domain and has been evolutionarily conserved (Fig. 1D; Burd and Dreyfuss 1994). Homologs for Tsunagi exist from the fission yeast Schizosaccharomyces pombe to humans (Fig. 1D). As with Mago and its homologs, a Tsunagi-related protein is not detectable within the genome of the budding yeast Saccharomyces cerevisiae. The primary structure of Tsunagi is reminiscent of the SR family of splicing factors. Classic SR proteins contain at least one RNP domain followed by carboxy-terminal arginine/serine (RS) dipeptide residues (Fu 1995). Each of the presumed Tsunagi homologs exhibits a series of basic residues at their carboxyl terminus, and within this region the vertebrate and yeast Tsunagi contain at least one RS-dipeptide repeat. However, the D. melanogaster and Caenorhabditis elegans proteins do not contain any RS sequences among the basic residues at their carboxyl termini.
Drosophila Tsunagi is 60% identical and 72% similar to the human RNA-binding motif protein 8 (RBM8). Kataoka et al. (2000) used RBM8 cDNAs to recover clones encoding the Xenopus laevis protein Y14 (Kataoka et al. 2000). Results from several laboratories suggest that Y14 is complexed with mRNAs and proteins to form ribonucleoprotein particles (mRNPs) that are preferentially exported from the nucleus to the cytoplasm (Kataoka et al. 2000; Le Hir et al. 2000; Kim et al. 2001). Therefore, in addition to having an RNA-binding motif, there is experimental evidence suggesting that Tsunagi-related proteins bind RNA.
Tsunagi and Mago form a complex and colocalize within the oocyte posterior pole
Although the results from yeast two-hybrid analysis show that Tsunagi and Mago can interact physically in a heterologous system, they do not indicate that this is a bona fide in vivo interaction. To investigate whether the proteins form a complex in vivo, coimmunoprecipitation experiments were undertaken. Immunoprecipitating α-Mago antibodies are not available; therefore, we made use of transgenic flies that produce a Myc–Mago fusion protein (p[myc–mago+]) expressed under the control of the endogenous mago promoter (Newmark et al. 1997). Flies carrying the transgene and homozygous or hemizygous for lethal or female sterile alleles of mago are phenotypically wild type, indicating that Myc–Mago function is indistinguishable from endogenous Mago function (Newmark et al. 1997).
Soluble extracts generated from transgenic flies synthesizing Myc–Mago were immunoprecipitated with the monoclonal antibody (Mab) 9E10, which recognizes the Myc epitope (α-Myc; Evan et al. 1985). When α-Myc immunoprecipitated proteins from p[myc–mago+] transgenic flies are immunoblotted and reacted with affinity-purified α-Tsunagi antisera, a protein with an apparent molecular weight of ∼19 kD is observed (Fig. 2B). Myc–Mago is not detectable when α-Myc is employed to precipitate proteins from wild-type control flies lacking the p[myc–mago+] transgene (Fig. 2B). A green fluorescent protein–Mago (GFP–Mago) fusion also provides Mago+ function (Newmark et al. 1997). An α-GFP antibody was used to perform immunoprecipitations from protein extracts of p[GFP–mago+] transgenic flies. Tsunagi also coimmunoprecipitates with GFP–Mago (data not shown). Together, these experimental results indicate that a Mago–Tsunagi complex is formed in vivo.
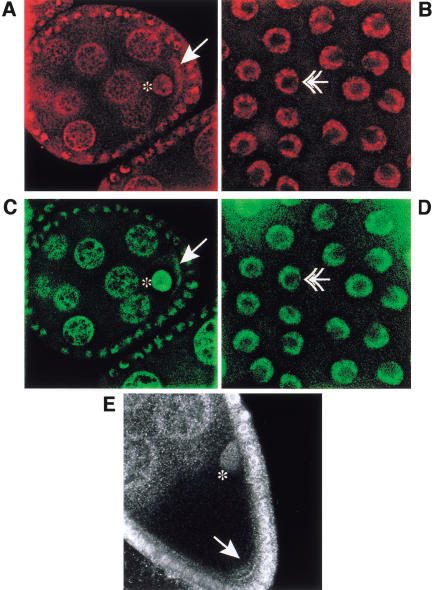
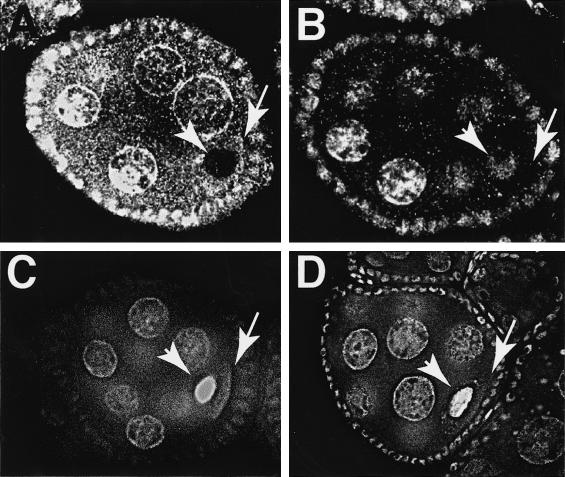
However, it does not indicate whether Tsunagi and Mago colocalize in all or a subset of the subcellular regions in which Mago can be detected in the ovary. The distribution of Tsunagi was examined using affinity-purified α-Tsunagi sera on whole-mount preparations of ovaries from wild-type females and females bearing the p[GFP–mago+] transgene. These studies reveal that during oogenesis Tsunagi and Mago are distributed nearly identically (Fig. 3; Newmark et al. 1997). The Tsunagi protein is detectable in nuclei of all cells, including germ-line cells (Fig. 3A) and somatic follicle cells (Fig. 3B). As illustrated in Figure 3, A and B, like Mago, Tsunagi is excluded from the prominent nucleoli of nurse cells and follicle cells (the absence of detectable Tsunagi and Mago in the nucleoli is most evident in Fig. 3B and 3D, respectively). Unlike Mago, Tsunagi is absent from the oocyte nucleus at stage 2, even though it is concentrated in the oocyte cytoplasm at this stage (Fig. 4A). During stages 1–5 and 8–9 of oogenesis, Tsunagi is enriched in the posterior pole cytoplasm (Fig. 3A,E). After stage 9 Tsunagi can no longer be detected within the posterior pole but is observed within the nuclei of germ-line cells and somatic follicle cells. Thus, Tsunagi and Mago colocalize in germ-line and follicle cell nuclei and within the posterior pole cytoplasm of the oocyte (Fig. 3, cf. A–D).
Figure 3.
Tsunagi and Mago colocalize during oogenesis. In all panels the oocyte nucleus is marked with an asterisk, protein localized in the posterior pole by an arrow, and follicle cell nuclei by double arrowheads. The anterior sides of the egg chambers are to the left and the posterior to the right in A–D. Tsunagi is in red and GFP–Mago in green. (A) The distribution of Tsunagi (red) in a stage-4 egg chamber. (B) Tsunagi in the follicle-cell nuclei of a stage-10 egg chamber. The spot within the follicle-cell nucleus without detectable Tsunagi is the nucleolus. (C–D) The distribution of GFP–Mago in the same egg chambers as in A and B. (E) Confocal image of a stage-9 egg chamber illustrating the distribution of Tsunagi and posterior at the bottom.
Figure 4.
Posterior pole localization of Tsunagi requires mago+ function. In all panels arrowheads designate the position of the oocyte nucleus and arrows the posterior pole. Egg chambers are oriented with the anterior to the left and posterior to the right. (A) Confocal image of a wild-type stage-2 egg chamber. At stage 2 Tsunagi is detected within the posterior pole but not within the oocyte nucleus. (B) A confocal image of a stage-2 egg chamber from a hemizygous mago1 female. Tsunagi is detected in the oocyte nucleus and is not observed above background in the cytoplasm. (C) Epifluorescence of GFP–Mago in a wild-type stage-4 egg chamber. (D) An epifluorescence image of GFP–Mago in a stage-4 egg chamber from a tsu7 homozygous female. The distribution of Mago is indistinguishable from its distribution in wild-type egg chambers.
Cytoplasmic localization of Tsunagi is dependent on posterior pole localization of Mago
The mago1 mutation (Gly19 → Arg) disrupts the posterior pole localization of Mago during oogenesis (Newmark et al. 1997). If cytoplasmic localization of Tsunagi were dependent on sequestration of Mago within the posterior pole of the oocyte, it would be expected that the distribution of Tsunagi within the oocyte cytoplasm would be altered in egg chambers from mago mutant females. The α-Tsunagi antiserum was used to detect Tsunagi in ovaries from mago1 mutant females. In egg chambers from mago1/Df(2R)F36 females, Tsunagi appears to be predominantly nuclear both in nurse cells and within the oocyte (Fig. 4, cf. A and B). Although Tsunagi is normally absent from the oocyte nucleus at stage 2 (as determined by its position within the ovariole, lack of a karyosome, and absence of the endobody, a nuclear structure first observed at stage 3), Tsunagi remains in the oocyte nucleus throughout oogenesis in egg chambers from mago1 hemizygous females (Fig. 4B). These data show that the immunofluorescence observed with α-Tsunagi in wild-type nurse cell and oocyte cytoplasm is not caused by nonspecific staining (see below for additional evidence). Furthermore, these data (1) indicate that accumulation of Tsunagi within the cytoplasm of both nurse cells and the oocyte is dependent on mago+ function and (2) suggest that posterior pole localization of Tsunagi within the oocyte is dependent on Mago localization within the oocyte posterior cytoplasm.
The distribution of Mago in mutant tsu egg chambers was examined using transgenic flies carrying a single copy of the p[GFP–mago+] transgene. In mutant tsu egg chambers, Mago is detected both within nuclei and concentrated within the posterior pole cytoplasm (see Fig. 4C,D). These results indicate that neither the nuclear nor the cytoplasmic localization of Mago is dependent on tsu+.
Isolation of lethal and female sterile alleles of tsu
Phenotypic analysis of tsu mutant ovaries provides a means of addressing the role of tsu in oogenesis, and determining whether Mago and Tsunagi are likely to interact to regulate similar oogenic events. Three strategies were used to recover mutations in tsu. First, the sequence of genomic DNA flanking inviable P-element insertions in the 45A region was compared to tsu genomic sequence to determine if the insertions are in close proximity to tsu or disrupt tsu sequence. One lethal P-element insertion, EP(2)0567, is within the 5′ untranslated region (UTR) of tsu, 72 bp upstream of the first translation initiation site (Fig. 1B,C) and has been renamed tsu1. Second, a screen was undertaken for EMS-induced mutations that fail to complement a deletion that removes tsu and flanking genes, Df(2R)Np3 [44D2-E1;45B8-C1] (S.E. Mohr and R.E. Boswell, in prep.). One EMS-induced mutation in tsu, tsu2, was identified on the basis that tsu2 fails to complement Df(2R)Np3, Df(2R)Np5 [44F11;45D9-E1], and tsu1. Third, P-element mobilization was used to induce excisions of the insertion in the tsu1 allele (Robertson et al. 1988). The mobilization events fall into the following three classes: (1) homozygous inviable mobilization events (e.g., tsu3, tsu4, and tsu5); (2) homozygous viable, female-sterile mobilization events (e.g., tsu6 and tsu7); and (3) complete revertants. The following heteroallelic mothers are viable and female sterile: tsu5/tsu6, tsu5/tsu7, and tsu6/tsu7. All other allelic combinations are inviable. When tsu6 and tsu7 are in trans to Df(2R)Np5, the mutant flies are inviable, suggesting that these are hypomorphic alleles of tsu.
A transgene that contains the entire tsu gene but with nonfunctional flanking genes, p[tsu+] (Fig. 1A), was used to verify that mutations recovered by the methods described above were alleles of tsu and to determine whether secondary mutations are associated with these alleles. The p[tsu+] transgene was introduced into flies by P-element-mediated transformation and tested for its ability to complement potential mutations in tsu. A single copy of the p[tsu+] transgene is sufficient to alleviate the zygotic lethality of inviable tsu alleles. The transgene also complements the female sterility observed in tsu5/tsu6, tsu5/tsu7, and tsu6/tsu7 heteroallelic mothers, and in homozygous tsu6 and tsu7 females.
Molecular lesions in the tsu2 and tsu5 mutant alleles were identified (Fig. 1B,C). Mobilization of the P element in tsu1 resulted in excision of all but 35 bp of the transposable element from the 5′ UTR of tsu to produce the tsu5 allele. The tsu2 mutation is a single base-pair change predicted to encode a nonsense codon resulting in premature termination of the synthesis of Tsunagi (W148 → STOP; see Fig. 1B,C). Therefore, genetic and molecular analyses indicate that tsu is an essential gene and that hypomorphic alleles of tsu can result in female sterility.
tsu+ is required in the germ line to form the major body axes and for germ cell determination
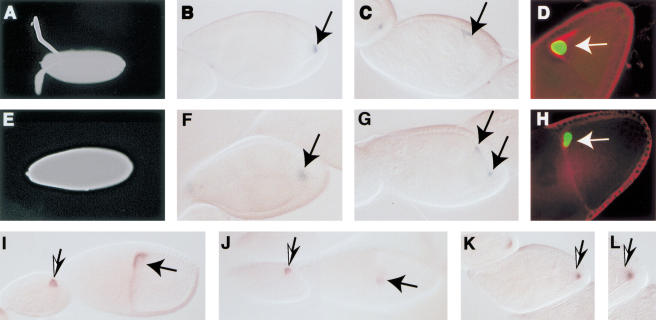
Given that Mago and Tsunagi form a complex and colocalize within the posterior pole plasm, we investigated whether mutations in tsu disrupt the same developmental events altered in mutant mago egg chambers. The shells of eggs collected from tsu mutant females were examined to determine whether ventralized eggs resembling those collected from hemizygous mutant mago females could be observed (Newmark et al. 1997). Of the eggs ∼17% (n = 424) from tsu7 homozygous females and ∼24% (n = 218) from tsu6/tsu7 females display a ventralized phenotype similar to that observed with eggs from hemizygous mago1 females (Fig. 5A,E). A similar phenotype is observed when eggs are collected from females mutant for gurken (grk), EGFr, and cornichon (González-Reyes et al. 1995; Roth et al. 1995).
Figure 5.
The wild-type function of tsu is required within the germ line for DV patterning and osk mRNA localization during oogenesis. In all panels anterior is to the left and posterior to the right. (A,E) Micrographs of wild-type and mutant tsu (from tsu6/tsu7 females) eggshells, respectively. (B,F) The distribution of Kinesin::β-gal in wild-type and mutant tsu stage-9 egg chambers (from tsu6/tsu7 females), respectively. (C,G) The distribution of Nod::β-gal in wild-type and mutant tsu stage-9 egg chambers (from tsu6/tsu7 females), respectively. (D,H) The position of the oocyte nucleus (green) and the distribution of Gurken protein (red) in stage-10 egg chambers from wild-type and tsu6/tsu7 females. In each panel the position of the oocyte nucleus is indicated by the arrow and was determined using α-cyclin E. (I) Distribution of grk mRNA in wild-type stage-8 (left) and stage-10 (right) egg chambers. (J) In egg chambers from tsu6/tsu7 females, grk mRNA is clearly detectable at stage 9 (left) but by stage 10 (right) it is significantly reduced relative to wild type in ∼70% (n = 70) of the chambers. (K,L) In stage-9 and stage-10 egg chambers the oocyte nucleus is mislocalized in ∼20% (n = 114) of the egg chambers. The micrographs show the position of the oocyte nucleus in a stage-9 egg chamber from a tsu7 homozygous mother and the distribution of grk mRNA. (L) A high magnification image of the posterior of the stage-9 egg chamber illustrated in K to show the oocyte nucleus.
The majority of the eggs (>60%) from females homozygous for tsu7 or heteroallelic for the alleles described above are abnormal. The eggs have defective dorsal appendages (fused and/or shorter than wild type) but normal aeropyles, an eggshell structure formed by the posterior follicle cells. None of the embryos from these mutant females hatch, regardless of the genotype of the embryo. Embryos from tsu6/tsu7 mutant females were stained with DAPI and ∼60% of the embryos appear to be unfertilized. The remaining embryos from such females develop to the cellular blastoderm stage, but primordial germ cells are not formed. Examination of cuticles of the embryos reveals a range of defects in cuticular patterning without a specific predominant phenotype.
Mutant females carrying mutations in genes encoding components of the pathway that responds to the posterior follicle cell-to-oocyte signal produce egg chambers with the following characteristics: First, because the oocyte-to-follicle cell signal (Gurken/EGFr) is not disrupted, the fates of posterior follicle cells are indistinguishable from wild type (Micklem et al. 1997; Newmark et al. 1997). Second, because the posterior follicle cell-to-oocyte signal is critical for disassembly of the MTOC that is within the posterior, it is expected that microtubule organization is aberrant in some fraction of the mutant egg chambers (Micklem et al. 1997; Newmark et al. 1997). Failure to disassemble the MTOC will also disrupt migration of the oocyte nucleus from a posterior to an anterior position.
The fates of follicle cells can be examined using the enhancer trap lines BB127 and 5A7, which in wild-type egg chambers produce β-galactosidase in centripetal follicle cells and border cells, respectively (Roth et al. 1995). In mutant egg chambers where the Gurken/EGFr signaling pathway has been disrupted, the posterior follicle fates are not established, and these cells develop as anterior follicle cells. In such egg chambers BB127 and 5A7 produce β-galactosidase in follicle cells within the posterior pole of the egg chamber, an indication that these cells have adopted anterior fates. Mutant tsu egg chambers (from tsu7/tsu7 and tsu5/tsu7 mothers) expressing BB127 and 5A7 only produce β-galactosidase in anterior follicle cells, suggesting that posterior follicle cell fate induction is unaffected in tsu mutant ovaries (data not shown).
In contrast, defects in oogenesis consistent with disruption of cytoskeletal organization are detected in tsu mutant ovaries. Kinesin::β-gal is localized to the posterior pole of wild-type stage 8–10 oocytes (Fig. 5B; Clark et al. 1994). In tsu mutant egg chambers Kinesin::β-gal (a plus-end-directed motor protein) is often mislocalized within the oocyte (∼40%–60%, dependent on the genotype), suggesting that microtubule organization is abnormal in these egg chambers (Fig. 5F). Nod::β-gal (a minus-end-directed motor protein) localizes to the anterior pole of stage 8–10 wild-type oocytes (Fig. 5C; Clark et al. 1997); however, in tsu mutant egg chambers the fusion protein is detectable both at the anterior margin and within the posterior pole cytoplasm (Fig. 5G). Migration of the oocyte nucleus is also abnormal in mutant tsu egg chambers; for example, the oocyte nucleus is not within the anterior pole in ∼20% (n = 114) of the stage 9 and 10 chambers from homozygous tsu7 mothers (Fig. 5K,L). Therefore, although follicle cell fates do not appear to be altered, rearrangement of the microtubules is aberrant in many tsu mutant egg chambers.
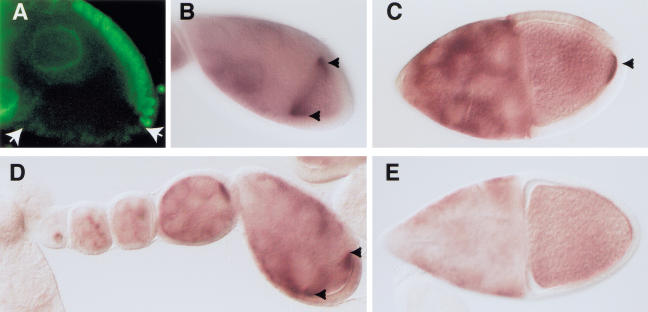
The phenotype of egg chambers and embryos from tsu mutant females is consistent with a role for tsu in axis formation and germ cell determination. To investigate whether tsu is required in the follicle cells or in the germ line, mosaic females with clones of tsu1 mutant cells in the follicle cells or in the germ line were induced using the FRT/FLP system for induction of mitotic recombination (Chou and Perrimon 1996; Duffy et al. 1998). The production of tsu1/tsu1 follicle-cell clones resulted in clusters of somatic follicle cells lacking detectable Tsunagi (Fig. 6A), providing further evidence of the specificity of the antisera. As expected for a gene that is required within the germ line, despite the production of patches of follicle cells without detectable Tsunagi, the eggs and embryos collected from the females were indistinguishable from wild type (Table 1).
Figure 6.
Germ-line and follicle-cell clonal analysis of tsu. Egg chambers are oriented with anterior to the left and posterior to the right. In B–E the arrowheads indicate the accumulation of osk mRNA. (A) Immunolocalization of Tsunagi in a stage-9 egg chamber showing a tsu1/tsu1 follicle-cell clone with its boundaries delineated by the arrows. (B,C) The distribution of osk mRNA in wild-type stage-9 and stage-10 egg chambers, respectively. (D,E) The distribution of osk mRNA in an ovariole derived from a germ-line mosaic female. As seen in D, osk mRNA accumulates in the anterior pole of stage-9 mosaic egg chambers. Posterior pole accumulation of osk mRNA is not detected in mosaic stage 10 (E) egg chambers.
Table 1.
Germ line and follicle cell analysis of tsu1
|
|
Percent defective eggshells
|
Percent unhatched embryos
|
|---|---|---|
| tsu+ follicle cell clonesa | 0 (n = 544) | 4% (n = 544) |
| tsu1 follicle cell clonesa | 0 (n = 707) | 3% (n = 707) |
| tsu+ germ line clones | 2% (n = 1237) | 28% (n = 895) |
| tsu1 germ line clones | 95% (n = 253)b | 100% (n = 268) |
Follicle cell clones were induced as described in Liu and Montell (1999).
The range of eggshell phenotypes is similar to that observed when females are transheterozygous or homozygous for tsu alleles.
Conversely, homozygosity for the tsu1 mutation in the germ line had a dramatic effect on oogenesis. Germ-line mosaic tsu1 females lay defective eggs and embryos that fail to hatch (Table 1). The eggshells from tsu1 germ-line mosaic females have defects similar to eggs laid by females homozygous or transheterozygous for alleles of tsu. The early accumulation (stages 1–8) of grk RNA and protein are unaffected when tsu+ is removed from the germ line in mosaic ovaries and in egg chambers from homozygous or transheterozygous females. During later oogenesis, stages 9–10, there is a marked reduction in the amount of detectable grk RNA (Fig. 5I,J) and protein (Fig. 5D,H). Therefore, although removing wild-type tsu function from the germ line does not appear to affect their accumulation during early oogenesis, both grk RNA and protein are absent or reduced at stages critical for specification of dorsal fates in anterior follicle cells.
Colocalization of Tsunagi with Mago in the posterior pole cytoplasm at stages 8–9 suggests that, like Mago, Tsunagi might be required for posterior osk mRNA localization. The distribution of osk mRNA during early oogenesis is unaffected in tsu1 germ-line mosaic females, that is, osk mRNA accumulates in the oocyte and osk mRNA is anteriorly localized at stages 8–9 (Fig. 6B,C). However, posterior localization of osk mRNA is disrupted in stage 9 and later egg chambers from tsu1 germ-line mosaic females (Fig. 6D,E) but not in egg chambers from control tsu+ germ-line mosaic females or tsu1 follicle-cell mosaic females (Fig. 6C). Collectively, with the data presented above, these results are consistent with a model in which tsu+ functions within the oocyte to interpret the follicle cell-to-oocyte signal to establish the major axes and to localize germ-plasm components.
Discussion
In this study we have characterized a gene (tsu) encoding a Drosophila RNA-binding protein, Tsunagi. It is increasingly clear that RNA-binding proteins shuttle between the nucleus and cytoplasm to coordinate specific aspects of mRNA metabolism. RNA-binding proteins have been shown to regulate mRNA export from the nucleus, mRNA translation, mRNA turnover, and cytoplasmic localization of mRNA (Shyu and Wilkinson 2000). Homologs of Tsunagi interact with mRNA in a splicing-dependent manner to mediate shuttling of the mRNAs from the nucleus to the cytoplasm (Kataoka et al. 2000; Kim et al. 2001; Le Hir et al. 2000). Our results show that a Mago–Tsunagi complex is localized within the posterior pole cytoplasm, where it functions to (1) interpret the posterior follicle cell-to-oocyte signal that delineates the spatial coordinates of the oocyte (stages 1–5) and (2) assemble and/or maintain components necessary for germ-cell determination (stages 8–9 of oogenesis).
Mago and Tsunagi interact to localize osk mRNA
The Mago–Tsunagi complex localizes within the posterior pole of the oocyte during stages 8 and 9 of oogenesis. Posterior pole localization of osk mRNA is first detected during stage 9 of oogenesis (Ephrussi et al. 1991; Kim-Ha et al. 1991). Previous studies revealed that osk mRNA accumulates within the oocyte but fails to localize within the posterior pole of egg chambers from mago1 mutant females, owing specifically to the inability of the Mago1 protein to localize within the posterior pole plasm (Newmark et al. 1997). In mago1 mutant egg chambers Tsunagi protein fails to localize within the posterior end of the oocyte. The results establish that the Mago–Tsunagi complex is detected within the posterior pole prior to and during the time when osk mRNA is initially sequestered within this discrete cytoplasmic region of the oocyte, and that detection of the complex within the posterior pole is dependent on wild-type mago function.
Mutational analysis of tsu reveals that mothers homozygous or heteroallelic for tsu alleles can produce egg chambers in which osk mRNA fails to localize within the posterior pole. Consistent with this observation, embryos from tsu mutant females that survive to the time of cellularization lack primordial germ cells. Although posterior pole localization of osk mRNA is not detected in tsu mutant oocytes, its anterior pole accumulation at stage 8 of oogenesis appears normal. A similar result is observed when germ-line clones lacking tsu+ function are examined for the presence and distribution of osk mRNA. However, when follicle-cell clones are induced no apparent abnormalities in osk mRNA localization are detected. The distribution of Mago protein within mutant tsu egg chambers is indistinguishable from wild type, suggesting that posterior pole localization of Mago protein occurs independently of Tsunagi function. These results show that (1) in mutant tsu egg chambers osk mRNA is transcribed and deposited into the oocyte, (2) anterior localization of osk mRNA does not require tsu+ function, (3) posterior pole localization of Mago protein is independent of tsu+ function, and (4) posterior pole accumulation of osk mRNA is dependent on tsu+ germ-line function.
What role might the molecular interaction between Mago and Tsunagi serve in the posterior localization of the Staufen protein/osk mRNA complex? Several lines of evidence indicate that Mago protein is required to anchor/localize components within the posterior pole that mediate the localization of the Staufen protein/osk mRNA complex within the pole during stage 9. First, Mago colocalizes with the Staufen protein/osk mRNA complex (Newmark and Boswell 1994). Second, Mago protein is mislocalized to the same ectopic site as the Staufen protein/osk mRNA complex in mutants in which oocyte polarity is disrupted; for example, gurken (González-Reyes et al. 1995; Roth et al. 1995; Newmark et al. 1997). Third, localization of Mago within the posterior pole (during stage 8) precedes the posterior pole accumulation of the Staufen/osk mRNA complex and is not dependent on localization of the complex (Newmark and Boswell 1994; Newmark et al. 1997). The evidence discussed above and the experimental results presented in this paper indicate that posterior pole localization of Mago occurs independently from the Staufen protein/osk mRNA complex and Tsunagi protein. Mago's ability to interact with a particular cytoplasmic location independently of the components that it localizes suggests that it may serve as an adaptor that recognizes a specific site within the posterior pole of the oocyte.
Given its interaction with Mago protein, it is likely that Tsunagi is required for anchoring the Staufen protein/osk mRNA complex within the posterior pole but not for its transport to this cytoplasmic destination. In agreement with this conclusion, the distribution of Tsunagi protein during multiple stages of oogenesis is indistinguishable from that of Mago protein. If Tsunagi protein were involved in transporting the Staufen protein/osk mRNA complex to the posterior pole, its subcellular distribution would more closely reflect that of Staufen protein and osk mRNA, which mirror one another during oogenesis (St Johnston et al. 1991). In addition, RNA-binding experiments failed to reveal interaction between Tsunagi protein and osk mRNA (S.T. Dillon and R.E. Boswell, unpubl.).
At least two simple models can be presented to explain the results obtained to date. In the first model, Mago protein might act as an adaptor molecule to anchor Tsunagi protein and associated mRNAs within the posterior cytoplasm of the oocyte. Although this model is appealing, it does not take into consideration the colocalization of Mago and Tsunagi proteins within nuclei. Another model is that Mago and Tsunagi proteins are assembled onto mRNP complexes to mark them for transport to and localization within the posterior pole. This alternative model takes into account the nuclear and cytoplasmic colocalization of the proteins but does not exclude the possibility that the Mago protein can act as an adaptor, linking mRNPs to a specific subcellular site. Importantly, this latter model is in agreement with experimental results showing that Y14 (a Xenopus laevis homolog of Tsunagi) shuttles from the nucleus to the cytoplasm in association with mRNAs and is retained on the mRNAs when they are exported to the cytoplasm (Kataoka et al. 2000; Le Hir et al. 2000; Kim et al. 2001). Therefore, our studies establish a role for Tsunagi protein in anchoring mRNAs within the posterior pole of the oocyte when associated with Mago protein, suggesting that homologs of Tsunagi in other organisms may also be involved in RNA localization. Regardless of the model, once mRNPs are localized within the posterior pole, translation of the complexed RNAs would result in the production of proteins essential for the assembly of germ-plasm-specific components, such as Staufen protein/osk mRNA and as yet unidentified mRNAs.
Establishment of the AP polarity of the oocyte requires tsu+ function in the germ line
Given the role of the Mago–Tsunagi complex in the localization of osk mRNA and Staufen protein, it is possible to envision a similar function for the complex during early oogenesis (stages 1–5). That is, during early oogenesis a Mago–Tsunagi complex is likely to be required for localizing mRNAs that encode components necessary to interpret the posterior follicle cell-to-oocyte signal.
Aberrant AP axis formation during oogenesis in tsu mutant egg chambers is revealed by two phenotypes. First, the migration of the oocyte nucleus from a posterior to an anterior position can be abnormal in tsu mutant egg chambers. Second, markers of microtubule organization such as Kinesin::β-gal and Nod::β-gal are found at ectopic sites within tsu mutant oocytes. These are phenotypes that are reminiscent of defects observed in mutants in which Gurken signaling is disrupted (Ruohola et al. 1991; González-Reyes et al. 1995; Roth et al. 1995). When Gurken signaling is blocked, disassembly of the MTOC at the posterior pole is inhibited and the oocyte nucleus fails to migrate to the anterior pole. Anomalies in nuclear migration and in the distribution of markers used to assess the integrity of the microtubule network are also detected in mago mutant egg chambers (Micklem et al. 1997; Newmark et al. 1997). Disassembly of the MTOC has not been examined in tsu mutant egg chambers but is aberrant in mago mutant oocytes (Micklem et al. 1997). The fact that Mago and Tsunagi proteins cooperate to establish the AP axis of the oocyte suggests that in oocytes from tsu mutant mothers the MTOC at the posterior pole may not disassemble.
Mutations that disrupt Gurken signaling also alter the fates of follicle cells, causing posterior follicle cells to develop as anterior follicle cells (Ruohola et al. 1991; González-Reyes et al. 1995; Roth et al. 1995). In egg chambers derived from tsu mutant females the accumulation and function of grk mRNA and protein during stages 1–6 (the time when posterior follicle cell fates are specified) are apparently normal. This is also evident from the fact that follicle cell fates are properly specified in tsu mutant egg chambers, as determined by the formation of the aeropyle (a structure produced by the posterior follicle cells) and by monitoring egg chambers with follicle cell markers that reveal their fates. Importantly, germ-line and follicle-cell clonal analysis indicates that tsu+ function is necessary within the germ line but not in follicle cells. Therefore, it appears that the patterning abnormalities detected when egg chambers lack wild-type tsu function arise from a requirement for Tsunagi protein in mediating the response of the oocyte to the posterior follicle cell-to-oocyte signal.
Posttranscriptional regulation of gurken requires tsu+ for DV patterning
In situ hybridization and immunolocalization reveal that grk mRNA and protein are altered during stages 9–10 in oocytes from tsu mutant females. The amount of detectable grk mRNA and protein is often reduced relative to wild type or undetectable above background. In contrast, the amount of grk mRNA and protein is indistinguishable from wild type in mago mutant egg chambers and they are both properly localized (Micklem et al. 1997; Newmark et al. 1997). The differences in grk mRNA and protein in mago mutant and tsu mutant egg chambers during stages 9–10 suggest that the DV pattern abnormalities in tsu mutant egg chambers are not caused solely by the altered migration of the oocyte nucleus, and although the two proteins cooperate in specific developmental processes, Mago and Tsunagi proteins are likely to function independently in other aspects of oocyte development.
DV patterning of the egg chamber occurs during midoogenesis from restriction of Gurken signaling to the future dorsal side of the egg chamber. The asymmetric localization of Gurken signaling is achieved by (1) migration of the oocyte nucleus to an anterior cortical position, (2) transport of grk mRNA from the oocyte nucleus to the dorsal-anterior corner of the oocyte, (3) anchoring of grk mRNA within the dorsal-anterior corner of the oocyte, and (4) translation of the spatially restricted grk mRNA (Koch and Spitzer 1983; Neuman-Silberberg and Schüpbach 1993; González-Reyes et al. 1995; Roth et al. 1995; Norvell et al. 1999; Saunders and Cohen 1999). Squid protein, a heterogeneous nuclear RNA-binding protein (hnRNP), has been implicated in the export of grk mRNA from the nucleus and in its delivery to an anchor within the cytoplasm (Norvell et al. 1999).
The Tsunagi homolog Y14 was originally identified through its interaction in a yeast two-hybrid screen with Ran-binding protein 5 (RanBP5; Kataoka et al. 2000). RanBP5 is related to the nuclear transporter receptor proteins importin-β and transportin (Deane et al. 1997; Yaseen and Blobel 1997), proteins that are part of the nuclear import/export machinery (Görlich and Kutay 1999; Nakielny and Dreyfuss 1999). Like Squid protein, RanBP5 has been shown to interact physically with transportin (Jäkel and Görlich 1998; Norvell et al. 1999). Based on the association of Squid protein and Tsunagi homologs with cellular components required in nuclear import and export, biochemical evidence indicating that the two proteins function in RNA export and the genetic evidence provided here, it is reasonable to propose that Tsunagi protein and Squid protein may interact with the nucleocytoplasmic transport machinery to regulate export and/or anchoring of grk mRNA within the dorsal-anterior corner of the oocyte.
Several roles for Tsunagi protein in the export and/or anchoring of grk mRNA are consistent with the data presented and the known biochemical roles of Tsunagi homologs. First, Tsunagi protein may be necessary to maintain the stability of grk mRNA during midoogenesis. Second, splicing-dependent export of grk mRNA from the oocyte nucleus may require Tsunagi+ protein. The RNA may not be detectable within the nucleus owing to rapid degradation of unspliced pre-mRNAs, as has been shown to occur in yeast (Bousquet-Antonelli et al. 2000), or it may be exported from the nucleus by a generalized export mechanism. Third, the protein may be required for the export and/or localization of grk mRNA within the dorsal-anterior cortex. Further experimentation will be necessary to determine the molecular function of Tsunagi protein in the stability, export, and/or anchoring of grk mRNA during midoogenesis.
The molecular and genetic analysis of tsunagi has revealed that the encoded protein functions in at least three distinct processes during Drosophila oogenesis. In early oogenesis (stages 1–5), Tsunagi protein forms a complex with Mago protein that is critical for interpreting the posterior follicle cell-to-oocyte signal. During stages 8 and 9 of oogenesis, Tsunagi protein cooperates with Mago protein to localize components necessary for anchoring osk mRNA within the posterior pole. Although other interpretations are possible, a simple model suggests that both during stages 1–5 and 8–9 the Mago–Tsunagi complex localizes RNAs encoding proteins that are essential for mediating axis formation and assembly of the germ plasm. At stages 9–10, Tsunagi protein has a function independent of Mago protein that is crucial for the export and/or localization of grk mRNA. In this paper we have focused on the role of Tsunagi during axis formation and germ plasm assembly during oogenesis. Our results indicate that Tsunagi interacts with a component of the cellular localization machinery, suggesting that homologs of Tsunagi may also be involved in RNA localization.
Materials and methods
Yeast two-hybrid screen against Mago Nashi
Full-length D. melanogaster mago coding sequence was subcloned into the interaction trap LexA fusion bait vector pEG202 (Gyuris et al. 1993) to create the plasmid pEG-mago. Standard interaction trap methods were used to screen, select, and isolate interacting cDNA clones (Golemis et al. 1996). First, it was determined that the protein product from pEG-mago, LexA-Mago, does not activate transcription by itself in the interaction trap. LexA-Mago was then used as bait against the D. melanogaster ovarian cDNA library RFLY3 (gift of R. Finley, Wayne State University) to select for proteins that interact with Mago. Tsunagi showed no interaction with a number of LexA fusion proteins, indicating that Tsunagi was binding specifically to Mago and not to LexA in the interaction trap.
Antibody generation and purification
A tsu cDNA recovered in the interaction trap screen that contains the entire tsu open reading frame was subcloned into the EcoRI and XhoI sites in the glutathione-S-transferase (GST) fusion vector pGEX-4T-1 (Pharmacia). This generated a GST–Tsunagi fusion protein that when expressed in the Escherichia coli strain BL21 using standard induction conditions and purification methods resolves at 45 kD on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE; Smith and Corcoran 1994).
Polyclonal antibodies were raised in rabbits (Cocalico Biologicals) against bacterially expressed and purified GST–Tsunagi. Antibodies specific for Tsunagi were affinity-purified from whole rabbit antisera against a maltose-binding protein (MBP)–Tsunagi fusion protein using an immunoblot purification protocol (Olmsted 1986). MBP–Tsunagi fusion protein was created by cloning the identical EcoRI and XhoI tsu cDNA fragment used to generate GST–Tsunagi fusion protein into the EcoRI and XbaI sites of the pMAL-c2 MBP fusion vector (New England Biolabs, NEB). MBP–Tsunagi was then expressed in and purified from BL21 following the protocol of Riggs (1994). MBP–Tsunagi resolves at 52 kD on SDS-PAGE.
SDS-PAGE and immunoblotting
All proteins were resolved using the Laemmli SDS-PAGE gel system (Laemmli 1970). Immunoblotting was performed with a semidry electroblotter (Bio-Rad), and protein was transferred onto nitrocellulose membranes (Schleicher & Schuell) following standard protocols (Towbin et al. 1979). To detect the Myc epitope on immunoblots, tissue culture supernatant from the monoclonal antibody cell line 9E10 (Evan et al. 1985) was used at a 1:2–1:10 dilution in 1× Tris buffered saline (TBS) containing 0.1% Tween-20 and 5% nonfat dry milk powder. A 1:100 dilution of affinity-purified α-Tsunagi antibody was used to probe for Tsunagi. Goat anti-rabbit horseradish peroxidase (Pierce) was the secondary antibody employed, and immunoblots were developed using enhanced chemiluminescence (SuperSignal, Pierce).
Generation of D. melanogaster protein extracts
Staged embryos and larvae were collected following previously published protocols (Mahowald 1994). Adult flies, embryos, and larvae were each homogenized in 2 volumes of lysis buffer (50 mM Tris at pH 7.5, 150 mM NaCl, 0.5 mM DTT, 0.05% NP-40, 0.2% Triton X-100, 1 mM MgCl2, 1 mM EGTA, 10 μM benzamidine-HCl, 10 μg/mL aprotinin, 10 μg/mL leupeptin, 10 μg/mL pepstatin A, 1 mM PMSF) using a motor-driven ice-cold homogenizer. All extracts were immediately snap-frozen in a dry ice/ethanol bath and stored at −80°C until use. Protein concentrations were determined using a BCA protein assay kit (Pierce).
Immunoprecipitations
Adult fly whole-tissue homogenates used in immunoprecipitation experiments were first centrifuged at 10,000g at 4°C for 10 min. Supernatant containing 5.0 mg of protein was diluted to a final volume of 0.5 mL with freshly made lysis buffer and then mixed with 40 μL of a 50% slurry of protein A agarose beads (Pierce) preloaded with the 9E10 monoclonal antibody. After incubation at 4 °C overnight with constant mixing, the beads were washed 3 times with 1.0 mL of freshly made lysis buffer. After 2× Laemmli sample buffer (50 μL) was added to the beads, half of each immunoprecipitate was then immediately loaded onto a 10%–20% SDS-PAGE gradient gel.
Whole-mount protein localization and in situ hybridization
Ovaries for immunolocalization were dissected in PBSTX (1× PBS, 0.1% Triton X-100), fixed for 20 min in PBSTX with 4% formaldehyde, and blocked in PBSTX with 1% BSA. Alternatively, ovaries were dissected in PBT (1× PBS, 0.1% Tween 20), fixed in PBT with 4% formaldehyde, and treated as described in Zhang and Kalderon (2000). The anti-Tsunagi antisera were diluted 1:10 in PBSTX or PBT. Alexa 488 or Alexa 594 (Molecular Probes) conjugated to goat anti-rabbit secondary antibodies diluted 1:250 in PBSTX were used to detect proteins. Goat anti-rat secondary antibodies conjugated to Texas Red (Vector) were diluted 1:250 in PBSTX or PBT. In situ hybridizations were performed using the protocol of Klingler and Gergen (1993). The Gurken protein and β-galactosidase fusion proteins were visualized as described in Newmark et al. (1997).
Analysis of the tsu genomic region and construction of the p[tsu+] transgene
The genomic location of tsu was determined by the hybridization of digoxigenin-labeled cDNA probes to polytene chromosomes, as described in Johnson-Schlitz and Lim (1987). An ∼9-kb NheI genomic fragment was isolated from the P1 phagemid DS02099 (BDGP) and cloned into pLITMUS 38 (NEB), resulting in pL38P1I. The pL38P1I fragment and the cDNA clone LD43647 from Pmm45A and LD33051 from Mys45A were sequenced using an ABI Prism 377 DNA Sequencer (Perkin Elmer). The accession numbers are as follows: Pmm45 (AF173548), Mys45A (AF173549), and tsu (AF173550).
The location of the lesions in tsu1, tsu2, and tsu5 was determined by preparation of genomic DNA from adult flies of the appropriate strain, PCR amplification of the tsu coding region, and sequencing of the PCR products using an ABI Prism 377 DNA Sequencer (Perkin Elmer) and analyzed as described by Schlag and Wassarman (1999).
A transgene containing an NheI–EagI genomic subfragment of pL38P1I was constructed by taking advantage of an EagI site in the vector flanking the genomic NheI site to produce p[tsu+]. Because the NheI site disrupts the first exon of the Pmm45A gene just 3′ of the predicted ATG start codon and the EagI site similarly disrupts the Mys45A gene just 3′ of the predicted ATG start codon, the fragment is able to provide only wild-type tsu activity.
Genetic analyses
New tsu alleles were produced by ethylmethane sulfonate mutagenesis (see S.E. Mohr and R.E. Boswell, unpubl.) or by P-element excision (Robertson et al. 1988).
Follicle cell and germ-line clones were induced by construction of recombinant lines containing both p[mini w+; FRT]2R-G13 and tsu1. Follicle-cell clones were induced as described by Liu and Montell (1999). Germ-line clones were generated using the FLP-DFS system (Chou and Perrimon 1996). Germ-line and follicle-cell clones were also induced in a background containing p[mini w+; FRT 2x ubi-nls-GFP]2R-G13 (kindly provided by S. Luschnig and B. Moussian, Max-Planck-Institut für Entwicklungsbiologie, Tübingen, Germany), and tsu1 homozygous clones identified by the absence of both nls-GFP and immunoreactive Tsunagi protein. Hatching frequency of embryos from mosaic females was investigated as described in Newmark et al. (1997).
Acknowledgments
We wish to thank David H. Parma, Esther Van de Vosse, and P.J. Bennett for helpful discussions and critical reading of the manuscript. The research was supported by NIH training grant GM07135 to S.E.M., NIH (GM17954) postdoctoral fellowship to S.T.D., and a grant from the NIH (GM57989) to R.E.B.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL boswell@spot.colorado.edu; FAX (303) 492-6465.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.927001.
References
- Bashirullah A, Cooperstock RL, Lipshitz HD. RNA localization in development. Ann Rev Biochem. 1998;67:335–394. doi: 10.1146/annurev.biochem.67.1.335. [DOI] [PubMed] [Google Scholar]
- Boswell RE, Prout ME, Steichen JC. Mutations in a newly identified Drosophila melanogaster gene, mago nashi, disrupt germ cell formation and result in the formation of mirror-image symmetrical double abdomen embryos. Development. 1991;113:373–384. doi: 10.1242/dev.113.1.373. [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark I, Giniger E, Ruohola-Baker H, Jan LY, Jan YN. Transient posterior localization of kinesin fusion protein reflects anteroposterior polarity of the oocyte. Curr Biol. 1994;4:289–300. doi: 10.1016/s0960-9822(00)00068-3. [DOI] [PubMed] [Google Scholar]
- Clark IE, Jan LY, Jan YN. Reciprocal localization of Nod and kinesin fusion proteins indicates microtubule polarity in the Drosophila oocyte, epithelium, neuron and muscle. Development. 1997;124:461–470. doi: 10.1242/dev.124.2.461. [DOI] [PubMed] [Google Scholar]
- Deane R, Schafer W, Zimmermann HP, Mueller L, Gorlich D, Prehn S, Ponstingl H, Bischoff FR. Ran-binding protein 5 (RanBP5) is related to the nuclear transport factor importin-β but interacts differently with RanBP1. Mol Cell Biol. 1997;17:5087–5096. doi: 10.1128/mcb.17.9.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy JB, Harrison DA, Perrimon N. Identifying loci required for follicular patterning using directed mosaics. Development. 1998;125:2263–2271. doi: 10.1242/dev.125.12.2263. [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Lehmann R. Induction of germ cell formation by oskar. Nature. 1992;358:387–392. doi: 10.1038/358387a0. [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R. oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu FD. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- Gatignol A, Buckler C, Jeang KT. Relatedness of an RNA-binding motif in human immunodeficiency virus type 1 TAR RNA-binding protein TRBP to human P1/dsI kinase and Drosophila staufen. Mol Cell Biol. 1993;13:2193–2202. doi: 10.1128/mcb.13.4.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis EA, Gyuris J, Brent R. Interaction trap/two-hybrid system to identify interacting proteins. In: Ausubel FM, Brent R, Kingston RE, Moore DM, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. 1996. , Unit 20.1. Wiley and Sons, New York, NY. [Google Scholar]
- González-Reyes A, St Johnston D. Patterning of the follicle cell epithelium along the anterior–posterior axis during Drosophila oogenesis. Development. 1998;125:2837–2846. doi: 10.1242/dev.125.15.2837. [DOI] [PubMed] [Google Scholar]
- González-Reyes A, Elliott H, St Johnston D. Polarization of both major body axes in Drosophila by gurken–torpedo signalling. Nature. 1995;375:654–658. doi: 10.1038/375654a0. [DOI] [PubMed] [Google Scholar]
- Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Ann Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- Jäkel S, Görlich D. Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Schlitz D, Lim JK. Cytogenetics of Notch mutations arising in the unstable X chromosome Uc of Drosophila melanogaster. Genetics. 1987;115:701–709. doi: 10.1093/genetics/115.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- Kim VN, Yong J, Kataoka N, Abel L, Diem MD, Dreyfuss G. The Y14 protein communicates to the cytoplasm the position of exon–exon junctions. EMBO J. 2001;20:2062–2068. doi: 10.1093/emboj/20.8.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J, Smith JL, Macdonald PM. oskar messenger RNA is localized to the posterior pole of the Drosophila oocyte. Cell. 1991;66:23–34. doi: 10.1016/0092-8674(91)90136-m. [DOI] [PubMed] [Google Scholar]
- Klingler M, Gergen PJ. Regulation of runt transcription by Drosophila segmentation genes. Mech Dev. 1993;46:3–19. doi: 10.1016/0925-4773(93)90019-t. [DOI] [PubMed] [Google Scholar]
- Koch EA, Spitzer RH. Multiple effects of colchicine on oogenesis in Drosophila: Induced sterility and switch of potential oocyte to nurse-cell developmental pathway. Cell Tiss Res. 1983;228:21–32. doi: 10.1007/BF00206261. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of the structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie YS, Macdonald PM. Apontic binds the translational repressor Bruno and is implicated in regulation of oskar mRNA translation. Development. 1999;126:1129–1138. doi: 10.1242/dev.126.6.1129. [DOI] [PubMed] [Google Scholar]
- Liu YR, Montell DJ. Identification of mutations that cause cell migration defects in mosaic clones. Development. 1999;126:1869–1878. doi: 10.1242/dev.126.9.1869. [DOI] [PubMed] [Google Scholar]
- Mahowald AP. Mass isolation of fly tissues. Methods Cell Biol. 1994;44:129–142. doi: 10.1016/s0091-679x(08)60910-3. [DOI] [PubMed] [Google Scholar]
- Micklem DR, Dasgupta R, Elliot H, Gergely F, Davidson C, Brand A, González-Reyes A, St. Johnston D. The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr Biol. 1997;7:468–478. doi: 10.1016/s0960-9822(06)00218-1. [DOI] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg FS, Schüpbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF α-Like protein. Cell. 1993;75:165–174. [PubMed] [Google Scholar]
- ————— The Drosophila TGFα-like protein Gurken: Expression and cellular localization during Drosophila oogenesis. Mech Dev. 1996;59:105–113. doi: 10.1016/0925-4773(96)00567-9. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Boswell RE. The mago nashi locus encodes an essential product required for germ plasm assembly in Drosophila. Development. 1994;120:1303–1313. doi: 10.1242/dev.120.5.1303. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Mohr SE, Gong L, Boswell RE. mago nashi mediates the posterior follicle cell-to-oocyte signal to organize axis formation in Drosophila. Development. 1997;124:3197–3207. doi: 10.1242/dev.124.16.3197. [DOI] [PubMed] [Google Scholar]
- Norvell A, Kelley RL, Wehr K, Schüpbach T. Specific isoforms of Squid, a Drosophila hnRNP, perform distinct roles in Gurken localization during oogenesis. Genes & Dev. 1999;13:864–876. doi: 10.1101/gad.13.7.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted JB. Analysis of cytoskeletal structures using blot-purified monospecific antibodies. Methods Enzymology. 1986;134:467–472. doi: 10.1016/0076-6879(86)34112-0. [DOI] [PubMed] [Google Scholar]
- Pokrywka NJ, Stephenson EC. Microtubules mediate the localization of bicoid RNA during Drosophila oogenesis. Development. 1991;113:55–66. doi: 10.1242/dev.113.1.55. [DOI] [PubMed] [Google Scholar]
- ————— Microtubules are a general component of messenger-RNA localization systems in Drosophila oocytes. Dev Biol. 1995;167:363–370. doi: 10.1006/dbio.1995.1030. [DOI] [PubMed] [Google Scholar]
- Ramos A, Grunert S, Adams J, Micklem DR, Proctor MR, Freund S, Bycroft M, St Johnston D, Varani G. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 2000;19:997–1009. doi: 10.1093/emboj/19.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs P. Expression and purification of maltose-binding protein fusions. In: Ausubel FM, Brent R, Kingston RE, Moore DM, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. 1994. , Unit 16.6. Wiley and Sons, New York, NY. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S, Neuman-Silberberg FS, Barcelo G, Schüpbach T. cornichon and the EGF receptor signaling process are necessary for both anterior–posterior and dorsal–ventral pattern formation in Drosophila. Cell. 1995;81:967–978. doi: 10.1016/0092-8674(95)90016-0. [DOI] [PubMed] [Google Scholar]
- Ruohola H, Bremer KA, Baker D, Swedlow JR, Jan LY, Jan YN. Role of neurogenic genes in establishment of follicle cell fate and oocyte polarity during oogenesis in Drosophila. Cell. 1991;66:433–449. doi: 10.1016/0092-8674(81)90008-8. [DOI] [PubMed] [Google Scholar]
- Saunders C, Cohen RS. The role of oocyte transcription, the 5′ UTR, and translation repression and derepression in Drosophila gurken mRNA and protein localization. Mol Cell. 1999;3:43–54. doi: 10.1016/s1097-2765(00)80173-2. [DOI] [PubMed] [Google Scholar]
- Schlag EM, Wassarman DA. Identifying mutations in Drosophila genes by direct sequencing of PCR products. Biotechniques. 1999;27:262–264. doi: 10.2144/99272bm09. [DOI] [PubMed] [Google Scholar]
- Shyu AB, Wilkinson MF. The double lives of shuttling mRNA binding proteins. Cell. 2000;102:135–138. doi: 10.1016/s0092-8674(00)00018-0. [DOI] [PubMed] [Google Scholar]
- Smith DB, Corcoran LM. Expression and purification of glutathione-S-transferase fusion proteins. In: Ausubel FM, et al., editors. Current protocols in molecular biology. New York, NY: John Wiley and Sons; 1994. pp. 16.7.1–16.7.7. [DOI] [PubMed] [Google Scholar]
- Spradling AC. Development genetics of oogenesis. In: Bate M, Martinez-Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 1–70. [Google Scholar]
- St Johnston D, Driever W, Berleth T, Richstein S, Nüsslein-Volhard C. Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development. 1989;107 Suppl:13–19. doi: 10.1242/dev.107.Supplement.13. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Beuchle D, Nüsslein-Volhard C. staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- Theurkauf WE, Alberts BM, Jan YN, Jongens TA. A central role for microtubules in the differentiation of Drosophila oocytes. Development. 1993;118:1169–1180. doi: 10.1242/dev.118.4.1169. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehlin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eeden F, St Johnston D. The polarisation of the anterior–posterior and dorsal–ventral axes during Drosophila oogenesis. Curr Opin Genet Dev. 1999;9:396–404. doi: 10.1016/S0959-437X(99)80060-4. [DOI] [PubMed] [Google Scholar]
- Wilhelm JE, Mansfield J, Hom-Booher N, Wang S, Turck CW, Hazelrigg T, Vale RD. Isolation of a ribonucleoprotein complex involved in mRNA localization in Drosophila oocytes. J Cell Biol. 2000;148:427–439. doi: 10.1083/jcb.148.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaseen NR, Blobel G. Cloning and characterization of human karyopherin β3. Proc Natl Acad Sci. 1997;94:4451–4456. doi: 10.1073/pnas.94.9.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kalderon D. Regulation of cell proliferation and patterning in Drosophila oogenesis by Hedgehog signaling. Development. 2000;127:2165–2176. doi: 10.1242/dev.127.10.2165. [DOI] [PubMed] [Google Scholar]