Abstract
We have identified a Drosophila homolog of the DNA replication initiation inhibitor Geminin (Dm geminin) and show that it has all of the properties of Xenopus and human Geminin. During Drosophila development, Dm Geminin is present in cycling cells; protein accumulates during S phase and is degraded at the metaphase to anaphase transition. Overexpression of Dm geminin in embryos inhibits DNA replication, but cells enter mitosis arresting in metaphase, as in dup (cdt1) mutants, and undergo apoptosis. Overexpression of Dm Geminin also induces ectopic neural differentiation. Dm geminin mutant embryos exhibit anaphase defects at cycle 16 and increased numbers of S phase cells later in embryogenesis. In a partially female-sterile Dm geminin mutant, excessive DNA amplification in the ovarian follicle cells is observed. Our data suggest roles for Dm Geminin in limiting DNA replication, in anaphase and in neural differentiation.
Keywords: Drosophila; cell cycle; DNA replication; mitosis, neurogenesis; Geminin
Strict regulation of DNA replication is essential for accurate propagation of the genetic material, as aberrant chromosome replication results in ploidy or mitotic defects, which can lead to tumorogenesis or cell death (for review, see Blow and Prokhorova 1999). Therefore, DNA replication must be restricted to once per cell cycle. To explain how DNA replication is limited to once per cell cycle, replication licencing factor (RLF) was proposed (Blow and Laskey 1988). RLF allows DNA to become replication-competent after mitosis, but once initiation occurs, RLF is inactivated until the next cell cycle. Biochemical purification of RLF revealed that it is composed of the Mcm complex (RLF-M) and RLF-B (Cdt1) (Thommes et al. 1997; Tada et al. 2001). Cdt1 together with Cdc6 promotes loading of the Mcm complex onto the Orc complex at replication origins in M/G1 phase to form the pre-replication complex (pre-RC) and is inactivated after replication origin firing in S phase (Maiorano et al. 2000; Nishitani et al. 2000; Whittaker et al. 2000; Tada et al. 2001).
DNA replication initiation is best described in Saccharomyces cerevisiae, which has well-defined DNA replication origins (for reviews, see Ekholm and Reed 2000; Ritzi and Knippers 2000; Lei and Tye 2001). Entry into S phase is driven by the regulated activity of G1 cyclin/Cdk protein kinases and Cdc7/Dbf4 (Ddk) protein kinases by triggering recruitment of Cdc45 to the pre-RC, which allows the binding of DNA polymerase α-primase. Conversely, phosphorylation of Cdc6 and Mcms by Cdks or Ddk results in their release from the origins and clearance from the nucleus. Re-replication is prevented in S phase by disassembly of the pre-RC and in G2/M phase by mitotic Cyclin B/Cdk activity, maintaining Mcms in a hyper-phosphorylated state and inhibiting new synthesis of Cdc6. At the completion of mitosis, inactivation of mitotic cyclin-associated Cdk activity marks the end of inhibitory signals that block new pre-RC formation and sets the cell into a pre-replicative state.
In more complex eukaryotes, replication initiation is similar to yeast, however, there are a number of differences in the regulatory mechanisms as well as additional tiers of regulation. In contrast to yeast pre-RC, bound Cdc6 and Mcms are not degraded nor exported from the nucleus after the initiation of DNA replication. An important regulation to prevent re-replication in both yeast and metazoans is high Cdk activity, although low Cdk activity by itself is not sufficient to allow the re-initiation of DNA replication in G2 in the Xenopus cell free system (Mahbubani et al. 1997; Sun et al. 2000). This additional regulation to prevent re-replication is provided by Geminin (McGarry and Kirschner 1998; Wohlschegel et al. 2000; Tada et al. 2001). Geminin was originally identified in Xenopus as a protein that undergoes ubiquitin-dependent degradation in mitosis and as an inhibitor of DNA replication initiation in vitro that functioned by preventing loading of Mcms onto chromatin (McGarry and Kirschner 1998). Recently two studies have revealed that Geminin prevents Mcm loading by binding to and inhibiting Cdt1 activity (Wohlschegel et al. 2000; Tada et al. 2001). Consistent with Geminin functioning to inhibit Cdt1 and prevent re-replication, Geminin is high in G2 cells but is degraded in mitosis thereby allowing Cdt1 function and re-assembly of the pre-RC in G1. Geminin is present in mammals but has not been identified in budding or fission yeast or Caenorhabditis elegans. Therefore, Geminin appears to be restricted to more complex eukaryotes where it acts as an additional layer of regulation to prevent re-replication. Geminin also appears to have a role in inducing neural differentiation (Kroll et al. 1998). Many questions, however, remain unanswered concerning the in vivo role of Geminin.
Drosophila is an ideal model organism in which to study the regulation of DNA replication because the cell division patterns have been well characterized during development (for reviews, see Edgar and Lehner 1996; Edgar and Orr-Weaver 2001). The first 14 S phases of embryogenesis occur in a syncitium (without cell membranes) and consist of very rapid S–M cycles driven by maternally-supplied cell cycle proteins. At G2 of cycle 14, cellularization occurs and the next three divisions are regulated in G2 by zygotic expression of the mitotic inducer Cdc25 (String). Most cells arrest in G1 of cycle 17 attributable to down-regulation of Cyclin E with the notable exception of nervous system cells that undergo several more divisions and the gut cells that undergo endoreplication. In larval stages, gut tissues, including the salivary glands, continue to undergo endoreplication, whereas imaginal discs undergo G1/G2 regulated cell cycles. During oogenesis, endoreplication of the nurse cells and follicle cells that surround the egg chamber also occurs.
In this study we characterize the in vivo function of a Drosophila geminin-related gene in the cell cycle and neural differentiation during development by analyzing Dm geminin mutants, the effects of overexpression, and the cell cycle distribution of the protein. These studies provide evidence that during Drosophila development, geminin has roles in limiting DNA replication, in anaphase and in neural differentiation.
Results
Identification of a Drosophila Geminin homolog
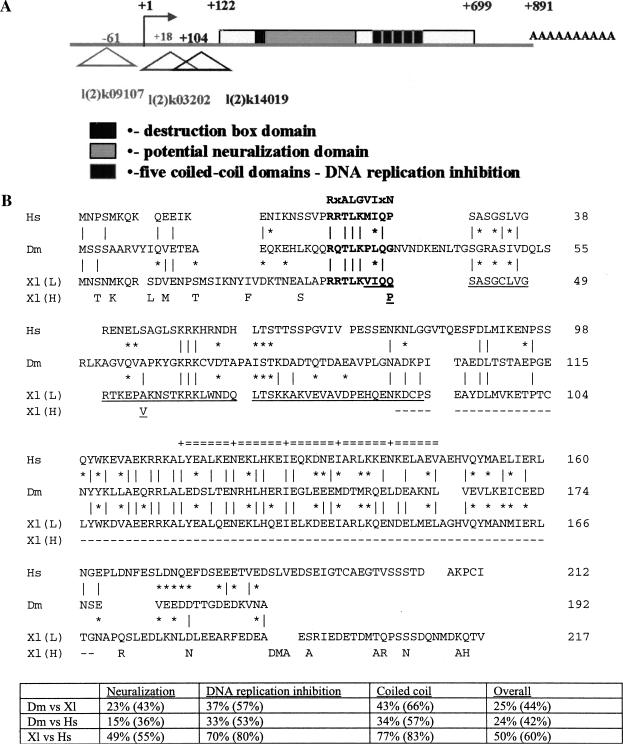
From analysis of the Berkeley Drosophila Genome Project (BDGP) database (Rubin et al. 2000), we identified a sequence (CG3183) at 42B3 on chromosome 2R that shows weak, but significant, similarity to Geminin from Xenopus (Xl Geminin) and Human (Hs Geminin) (Fig. 1). By DNA sequencing of cDNA clones, we determined the largest cDNA to be 891 bp, encoding a 192-amino-acid protein (GenBank accession no. AF407275). Analysis of the gene structure revealed that it was an intronless gene. Three P element alleles l(2)k09107, l(2)k14019, and l(2)k03202 were found to be inserted within or 5′ to the transcription unit (Fig. 1A).
Figure 1.
Drosophila Geminin and comparison with Xenopus and human Geminin. (A) Drosophila (Dm) geminin gene structure showing location of P-element alleles relative to the transcriptional unit. (B) Sequence comparison with Xenopus (Xl) and human (Hs) Geminin. The sequences were aligned using the GCG Gap program. Identical amino acids are marked with a vertical line and conservative changes are indicated (*). Differences between Xl H and L forms are indicated. The putative destruction box is in boldface type. The neuralization domain is underlined and the DNA replication inhibition domain is indicated by the broken underline. The double line above the sequence alignment shows the coiled-coil region. The degree of identity and similarity (in parentheses) between Xl Geminin L, Hs and Dm Geminin sequences is indicated for each domain.
The Drosophila Geminin-related sequence showed only limited identity overall with Xl and Hs Geminin, although when specific functional regions were compared, the level of identity was much greater (Fig. 1B). Geminin can be divided into two functional domains—the DNA replication inhibition domain containing a coiled-coil motif at the carboxyl terminus and a neuralization domain at the amino terminus (Kroll et al. 1998; McGarry and Kirschner 1998; Fig. 1B). The Drosophila Geminin-related sequence showed weak sequence conservation with Xl and Hs Geminin in the neuralization domain but higher levels of homology were observed with the DNA replication inhibition domain and with the coiled-coil motif (Fig. 1B). The Drosophila Geminin-related sequence has a potential destruction box for ubiquitin-mediated degradation in mitosis (RxALGVIxN) at its amino terminus, which shares a 5/9 match with the Xl Geminin H sequence shown to mediate degradation of the protein (McGarry and Kirschner 1998). Therefore, although the overall sequence similarity of the Drosophila Geminin-related protein is considerably lower than that observed between Xl and Hs Geminin, the basic arrangement of the protein is conserved.
The Drosophila Geminin homolog inhibits DNA replication by preventing binding of Mcms to chromatin
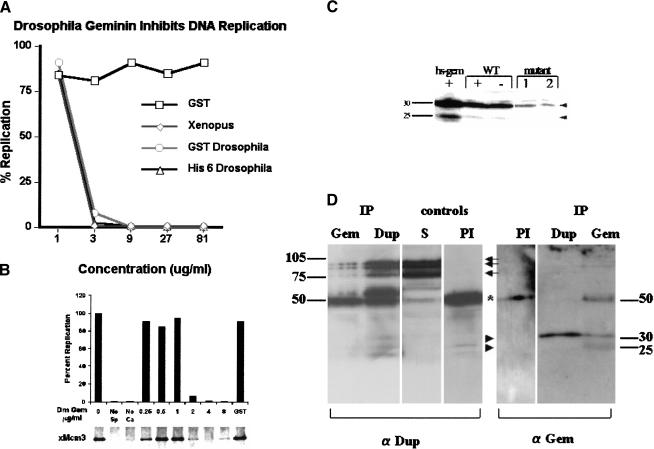
To confirm that the Drosophila Geminin-related protein was in fact the Drosophila homolog of Geminin, its effect on DNA replication was determined using a cell-free replication extract from unfertilized Xenopus eggs. In this system, the template for replication is Xenopus sperm head DNA and the reaction is started by calcium addition that mimics fertilization. Addition of bacterially produced Drosophila Geminin-related protein completely inhibited the incorporation of [α-32P]dCTP into DNA at concentrations of 1–4 μg/mL (50–150 nM) (Fig. 2A). The same concentration of Xl Geminin was required to inhibit replication in the extract.
Figure 2.
Drosophila Geminin inhibits DNA replication in Xenopus extracts by preventing Mcm loading and interacts with Drosophila Cdt1(Dup) in vivo. (A) Drosophila (Dm) Geminin inhibits DNA replication in Xenopus extracts. Purified Xl Geminin (⋄), GST-Dm Geminin (○), His-6 Dm Geminin (▵), or GST (□) were added at the concentrations shown to a Xenopus DNA replication in vitro assay system. Replication assays were carried out as described previously (McGarry and Kirschner 1998). (B) Dm Geminin inhibits Mcm loading. The indicated amounts of Dm Geminin were added to the Xenopus in vitro assay system and DNA replication measured (top) or the chromatin binding of xMcm3 determined (bottom). Concentrations of Dm Geminin that inhibit DNA replication (top) strongly inhibited xMcm3 chromatin binding. The GST control added at a concentration of 8 μg/mL had no effect on DNA replication or xMcm3 binding. In the control tracks (no sperm DNA or no calcium), DNA replication and xMcm3 binding were not observed. (C) Anti-Dm Geminin Western analysis. Dm Geminin bands are indicated (arrowheads). The faint band above 30 kD is a background band caused by the secondary antibody (data not shown). No Dm Geminin antibody reacting bands were detected elsewhere on the gel (data not shown). (WT) Wild type; (Hs-Gem) heat shocked hsp70–GAL4 UAS–Dm geminin embryos; (mutant 1) l(2)k14019/CyO; and (mutant 2) l(2)k03202/CyO. (+) Heat shocked. Equal amounts of protein were loaded per track. (D) Dm Dup(Cdt1) and Dm Geminin form a complex in Drosophila embryos. Dup or Dm Geminin were immunoprecipitated from embryonic extracts and the precipitate analyzed by immunoblotting with the Dup antibody (left) or Dm Geminin antibody (right). (Left) Dup bands (arrows); Dm Geminin bands (arrowheads); and IgG heavy chain (*) are indicated. Controls, (S) Supernatant; (PI) preimmune sera.
To determine whether Drosophila Geminin-related protein would inhibit binding of Mcms onto the pre-RC, chromatin was pelleted from the replication assays and the amount of Mcm complex bound to the chromatin was determined by immunoblotting (Fig. 2B, bottom). Drosophila Geminin-related protein inhibited Mcm binding at concentrations >2 μg/mL, which also inhibited DNA replication (Fig. 2B, top). The protein had no effect on the binding of Cdc6 or Orc complex to chromatin (data not shown). We conclude that the Drosophila Geminin-related protein, like Xl Geminin, inhibits DNA replication by preventing Mcm binding to chromatin.
Drosophila Geminin forms a complex with Cdt1(Dup) in vivo
Recent studies have shown that Geminin acts to inhibit DNA replication by binding to and preventing Cdt1 from loading Mcms onto the pre-RC complex (Wohlschegel et al. 2000; Tada et al. 2001). We therefore investigated whether Drosophila Geminin-related protein could form a complex with Drosophila Cdt1 homolog Doubleparked (Dup) in vivo. Western analysis of Drosophila embryonic extracts using antisera to the Drosophila Geminin-related protein specifically detected two bands at 25 kD and 30 kD (slightly larger than the predicted size of ∼22 kD), the abundance of which were increased in extracts after heat shock-induced expression of the Drosophila Geminin-related protein and decreased in extracts made from embryos from l(2)k14019 or l(2)k03202 mutant flies, where one-half of the wild-type level is expected (Fig. 2C). We conclude from these data that both bands are the Drosophila Geminin-related protein.
Immunoprecipitation–Western analysis of embryonic extracts using antibodies to the Drosophila Geminin-related protein and to Dup (obtained from Dr. T. Orr-Weaver; Whittaker et al. 2000) revealed that these proteins form a complex in vivo (Fig. 2D). When the Geminin-related antibody was used in the immunoprecipitation, Dup (which gives three bands >75 kD; J. Claycomb and T. Orr-Weaver, pers. comm.) was coprecipitated (Fig. 2D, left panel). In the converse experiment, immunoprecipitation with the Dup antibody coprecipitated the 30-kD Geminin-related protein but not the 25-kD band (Fig. 2D, right panel). We conclude that the 30-kD Drosophila Geminin-related protein forms a complex with Dup in vivo. Therefore, the Drosophila Geminin-related protein behaves similarly to Xl and Hs Geminin by two different criteria—the inhibition of DNA replication by preventing the loading of Mcms and complex formation with Cdt1(Dup). Given these data, we will henceforth refer to the Drosophila melanogaster Geminin-related protein as Dm Geminin.
Distribution of Dm Geminin throughout Drosophila development
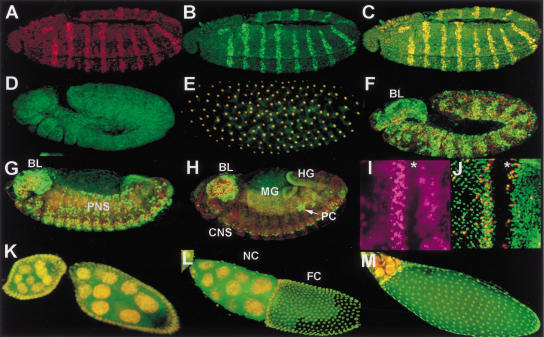
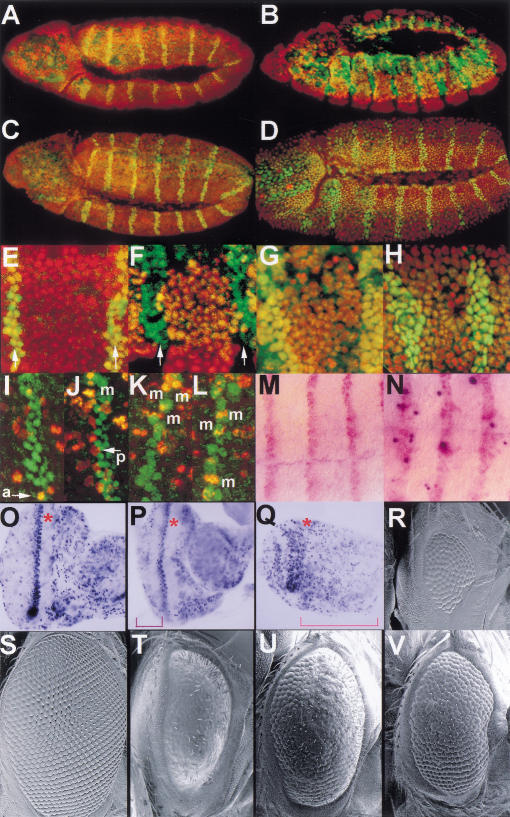
To explore the distribution of Dm Geminin during Drosophila development, we stained embryos and larval tissue with the anti-Dm Geminin antibody (Fig. 3). To demonstrate that the antibody specifically recognized Dm Geminin in vivo, we examined embryos ectopically expressing Dm geminin under control of the engrailed (en) driver. En is expressed in stripes and Dm Geminin protein was dramatically elevated in stripes coincident with the En pattern (Fig. 3A–C). In contrast, homozygous Dm geminin mutant embryos showed considerably reduced staining with anti-Dm Geminin antibody compared with wild type (Fig. 3D,E). Taken together, these results demonstrate that the anti-Dm Geminin antibody is specific for Dm Geminin in vivo.
Figure 3.
Dm Geminin protein is present in mitotic and endoreplicating cells during development. (A) en–GAL4 UAS–Gem stage-11 embryo stained with the anti-En antibody and propidium iodide PI (both in red) showing the En stripes. (B) en–GAL4 UAS–Gem embryo shown in A stained with the anti-Dm Geminin antibody (in green throughout). (C) Merge of A and B showing high Dm Geminin protein levels in the En stripes (appears yellow). (D) A stage-11 Dm geminin mutant l(2)03202/Df stained with anti-Dm Geminin showing low levels of Dm Geminin protein compared with F. (E) Merge of syncitial stage embryo stained with anti-Dm Geminin and PI (in red throughout) showing the nuclear localization of Dm Geminin. (F–H) Merges of Dm Geminin and PI in embryos. (F) A stage-11 embryo showing epidermal expression. (G) A stage-13 embryo showing Dm Geminin in PNS, CNS, and hindgut (HG) cells. (H) A stage-14 embryo showing Dm Geminin in the CNS, pole cells (PC arrow), and the gut. (BL) Brain lobe; (HG) hindgut; (MG) midgut. Embryos are oriented with anterior to the left and ventral side down in this and all other figures. (I) A larval eye disc labeled with BrdU (purple) showing domains of S-phase cells. (J) Merge of Dm Geminin and PH3 (red) in a larval eye disc. (I,J) The morphogenetic furrow, where cells are arrested in G1, is indicated (*). Eye discs are orientated anterior to the right in this and subsequent Figures. (K–M) Ovaries stained with anti-Dm Geminin and PI. (K) Merge of Dm Geminin and PI in egg chambers at stages 7 (left) and 8 (right) showing Dm Geminin in the nurse cell nuclei (in the interior) and follicle cell nuclei (at the perimeter). (L) Merge of Dm Geminin and PI in an egg chamber at stage 10B showing general nuclear localization of Dm Geminin in the follicle cells. (NC) Nurse cells; (FC) follicle cells. (M) Merge of Dm Geminin and PI in an egg chamber at stage 12. Ovaries are oriented anterior to the left in this and subsequent figures.
Staining of Drosophila embryos with the anti-Dm Geminin antibody and with propidium iodide to stain DNA showed that Dm Geminin is located to the nucleus (Fig. 3E). Therefore, like Xl Geminin (McGarry and Kirschner 1998), Dm Geminin is a nuclear localized protein. Throughout embryogenesis, Dm Geminin protein expression is correlated with mitotically dividing or endoreplicating cells (Fig. 3E–H). Early in embryogenesis during the rapid syncitial divisions, Dm Geminin is present at high levels irrespective of cell cycle stage (Fig. 3E; data not shown). In the G2 regulated cycles 14–16, Dm Geminin is present in a dynamic pattern similar to the domains of mitosis that occur at this stage (Fig. 3F). At the stage where cells of the peripheral nervous system (PNS) and central nervous system (CNS) divide, Dm Geminin was absent from the G1-arrested cells of the epidermis but present in the dividing neural cells (Fig. 3G). Later in development, Dm Geminin was absent in the PNS cells, which had stopped dividing, but present in the dividing CNS cells (Fig. 3H). Dm Geminin persisted in the pole (germ) cells (Fig. 3H) that are arrested in G2 at this stage (Su et al. 1998b). In addition, Dm Geminin was detected in the endoreplicating tissues of the gut (Fig. 3H).
In the eye imaginal disc, cell proliferation occurs in a spatial arrangement (for review, see Wolff and Ready 1991). Dm Geminin is present in the region of asynchronous dividing cells in the anterior region and in a band posterior to the morphogenetic furrow (MF), where the synchronous S phases occur, but not in the G1-arrested cells within the MF (Fig. 3, cf. J with I). Dm Geminin showed only partial overlap with cells staining with the mitotic marker anti-phospho histone H3 (PH3) that stains DNA in all stages of mitosis (Su et al. 1998a). In the posterior part of the eye imaginal disc where many cells are differentiating, Dm Geminin is present in a subset of cells that may represent the undifferentiated G2-arrested cells that are present in this region. These results show that Dm Geminin is present in proliferating cells and is at high levels in S–G2 phase cells, at low levels or absent in some mitotic cells, and absent from G1 cells.
Dm Geminin was also expressed during adult ovary development where endoreplication occurs in the nurse cells within the egg chamber and in the surrounding follicle cells (for review, see Edgar and Orr-Weaver 2001). The follicle cells undergo genomic endoreplication until stage 10A and switch to DNA replication amplification of specific foci including the chorion genes at stage 10B (Calvi et al. 1998). Early in oogenesis Dm Geminin is present in the nuclei of the endoreplicating nurse cells, but at later stages Dm Geminin is present in the nuclei and cytoplasm (Fig. 3K–M). The redistribution of Dm Geminin to the cytoplasm correlates with nurse-cell apoptosis before dumping of the nurse cell cytoplasm into the oocyte. Dm Geminin is present in the nuclei of the follicle cells throughout oogenesis and was not specifically localized to the amplification foci at stage 10B, as occurs with replication proteins Dup and Orc2 (Fig. 3K–M; Whittaker et al. 2000). In stage-12 egg chambers, when amplification becomes limited to the anterior follicle cells, Dm Geminin protein was observed in the nuclei of all follicle cells (Fig. 3M) and remained in all follicle cells until the end of oogenesis (data not shown). Therefore, Dm Geminin is present in mitotic and endoreplicating cells at different stages of development, consistent with a role in all replicative cycles throughout development.
Dm Geminin is degraded at the metaphase–anaphase transition
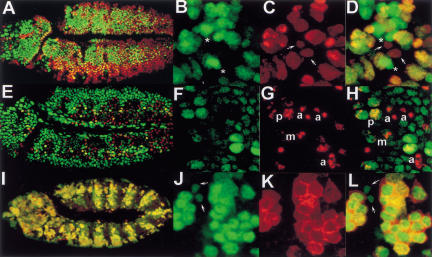
The dynamic pattern of Dm Geminin expression during embryogenesis is similar to that of Cyclin A or B, which accumulate during late S–G2 and are degraded during mitosis (Lehner and O'Farrell 1990). To explore the cell cycle distribution of Dm Geminin further, we examined anti-Dm Geminin staining relative to cell cycle markers of S phase (BrdU-labeling), mitosis (anti-PH3 antibody staining; Su et al. 1998a), and late S phase until the metaphase to anaphase transition (anti-Cyclin B staining; Lehner and O'Farrell 1990). Dm Geminin and BrdU-labeled cells showed partial overlap (Fig. 4A–D). Small, weakly BrdU-labeled cells (early S phase) did not contain Dm Geminin; strong BrdU-labeled cells showed higher levels of Dm Geminin; and large non-BrdU-labeled cells (G2/M) showed the highest levels of Dm Geminin (Fig. 4D), consistent with Dm Geminin accumulating during S–G2 phase. Co-localization of Dm Geminin with PH3 showed that as cells enter metaphase and DNA is stained strongly with anti-PH3, Dm Geminin is still present but reduced in level (Fig. 4E–H). In anaphase, when anti-PH3-stained chromosomes separate, Dm Geminin is undetectable (Fig. 4F–H). Dm Geminin protein distribution during the cell cycle was remarkably similar to Cyclin B although Dm Geminin was nuclear-localized and Cyclin B was cytoplasmic (Fig. 4I–L). Some small interphase cells (likely to be in early S phase) were observed to contain Dm Geminin but not Cyclin B, suggesting that Dm Geminin begins to accumulate before Cyclin B (Fig. 4L). Taken together, these data show that Dm Geminin, like Xl Geminin and Dm Cyclin B, accumulates during S phase and is degraded at the metaphase to anaphase transition.
Figure 4.
Dm Geminin accumulates in S/G2 and is degraded at anaphase. Stage-11 embryos with BrdU and Dm Geminin (A–D) or phosphohistone H3 (PH3) and Dm Geminin (E–H) or Cyclin B and Dm Geminin (I–L). (A) BrdU (red) and Dm Geminin (green throughout) merge. (B–D) High power of A showing that most BrdU-labeling cells also stain with Dm Geminin. (B) Dm Geminin. (C) BrdU. (D) Merge. (Arrows) Weak BrdU-labeled small nuclei likely to be in early S phase with no Dm Geminin. (*) Non-BrdU-labeled large interphase nuclei containing the highest levels of Dm Geminin. (E) PH3 (red) and Dm Geminin merge. (F–H) High power of E. (F) Dm Geminin. (G) PH3. (H) Merge showing Dm Geminin is abundant in prophase (p), still present in metaphase (m), and absent from anaphase cells (a). (I) Cyclin B (red) and Dm Geminin merge. (J–L) High power of I. (J) Dm Geminin. (K) Cyclin B. (L) Merge. Note the smaller interphase cells (S phase) have low levels of Dm Geminin but no Cyclin B (arrows).
Overexpression of Dm Geminin in Drosophila embryos inhibits DNA replication and leads to cell death
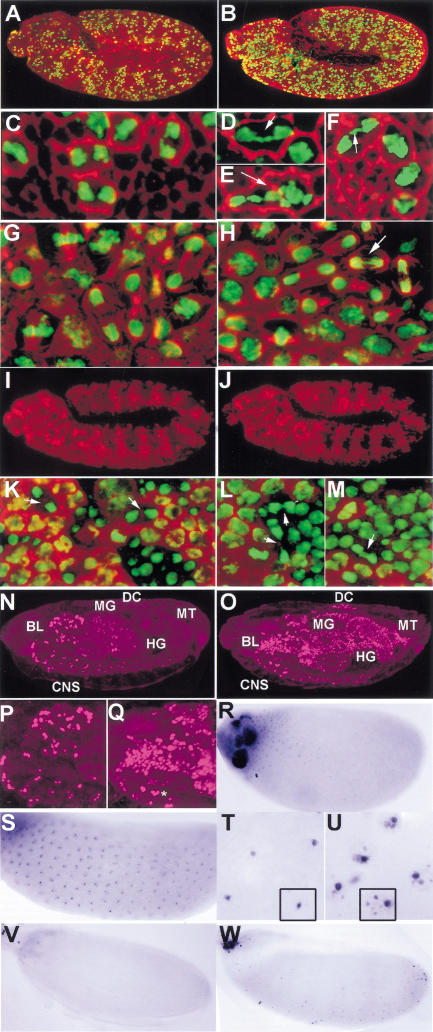
To determine whether overexpression of Dm Geminin acts to inhibit DNA replication in vivo, we generated transgenic flies that contain Dm Geminin under control of the S. cerevisiae UAS(GAL4) promoter. Ectopic overexpression of Dm Geminin during embryogenesis by heat shock induction of hsp70–GAL4 UAS–Dm geminin flies resulted in a general decrease in BrdU-labeling cells in mitotic and endoreplicating tissues (data not shown). To demonstrate this effect more clearly, we used the en–GAL4 driver to overexpress Dm Geminin in a striped pattern during embryogenesis (Fig. 3A–C). Ectopic overexpression of Dm Geminin resulted in a dramatic decrease in S-phase cells within the En stripe relative to surrounding cells (Fig. 5, cf. B and F with A and E). Propidium iodide staining of En–Dm Geminin-expressing cells revealed more condensed nuclei within the En stripes (Fig. 5, cf. D and H with C and G), suggesting that cells were attempting to enter mitosis. Staining with anti-PH3 to detect mitotic cells showed that many cells (4× as many as normal) in the En–Dm Geminin stripe were in mitosis (Fig. 5, cf. K and L with I and J). A similar phenotype is observed in Dup mutants despite the fact that they fail to replicate their DNA (Whittaker et al. 2000) and occurs presumably because the DNA replication checkpoint can only be triggered after the loading of DNA polymerase α onto the pre-RC (Michael et al. 2000). To determine the fate of these cells, we carried out TUNEL to detect apoptotic cells. Wild-type embryos at stage 11 normally show very little TUNEL staining, whereas the En–Dm Geminin stage-11 embryos showed numerous TUNEL-positive cells associated with the En stripes (Fig. 5, cf. N with M). These data show that ectopic overexpression of Dm Geminin results in inhibition in DNA replication, increased numbers of metaphase cells, and increased apoptosis.
Figure 5.
Over-expression of Dm Geminin during Drosophila development reduces DNA replication. (A) A stage-11 wild-type embryo with BrdU (red) and anti-En (green in A–L). (B) A stage-11 en–GAL4 UAS–Dm geminin embryo with BrdU (red) and anti-En showing decreased BrdU labeling in the En stripes. (C) A wild-type stage-11 embryo stained with anti-En and PI (red). (D) A stage-11 en–GAL4 UAS–Dm geminin embryo stained with anti-En and PI (red). (E) High magnification of A. (F) High magnification of B. Arrows indicate En stripes. (G) High magnification of C. (H) High magnification of D. (I,J) High magnification of stage-11 wild-type embryo with anti-En and anti-PH3 (red). (K,L) High magnification of stage-11 en–GAL4 UAS–Dm geminin embryos with anti-En and anti-PH3 showing more PH3 in En stripes. (I–L) Anaphase (a); prophase cells (p); and metaphase cells (m) within the En stripe are indicated. An average of 14.1±4.1 (SE) mitotic cells were present in the En stripe in the en–GAL4 UAS–Dm geminin embryo compared with an average of 3.5 ± 1.2 (SE) in an equivalent sized interstripe region (n = 13). Wild-type embryos contained a similar density of mitotic cells in En stripes compared with interstripe regions. (M) High magnification of TUNEL (blue) and En (red) in a wild-type stage-11 embryo. (N) High magnification of TUNEL (blue) and En (red) in an en–GAL4 UAS–Dm geminin stage-11 embryo showing more TUNEL-positive cells. (O–Q) BrdU labeling of eye imaginal discs from wild type (O), GMR–GAL4 UAS–Dm geminin (3 copies) (P), and ey–GAL4 UAS–Dm geminin (2 copies) (Q). In P and Q the brackets indicate the regions where Dm geminin is ectopically expressed. Wild-type eye discs contained an average of 119±14 (SE) (n = 3) compared with 63±6 (n = 4) S-phase cells for GMR–GAL4 UAS–Dm geminin eye discs within an equivalent region of the post-MF S-phase band. The MF is indicated (*). (R–V) Scanning electron micrograph (SEM) of adult eyes from (R) ey–GAL4 UAS–Dm geminin (2 copies), (S) wild type, (T) GMR–GAL4 UAS-Dm geminin (3 copies), (U) GMR–GAL4 UAS–Dm geminin (2 copies), (V) GMR–GAL4 UAS–Dm geminin (2 copies)/dupa1. Note in V that the eye is smaller and more disorganized than in U. Adult eyes are oriented anterior to the right.
Ectopic overexpression of Dm Geminin using the eyeless(ey)–GAL4 driver, which is expressed during the early proliferative phase of the eye–antennal imaginal disc, also resulted in a dramatic decrease in S phases and in the size of third instar larvae eye discs and the size of the adult eye (Fig. 5, cf. Q and R with O and S). Overexpression of Dm Geminin using the GMR–GAL4 driver, which is expressed posterior to the MF in the eye imaginal discs of third instar larvae, led to a 40%–50% decrease in S-phase cells within this region, but to severely rough adult eyes (Fig. 5, cf. P and T with O and S). Taken together, these data show that ectopic overexpression of Dm Geminin leads to an inhibition of S phases in both mitotic and endoreplicative cycles and at different developmental stages.
Dm Geminin genetically interacts with Dm Cdt1(Dup)
The GMR–Dm geminin rough eye phenotype represents a good phenotype in which to examine genetic interactions. This phenotype was responsive to the dose of Dm Geminin because two copies of UAS–Dm geminin resulted in a less severe phenotype compared with three copies of the transgene (Fig. 5, cf. U with T). In addition, reducing the dose of endogenous Dm Geminin by half using the strong P alleles resulted in a less severe phenotype (data not shown). To determine whether Dm geminin genetically interacted with dup(cdt1), we reduced the dosage of dup by half using a null allele (dupa1; Whittaker et al. 2000) in a GMR–GAL4 UAS–Dm geminin (two copies) background. Halving the dosage of dup enhanced the GMR–Dm geminin eye phenotype, leading to a smaller, rougher eye (Fig. 5, cf. V with U). Moreover, the dupa1 mutant embryonic cycle 16 S-phase defect was suppressed by a Dm geminin mutant (A. Herr and H. Richardson, unpubl.). Therefore, consistent with the biochemical interaction observed between Dm Geminin and Dup (Fig. 2D), Dm geminin genetically interacts with dup.
Dm geminin mutants exhibit mitotic defects in cycle-16 embryos
Of the three P element alleles that are inserted within or 5′ of the Dm geminin transcriptional unit, l(2)09107 is a partially female sterile allele, whereas l(2)k14019 and l(2)k03202 are embryonic/larval lethal (data not shown). The P element allele l(2)k14019 mutant phenotype has been described previously by Roch et al. (1998) and shown to be third instar larval lethal with reduced imaginal discs. When crossed to a deficiency of the region (Df(2R)ST1), 40% of l(2)k14019/Df and l(2)k03202/Df embryos died before hatching and the rest died during larval development. In agreement with the data of Roch et al. (1998), occasional l(2)k14019/Df trans-heterozygous third instar larvae were observed but no third instar l(2)k03202/Df larvae were detected, suggesting that l(2)k03202 is a stronger allele than l(2)k14019.
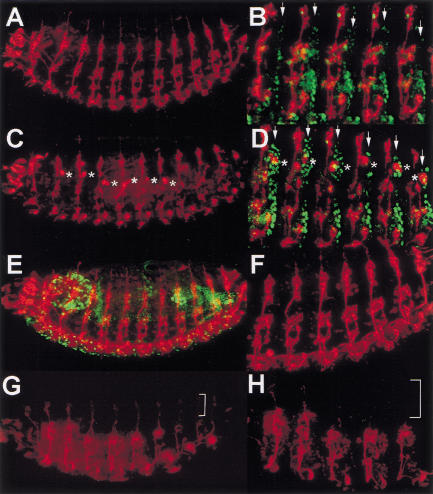
Because l(2)k03202 was the strongest allele, we focused on examining l(2)k03202/Df(2R)ST1 trans-heterozygous embryos for cell cycle defects (Fig. 6). During early embryogenesis, the overall S-phase patterns of the mutant appeared normal and cells exited from S phase at the appropriate developmental time (data not shown). To examine mitoses during cycle 14–16, we stained mutant embryos using anti-PH3 to detect mitotic chromosomes and anti-Actin to visualize cell outlines. Interestingly, l(2)k03202/Df(2R)ST1 mutant embryos at stage 11 undergoing cycle 16 exhibited an increased number of mitotic cells, suggesting that cells were either entering mitosis prematurely or being delayed in mitosis (Fig. 6, cf. B with A). Many of these mitotic cells (∼30%) showed anaphase defects (Fig. 6, cf. D–F with C). These included anaphase chromosome bridges (Fig. 6D,F) and chromosomes apparently severed by the cytokinetic contractile apparatus (“cut” chromosomes; Fig. 6E).
Figure 6.
Dm geminin mutants exhibit mitotic defects and late over-replication defects. (A–F) Stage-11 embryos with anti-Actin (red) and anti-PH3 (green). (A) Wild type. (B) l(2)03202/Df. (C) High-power wild type. (D–F) High-power l(2)03202/Df showing the mitotic defects: chromosome bridges (arrows in D,F); cut chromosomes (arrow in E). (G,H) High power of stage-11 embryos with anti-Tubulin (red) and PI (green). (G) Wild type. (H) l(2)03202/Df embryo showing a normal, appearing mitotic spindle but anaphase defects (arrow). (I,J) Stage-11 embryos with anti-Cyclin B (red). (I) Wild type. (J) l(2)03202/Df. (K) High power of I with Cyclin B (red) and PI (green) showing that Cyclin B is absent in anaphase (arrows) and telophase cells. (L,M) High power of l(2)03202/Df stage-11 embryos with Cyclin B (red) and PI (green) showing anaphase defects in the absence of Cyclin B (arrows). (N) Stage-16 wild-type embryo with BrdU showing S phase in the CNS, the midgut (MG), and some dorsal cells (DC). (O) Stage-16 l(2)03202/Df embryo with BrdU showing increased S-phase cells in the midgut (MG), dorsal cells (DC), hindgut (HG), and Malpighian tubule (MT) endoreplicating domains. (P) High magnification of the brain lobe and CNS from N. (Q) High magnification of the brain lobe and CNS from O showing more S-phase cells with more intense BrdU labeling and increased size (e.g., *). Dm geminin mutant embryos in this and other figures were picked by the absence of LacZ staining used to mark the balancer chromosome (see Materials and Methods). (R) Stage-12 wild-type ovarian follicle cells with BrdU showing chorion gene amplification foci. (S) Stage-12 l(2)k09102/l(2)k14019 ovarian follicle cells with BrdU. (T) High magnification of R. (U) High magnification of S. The boxes in T and U indicate single follicle cell nuclei. Note that whereas only anterior follicle cells label at only one focus in wild type (R,T) there are four labeling foci in all follicle cells in the mutant (S,U). (V) Stage-14 wild-type ovarian follicle cells with BrdU. (W) Stage-14 l(2)03202/Df ovary with BrdU. Note the continuation of DNA amplification in many follicle cells in the mutant (W) compared with wild type (V).
Anaphase defects can be caused by a defect in the anaphase promoting complex (APC) mediated proteolysis of chromosome cohesion proteins, which hold sister chromatid centromeres together (for review, see Biggins and Murray 1998). Because it was clear that the centromeric regions were separating, whereas the chromosome arms were not in Dm geminin mutant cells, chromosome cohesion defects were considered unlikely. Chromosome segregation defects can also arise because of a defective mitotic spindle (e.g., Inoue et al. 2000). In Dm geminin mutant embryos, however, cells with lagging anaphase chromosomes contained an apparently normal mitotic spindle as visualized using an anti-Tubulin antibody (Fig. 6, cf. H with G). The anaphase defects may also arise because or a failure to degrade Cyclin B (for review, see King et al. 1996). In Dm geminin mutant embryos, however, the overall pattern of Cyclin B appeared normal and in cells exhibiting anaphase defects Cyclin B had already been degraded (Fig. 6, cf. J,L, and M with I and K). Therefore the chromosome segregation defects of the Dm geminin mutant are not attributable to the inability to degrade Cyclin B, to defects in centric chromosome cohesion, or to any visible abnormalities of the mitotic spindle. Possible reasons to explain this phenotype are discussed below.
Dm geminin mutants exhibit over-replication defects late in embryogenesis and in oogenesis
Because Dm Geminin is maternally supplied and Dm Geminin is very low but not completely absent in cycle 16 Dm geminin mutants, we examined older Dm geminin mutant embryos for over-replication defects. In stage-16 wild-type embryos, endoreplication occurs in the midgut and the dorsal cells (Fig. 6N). In the Dm geminin mutant, endoreplication occurred in more cells in the midgut and dorsal cell domains and continued in the hindgut and Malpighian tubules domains where it should have ceased (Fig. 6O). This phenotype is consistent with over-replication occurring in these cells. Furthermore, greater numbers of S-phase cells were observed in the CNS than normal (Fig. 6, cf. Q with P). Many cells appeared to label more intensely with BrdU and some were increased in size compared with wild type.
We next examined DNA replication in the follicle cells during oogenesis in females trans-heterozygous for the weak, partially female sterile Dm geminin mutant and a strong P allele (l(2)k09107/l(2)k14019). In stage-10B ovaries, the follicle cells of the Dm geminin mutant had switched from general genomic endoreplication to the amplification cycles as normal (data not shown). In most stage-12 wild-type ovaries, DNA amplification was only observed in the anterior region in one focus per cell (Fig. 6R,T), whereas 100% (n = 20) of Dm geminin mutant stage-12 ovaries showed strong BrdU labeling of four amplification foci in all follicle cells (Fig. 6S,U). By stage 14, all follicle cells of wild-type ovaries had ceased amplification (n = 22), whereas many follicle cells from 50% of Dm geminin mutant ovaries (n = 55) were still continuing amplification (Fig. 6, cf. W with V). Therefore, reduced Dm Geminin function in the Dm geminin mutant leads to continued DNA amplification in follicle cells, suggesting that Dm Geminin has an important role in mediating cessation of these replicative cycles.
A role for Dm Geminin in neurogenesis
Xl geminin was also identified in a screen for genes that induced neural tissue when overexpressed (Kroll et al. 1998). To determine whether Dm Geminin also induced neural differentiation, we overexpressed Dm geminin in embryos using En–GAL4 UAS–Dm geminin and examined these for neural defects by staining with the axonal antibody 22C10. Ectopic overexpression of Dm Geminin in the En stripes (Fig. 7B,D) resulted in the formation of ectopic neuronal cells (Fig. 7, cf. C and D with A and B). These ectopic neural cells appeared more epidermal than the normal PNS neurons consistent with the expression of the En driven-Dm Geminin in the epidermal cells. Although some disruption in the normal PNS pattern was observed, the ectopic neurons are unlikely to be caused by inappropriate migration of normal neurons because En is not expressed in the PNS and these ectopic neurons were epidermal. Not every Dm Geminin overexpressing cell was induced to form a neuron, perhaps because of cell death, which we have shown to occur in some cells when Dm geminin is ectopically overexpressed using the En driver (Fig. 5N). These data, however, shows that overexpression of Dm Geminin is capable of inducing full neural differentiation in at least some cells. Therefore, like Xl Geminin, ectopic overexpression of Dm Geminin is capable of inducing neural differentiation.
Figure 7.
Dm Geminin overexpression and Dm geminin mutants affect neurogenesis. (A–D) Stage-14 embryos with 22C10 (red) and anti-En (green). (A) Wild type with 22C10. (B) High magnification of A with 22C10 and En (arrows). (C) En–GAL4 UAS–Dm geminin with 22C10. (D) High magnification of C with 22C10 and En. Note (in C,D) ectopic neural cells (*) are present within the En stripe (arrows in D). (E–H) Stage 14/15 embryos with 22C10 (red) and anti-Dm Geminin (green). (E) Wild type with Dm Geminin and 22C10. (F) High magnification of E with 22C10. (G) l(2)03202/Df with Dm Geminin and 22C10. (H) High magnification of G with 22C10. Note (in G,H) the decrease in 22C10 staining in the dorsal region (brackets) in the Dm geminin mutant. Dm geminin mutants were detected by reduced Dm Geminin levels and the absence of LacZ (data not shown).
Neural differentiation was then examined in l(2)k03202/Df(2R)ST1 mutant embryos by staining with the 22C10 antibody. Although many Dm geminin mutant embryos showed a mostly normal 22C10 staining pattern, a small percentage had a striking reduction in 22C10 staining of the dorsal-most peripheral neurons (Fig. 7, cf. G and H with E and F). The variability of this phenotype may be a consequence of maternal Dm Geminin depletion. Further analysis is required to determine whether these neural defects may be a secondary consequence of the cell cycle defects observed earlier in development or to a specific function for Dm Geminin in neural differentiation.
Discussion
In this paper we describe a Drosophila homolog of Xenopus (Xl) and human (Hs) Geminin, which has an important role in the block to re-replication by inhibiting Cdt1-mediated binding of Mcms to the pre-replication complex. Drosophila Geminin (Dm Geminin) exhibits all the functional characteristics described for Xl and Hs Geminin: (1) Inhibits DNA replication in a Xenopus in vitro assay by blocking Mcms loading onto chromatin and also when overexpressed in flies; (2) binds to Cdt1(Doubleparked, Dup) in vivo and genetically interacts with dup; (3) accumulates in S/G2 and is degraded at the metaphase to anaphase transition; and (4) when overexpressed, induces neural differentiation. In addition, the expression of Dm Geminin during development and the phenotype of Dm geminin mutants are consistent with Dm Geminin being a functional homolog of Xl and Hs Geminin.
Dm Geminin expression during development
Dm Geminin protein and mRNA (data not shown) expression is correlated with mitotically proliferating and endoreplicating cells. Dm Geminin is absent in G1/early S-phase cells, increases during S/G2 and is degraded at the metaphase to anaphase transition in mitosis, similar to Cyclin B. The similar cell cycle profile observed for Dm Geminin and Cyclin B suggests that these proteins may be degraded by the same mechanism. The destruction box motifs of Dm and Xl Geminin, however, show variations compared with the canonical destruction motifs of Xl or Dm Cyclin B (for review, see King et al. 1996). Furthermore, whereas Dm Cyclin B is degraded by ubiquitin-mediated proteolysis in Xenopus oocyte mitotic extracts (Glotzer et al. 1991), Dm Geminin is not (data not shown), suggesting that Dm Geminin and Cyclin B may be degraded by different mechanisms in Drosophila. Also, because Xl Geminin is degraded in Xenopus oocyte mitotic extracts (McGarry and Kirschner 1998) and Dm Geminin is not, there may be species-specific differences in the Geminin degradation pathway.
Dm Geminin as a neural differentiation factor
Ectopic expression of Dm Geminin in the embryo can induce some cells to ectopically differentiate as neural cells. The reason that not all cells expressing Dm Geminin differentiate into neural cells as occurs with Xl Geminin (Kroll et al. 1998) may be attributable to induction of apoptosis or to the requirement of other neuralizing factors. Ectopic neural differentiation may explain the severity of the eye phenotype observed when Dm Geminin is ectopically overexpressed using the GMR driver, which cannot simply be explained by an S-phase inhibitory effect. GMR-driven Dm Geminin resulted in only a 40%–50% decrease in S-phase cells in the eye imaginal disc, whereas the eye phenotype was more severe than that observed when post-MF S phases were almost completely ablated by GMR-human p21 (de Nooij and Hariharan 1995). GMR-driven expression of the apoptosis inhibitor p35 partially rescues the eye phenotype by increasing the size of the eyes, but they are still disorganized (A. Herr and H. Richardson, unpubl.), suggesting that apoptosis contributes to the rough eye phenotype but that other mechanisms, perhaps ectopic neural differentiation, may also be involved. Further analysis is required to determine whether Dm Geminin has a physiological role in neurogenesis during development. If Geminin is involved in neurogenesis, however, it must function at an early stage in neural differentiation as Geminin expression in Xenopus and Drosophila is specific to cells that are actively cycling (Kroll et al. 1998; Saka and Smith 2001; this study).
The role of Dm Geminin in preventing re-replication
Evidence of over-replication was observed in Dm geminin mutants in (1) endoreplicating tissues in stage 16 embryos; (2) the CNS in stage 16 embryos; and (3) DNA amplification cycles of ovarian follicle cells. These data are consistent with excessive DNA replication occurring in Dm geminin mutants. Together with the inhibition of DNA replication by Geminin overexpression, these data show that Dm Geminin has a role in limiting DNA replication. Because Dm Geminin is absent before S-phase initiation in the mitotic cycles, the presence of Dm Geminin may be sufficient to limit DNA replication. Dm Geminin is present throughout the endoreplication and amplification cycles, suggesting that other factors are required to activate Dm Geminin to limit DNA replication in these cycles.
Our inability to observe excessive DNA replication early in embryogenesis is most likely attributable to the maternal supply of Dm Geminin. In Xenopus, however, in vitro DNA replication assays, antibody ablation of >99% of endogenous Xl Geminin resulted in no apparent re-replication as measured by BrdU labeling and density gradient analysis (McGarry and Kirschner 1998). Furthermore, the terminal phenotype of Geminin depletion from Xenopus eggs causes abnormalities at the blastula stage that is different to the Drosophila geminin mutant phenotype (T.J. McGarry, in prep.). The failure to observe excessive replication in Geminin-ablated Xenopus embryos may indicate that DNA replication licensing is regulated differently depending on developmental times or in different organisms.
Functional analysis of Xl Geminin has separated the neuralizing and DNA replication inhibition domains, suggesting that Geminin is dual functional (Kroll et al. 1998; McGarry and Kirschner 1998). Dual functional proteins that can act as negative cell cycle regulators and differentiation inducers are not unique because Cdk inhibitors of the p21 family and the Rb family also function in this manner (for review, see Zhu and Skoultchi 2001). Interestingly, the Cdk inhibitors human p21CIP1 and Xenopus p27XIC1 also have separable cell cycle inhibition and differentiation functional domains (Ohnuma et al. 1999). We are presently investigating whether the neuralization and DNA replication inhibition functions of Dm Geminin are also functionally separable.
Geminin and anaphase defects
The anaphase defects observed in cycle 16 Dm geminin mutant embryos may indicate that Dm Geminin also has a role in anaphase. Given the evidence for Geminin in the block to re-replication in other organisms and our observation of over-replication defects later in development, however, the most likely reason for the anaphase defects in the Dm geminin mutant cycle 16 embryos is caused by prior S-phase defects. Although our BrdU-labeling studies did not reveal over-replication defects before mitosis of cycle 16, a low amount of re-replication would be difficult to detect. Over-replication defects that fail to elicit the DNA replication or damage checkpoints (for review, see Clarke and Gimenez-Abian 2000), allowing entry into mitosis, would result in drastic consequences. If chromosomes were partially over-replicated and the over-replicated portion included the centromere, then chromosome bridges would be expected to occur during anaphase. Therefore, the anaphase defects we observed may be a manifestation of over-replication in Dm geminin mutants. If this is the case, then Geminin has an important role in ensuring that DNA re-replication does not occur before chromosome segregation.
The anaphase defects of Dm geminin mutant embryos are similar to those observed in mutants of Drosophila lodestar, topI, and barren, which are involved in decatenization and unwinding of replicated DNA (Girdham and Glover 1991; Bhat et al. 1996; Zhang et al. 2000). Whether the defects observed in Dm geminin mutants are related to chromosome decatenization and whether lodestar, topI, and barren also show over-replication defects, requires further investigation.
In summary, this study describes the first analysis of the role of Drosophila geminin during development. Dm geminin mutants show over-replication defects, anaphase defects, and neural differentiation defects. Together with our analysis of the ectopic expression of Dm Geminin, our data suggest that Geminin has functions in preventing re-replication, in anaphase and in neural differentiation.
Materials and methods
Mutant analysis, generation of transgenic flies, and genetic interactions
The Dm geminin P-element alleles l(2)k03202, l(2)k14019, and l(2)k09107 were obtained from BDGP. l(2)k03203 was listed in the BDGP database as having a second P element on the X chromosome. By genetic crosses, the mutant X chromosome was replaced with a wild-type X chromosome. The lethality of l(2)k03202 is attributable to the P insertion in Dm geminin, as after P-element excision using Δ2–3 transposase, the lethality was reverted. For the analysis of mutant embryos, the Dm geminin mutants' second chromosome was balanced over a CyO wingless–lacZ chromosome, enabling homozygous embryos to be distinguished by LacZ antibody staining.
The Dm geminin cDNA sequence was determined by sequencing the cDNA clones GM06555, LD07974, and LD04903. The location of the l(2)k03202 P-element insertion was determined by inverse PCR of SspI digested and re-ligated genomic DNA from l(2)k03202/CyO flies using the following primers: Pry4, 5′-CAATCATATCGCTGTCTCACTCA-3′ and Pry1, 5′-GGAATGCATGTCCGTGGGGTTTGAAT-3′.
To generate the UAS–Dm geminin construct, full-length Dm geminin cDNA (LD07974) was subcloned into pUAST and transgenic flies were generated as described previously (Richardson et al. 1995). A UAS–Dm geminin transgene on the third chromosome was used for all experiments. Flies containing three copies of UAS–Dm geminin and one copy of GMR–GAL4 (GMR–GAL4 UAS–Dm geminin/CyO; UAS–Dm geminin/UAS–Dm geminin) were generated by creating recombinants of GMR–GAL4 and UAS–Dm geminin on the second chromosome, followed by crossing in the UAS–Dm geminin transgene on the third chromosome. All fly stocks were obtained from the Bloomington Stock Centre except the dupα1 mutant, which was provided by T. Orr-Weaver (Whittaker et al. 2000).
Generation of Dm Geminin antisera, coimmunoprecipitation, and immunoblotting
To express Dm Geminin in bacteria, the coding region of Dm geminin was PCR-amplified from the cDNA clone and cloned into pGEX-2T using Pfu polymerase and the following PCR primers: forward primer 5′-GCGGATCCATGTCTTCGAGC GCTGCCAGGGTC-3′, reverse primer 5′-GCGGATCCCTAG GCGTTGACCTTGTCCTCGTC-3′ (BamHI sites used for cloning are underlined). Bacterially expressed Dm Geminin protein was purified on a SDS-polyacrylamide gel and used to inoculate rats. After three boosts, the serum was harvested and used crude or after purification on a protein G sepharose column.
For Western analysis, hsp70–GAL4 UAS–Dm geminin embryos from a 4-h lay aged for 16 h at 18°C were heat shocked for 1 h and recovered for 1 h before collection. A similar time collect of wild-type embryos were either heat shocked as for the hsp70–GAL4 UAS–Dm geminin or aged for a further 2 h before collection. Embryos from heterozygous Dm Geminin adults were also from a 4-h lay aged for 16 h at 18°C (to decrease the levels of the maternally supplied product). Protein was quantitated using the BCA protein assay kit (Pierce).
For Immunoprecipitation extracts from a 0- to 16-h embryo collect from w1118, embryos were homogenized in HoB buffer (25 mM HEPES at pH 7.4, 150 mM NaCl, 5 mM NaF, 1 mM EDTA, 1 mM DTT, 0.1 mM Na3VO4, 0.1% Triton X-100, and one complete Mini-EDTA free Protease Inhibitor cocktail tablet [Boehringer Mannheim]). Dm Geminin (30 μL of 1:500) or Dm Dup antibodies (30 μL of 1:1000) bound to protein A Sepharose CL-4B beads were incubated with embryonic extracts, precipitated by centrifugation, and resuspended in protein sample buffer before loading on a 12% SDS-polyacrylamide gel. Western analysis was carried out using the Dm Geminin or Dup antibodies followed by horse radish peroxidase (HRP)-conjugated anti-rat or anti-guineapig antibodies, respectively. HRP was detected using the ECL-Plus kit (Amersham Pharmacia).
Antibody staining, BrdU labeling, and microscopy
BrdU labeling and antibody staining of Drosophila tissues or embryos were carried out as described previously unless otherwise indicated (Richardson et al. 1995; Secombe et al. 1998). For anti-Tubulin antibody staining, embryos were fixed in 5 μM Taxol (to stabilize the microtubules) and 50 mM EGTA for 1 h in 8% paraformaldehyde. For ovaries, virgin adult females were collected and aged in the presence of males for 36–48 h at 25°C before dissection.
Rat anti-Dm Geminin antibody was detected using an anti-rat-biotin-conjugated secondary antibody followed by streptavidin-Alexa 488 (Molecular Probes). Mouse monoclonal 22C10 (Developmental studies hybridoma bank) was detected using anti-mouse-rhodamine. To detect En, the anti-En monoclonal antisera (Developmental studies hybridoma bank), anti-mouse rhodamine (raised in sheep), and anti-sheep rhodamine were used. For double BrdU labeling and antibody staining, the antibody staining was carried out first on BrdU-labeled samples and detected by biotinylated secondary antibodies followed by streptavidin-Alexa 488 (Molecular Probes). Embryos were re-fixed in 8% paraformaldehyde/PBT before acid hydrolysis and detection of BrdU with the anti-BrdU monoclonal antibody (Becton Dickenson) followed by anti-mouse biotin and streptavidin-lissamine rhodamine. TUNEL staining was carried out using the Roche AP in situ cell death detection kit as described previously (Quinn et al. 2000). In the TUNEL-stained embryos, En was detected using the anti-En antibody as described above but detection was via streptavidin-alkaline phosphatase (AP) using the fast-red (Roche) substrate. All secondary antibodies were from Jackson Immunochemicals unless otherwise indicated. Other antibodies were mouse monoclonal anti-Tubulin (Developmental studies hybridoma bank), rabbit anti-phosphohistone H3 (Santa Cruz), and rabbit anti-LacZ (Rockland). All fluorescent-labeled samples were analyzed by confocal microscopy (Biorad MRC1000), whereas HRP or AP colormetrically detected samples were analyzed on the Zeiss Axiophot Photophot using Nomarski optics.
Scanning electron micrographs of adult eyes were carried out as described previously on a Field Emission Scanning Electron Microscope (Secombe et al. 1998).
DNA replication and chromatin-binding assays
To purify Dm Geminin for DNA replication and chromatin-binding assays, Dm geminin was expressed in bacteria in the pET28a vector. Dm geminin from pGEX-2T was amplified by PCR and ligated into pET28a using the following primers: forward 5′-GCGCGCCATATGTCTTCGAGCGCTGCC-3′; and reverse 5′-GCGCGCCTCGAGCTAGGCGTTGACCTTGTCC TC-3′ (NdeI and XhoI sites are underlined). His-tagged Dm Geminin was expressed and purified on nickel-agarose beads using standard techniques (QIAGEN). The Dm Geminin protein was dialyzed against 10 mM HEPES (pH 7.7), 200 mM NaCl before use.
DNA replication assays and chromatin-binding assays were performed from CSF-arrested Xenopus extracts as described previously (McGarry and Kirschner 1998). For chromatin-binding assays, replication assays were diluted into a buffer containing a non-ionic detergent and the chromatin was pelleted through a sucrose cushion.
Acknowledgments
We thank Matthew O'Connell and Patrick Humbert for critical comments on the manuscript. We are grateful to Dr. Terry Orr-Weaver for the gift of anti-Dup; Ron Laskey for the anti-Xl Mcm3; and Dr. David Glover for the anti-Dm Cyclin B antibodies. We thank Drs. Kris Kroll, Terry Orr-Weaver, and Hui Zhang for communication of unpublished data. We acknowledge the technical assistance of Tod Fullston, Michelle Coombe, and the staff at the Animal House at Adelaide University. This work was supported by the Wellcome Trust and previously by the Australian Research Council Special Research Center in the Molecular Genetics of Development at the University of Adelaide. H.R. is a Wellcome Senior Research Fellow in Medical Science.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL h.richardson@pmci.unimelb.edu.au; FAX 613-965-614-11.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.916201.
References
- Bhat MA, Philo AV, Glover DM, Bellen HJ. Chromosome segregation at anaphase requires the barren product, a novel chromosome associated protein that interacts with Topoisomerase II. Cell. 1996;87:1103–1114. doi: 10.1016/s0092-8674(00)81804-8. [DOI] [PubMed] [Google Scholar]
- Biggins S, Murray AW. Sister chromatid cohesion in mitosis. Curr Opin Cell Biol. 1998;10:769–775. doi: 10.1016/s0955-0674(98)80120-8. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988;332:546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Prokhorova TA. Saying a firm 'no' to DNA re-replication. Nat Cell Biol. 1999;1:E175–E177. doi: 10.1038/15611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi BR, Lilly MA, Spradling AC. Cell cycle control of chorion gene amplification. Genes & Dev. 1998;12:734–744. doi: 10.1101/gad.12.5.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke DJ, Gimenez-Abain JF. Checkpoints controlling mitosis. BioEssays. 2000;22:351–363. doi: 10.1002/(SICI)1521-1878(200004)22:4<351::AID-BIES5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- de Nooij JC, Hariharan IK. Uncoupling cell fate determination from patterned cell division in the Drosophila eye. Science. 1995;270:983–985. doi: 10.1126/science.270.5238.983. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Lehner CF. Developmental control of cell cycle regulators: A fly's perspective. Science. 1996;274:1646–1652. doi: 10.1126/science.274.5293.1646. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Orr-Weaver TL. Endoreplication cell cycles: More for less. Cell. 2001;105:297–306. doi: 10.1016/s0092-8674(01)00334-8. [DOI] [PubMed] [Google Scholar]
- Ekholm SV, Reed SI. Regulation of G1 cyclin dependent kinases in the mammalian cell cycle. Curr Opin Cell Biol. 2000;12:676–684. doi: 10.1016/s0955-0674(00)00151-4. [DOI] [PubMed] [Google Scholar]
- Girdham CH, Glover DM. Chrosomome tangling and breakage at anaphase results from mutations in lodestar, a Drosophila gene encoding a putative nucleoside triphosphate-binding protein. Genes & Dev. 1991;5:1786–1799. doi: 10.1101/gad.5.10.1786. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Inoue YH, do Carmo Avides M, Shiraki M, Deak P, Yamaguchi M, Nishimoto Y, Matsukage A, Glover DM. Orbit, a novel microtubule-associated protein essential for mitosis in Drosophila melanogaster. J Cell Biol. 2000;149:153–166. doi: 10.1083/jcb.149.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Salic AN, Evans LM, Kirschner MW. Geminin, a neuralizing molecule that demarcates the future neural plate at the onset of gastrulation. Development. 1998;125:3247–3258. doi: 10.1242/dev.125.16.3247. [DOI] [PubMed] [Google Scholar]
- Lehner CF, O'Farrell PH. The roles of Drosophila cyclins A and B in mitotic control. Cell. 1990;61:535–547. doi: 10.1016/0092-8674(90)90535-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Tye BK. Initiating DNA synthesis: from recruiting to activating the MCM complex. J Cell Sci. 2001;114:1447–1454. doi: 10.1242/jcs.114.8.1447. [DOI] [PubMed] [Google Scholar]
- Mahbubani HM, Chong JP, Chevalier S, Thommes P, Blow JJ. Cell cycle regulation of the replication licensing system: involvement of a Cdk-dependent inhibitor. J Cell Biol. 1997;136:125–135. doi: 10.1083/jcb.136.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano D, Moreau J, Mechali M. XCdt1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature. 2000;404:622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- Michael WM, Ott R, Flanning E, Newport J. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science. 2000;289:2133–2137. doi: 10.1126/science.289.5487.2133. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z, Nishimoto T, Nurse P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature. 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- Ohnuma S, Philpott A, Wang K, Holt CE, Harris WA. p27XIC1, a Cdk inhibitor, promotes the determination of glial cells in Xenopus retina. Cell. 1999;99:499–510. doi: 10.1016/s0092-8674(00)81538-x. [DOI] [PubMed] [Google Scholar]
- Quinn LM, Dorstyn L, Colussi PA, Chen P, Coombe M, Abrams J, Kumar S, Richardson H. An essential role for the caspase Dronc in developmentally programmed cell death in Drosophila. J Biol Chem. 2000;275:40416–40424. doi: 10.1074/jbc.M002935200. [DOI] [PubMed] [Google Scholar]
- Richardson HE, O'Keefe LV, Marty T, Saint R. Ectopic cyclin E expression induces premature entry into S phase and disrupts pattern formation in the Drosophila eye imaginal disc. Development. 1995;121:3371–3379. doi: 10.1242/dev.121.10.3371. [DOI] [PubMed] [Google Scholar]
- Ritzi M, Knippers R. Initiation of genome replication: Assembly and disassembly or replication-competent chromatin. Gene. 2000;245:13–20. doi: 10.1016/s0378-1119(00)00020-2. [DOI] [PubMed] [Google Scholar]
- Roch F, Serras F, Cifuentes FJ, Corominas M, Alsina B, Amoros M, Lopez-Varea A, Hernandez R, Guerra D, Cavicchi S, et al. Screening of larval/pupal P-element induced lethals on the second chromosome in Drosophila melanogaster: Clonal analysis and morphology of imaginal discs. Mol Gen Genet. 1998;257:103–112. doi: 10.1007/pl00008620. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Miklos GLG, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2218. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Smith JC. Spatial and temporal patterns of cell division during early Xenopus embryogenesis. Dev Biol. 2001;229:307–318. doi: 10.1006/dbio.2000.0101. [DOI] [PubMed] [Google Scholar]
- Secombe J, Pispa J, Saint R, Richardson H. Analysis of a Drosophila cyclin E hypomorphic mutation suggests a novel role for Cyclin E in cell proliferation. Genetics. 1998;149:1867–1882. doi: 10.1093/genetics/149.4.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TT, Sprenger F, DiGregogio P, Campbell SD, O'Farrell PH. Exit from mitosis in Drosophila syncitial embryos required proteolysis and cyclin degradation and is associated with localized dephosphorylation. Genes & Dev. 1998a;12:1495–1503. doi: 10.1101/gad.12.10.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TT, Campbell SD, O'Farrell PH. The cell cycle program in germ cells of the Drosophila embryo. Dev Biol. 1998b;196:160–170. doi: 10.1006/dbio.1998.8855. [DOI] [PubMed] [Google Scholar]
- Sun W, Hola M, Pedley K, Tada S, Blow JJ, Todorov IT, Kearsey SE, Brooks RF. The replication capacity of intact mammalian nuclei in Xenopus egg extracts declines with quiescence, but the residual DNA synthesis is independent of Xenopus MCM proteins J. Cell Sci. 2000;113:683–695. doi: 10.1242/jcs.113.4.683. [DOI] [PubMed] [Google Scholar]
- Tada S, Li A, Maiorano D, Mechali M, Blow J. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by Geminin. Nat Cell Biol. 2001;3:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thommes P, Kubota Y, Takisawa H, Blow JJ. The RLF-M component of the replication licensing system forms complexes containing all six MCM/P1 polypeptides. EMBO J. 1997;16:3312–3319. doi: 10.1093/emboj/16.11.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker AJ, Royzman I, Orr-Weaver TL. Drosophila Double parked: A conserved, essential replication protein that colocalizes with the origin recognition complex and links DNA replication with mitosis and the down-regulation of S phase transcripts. Genes & Dev. 2000;14:1765–1776. [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by Geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. The beginning of pattern formation in the Drosophila compound eye: The morphogenetic furrow and the second mitotic wave. Development. 1991;113:841–850. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- Zhang CX, Chen AD, Gettel NJ, Hsieh T. Essential functions of DNA topoisomerase I in Drosophila melanogaster. Dev Biol. 2000;222:27–40. doi: 10.1006/dbio.2000.9704. [DOI] [PubMed] [Google Scholar]
- Zhu L, Skoultchi AI. Coordinating cell proliferation and differentiation. Curr Opin Genet Dev. 2001;11:91–97. doi: 10.1016/s0959-437x(00)00162-3. [DOI] [PubMed] [Google Scholar]