Abstract
The RecQ DNA helicases, human BLM and yeast Sgs1, form a complex with topoisomerase III (Top3) and are thought to act during DNA replication to restart forks that have paused due to DNA damage or topological stress. We have shown previously that yeast cells lacking SGS1 or TOP3 require MMS4 and MUS81 for viability. Here we show that Mms4 and Mus81 form a heterodimeric structure-specific endonuclease that cleaves branched DNA. Both subunits are required for optimal expression, substrate binding, and nuclease activity. Mms4 and Mus81 are conserved proteins related to the Rad1–Rad10 (XPF/ERCC1) endonuclease required for nucleotide excision repair (NER). However, the Mms4–Mus81 endonuclease is 25 times more active on branched duplex DNA and replication fork substrates than simple Y-forms, the preferred substrate for the NER complexes. We also present genetic data that indicate a novel role for Mms4–Mus81 in meiotic recombination. Our results suggest that stalled replication forks are substrates for Mms4–Mus81 cleavage—particularly in the absence of Sgs1 or BLM. Repair of this double-strand break (DSB) by homologous recombination may be responsible for the elevated levels of sister chromatid exchange (SCE) found in BLM−/− cells.
Keywords: DNA replication, DNA helicase, endonuclease, topoisomerase, recombination
Efficient DNA replication requires that forks move unimpeded throughout the genome. However, even under optimal conditions, forks are blocked by transcription, DNA packaging proteins, and topological stress. Another significant cause of replication fork arrest is DNA damage. The process of re-establishing fork movement has been studied most productively in bacteria, in which connections have been identified between this process and recombinational repair enzymes (Cox 2001). Because of their role in restarting replication, mutations in recombination enzymes are often associated with DNA damage sensitivity, even though the repair systems themselves (e.g., excision repair) are fully functional (Courcelle and Hanawalt 2001). Moreover, failure to efficiently resume synthesis can lead to double-strand DNA breaks (DSBs), hyper-recombination, and genome rearrangements. One pathway for restoring replication forks requires both excision repair and recF function (Courcelle and Hanawalt 2001). recF is required to resume DNA synthesis following UV-induced DNA damage and to stabilize the nascent strands at stalled forks (Courcelle et al. 1997). Two other members of the recF pathway, the RecJ exonuclease and the RecQ helicase, are required for limited degradation of nascent strands at stalled replication forks (Courcelle and Hanawalt 1999). This degradation may be important for proteins to gain access to the fork so as to stabilize it and reload the replication machinery.
In eukaryotic cells, RecQ DNA helicases comprise a family of proteins required for genome stability and resistance to DNA-damaging agents (Chakraverty and Hickson 1999). The human diseases Bloom's syndrome, Werner's syndrome, and a subset of Rothmund-Thomson syndrome are caused by mutations in the RecQ helicase genes BLM, WRN, and RTS (RECQL4), respectively. These diseases are characterized by a predisposition to cancer and the mutant cell lines exhibit cytogenetic evidence of DNA rearrangements. The yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe each contain a single RecQ helicase, Sgs1 and Rqh1, respectively. Mutations in SGS1 result in increased rates of recombination, impaired sporulation, and an increased sensitivity to DNA-damaging agents as well as the DNA synthesis inhibitor hydroxyurea (HU) (Gangloff et al. 1994; Watt et al. 1996; Mullen et al. 2000). rqh1 mutants were identified in fission yeast on the basis of their sensitivity to HU and were shown to be defective in recovery from S-phase arrest (Stewart et al. 1997). These studies suggest that recovery from DNA synthesis arrest is a conserved function of RecQ DNA helicases.
The eukaryotic RecQ homologs described above share a helicase domain with Escherichia coli RecQ, but also contain an N-terminal extension. The WRN protein is unique among these enzymes in that its N-terminal domain displays a 3′–5′ exonuclease activity (Huang et al. 1998). WRN copurifies with a number of replication proteins including PCNA and DNA topoisomerase I (Top1) (Lebel et al. 1999). In contrast to WRN, BLM and the yeast enzymes are closely associated with DNA topoisomerase III (Top3). The yeasts require Top3 for optimal growth or viability, but this requirement is bypassed by mutations in SGS1 or rqh1+ (Gangloff et al. 1994; Goodwin et al. 1999). Top3 physically interacts with Sgs1 and BLM, and in both cases it binds the extreme N terminus of its cognate helicase (Gangloff et al. 1994; Bennett et al. 2000; Wu et al. 2000; Fricke et al. 2001; Hu et al. 2001). In the cell, BLM colocalizes with Top3α in PML bodies (Johnson et al. 2000; Wu et al. 2000; Hu et al. 2001). Interestingly, human cells lacking PML display elevated rates of sister-chromatid exchange (SCE), which is the primary genomic instability associated with Bloom's syndrome cells (Zhong et al. 1999). The interaction between BLM and Top3 may underlie its ability to functionally complement yeast sgs1 phenotypes relative to WRN (Heo et al. 1999).
Eukaryotic Top3 displays weak superhelical relaxing activity and, like Top1 from Escherichia coli, it requires access to ssDNA regions for this activity (Kim and Wang 1992). On the basis of its poor relaxing activity, it was suggested that Top3's in vivo function may involve recombination as opposed to the relaxation of superhelical stress (Wang et al. 1990). Interestingly, E. coli Top3 also displays poor relaxing activity but is particularly active in the resolution of replicated pBR322 molecules in an in vitro DNA replication system (DiGate and Marians 1988). Further, Top3 from yeast or E. coli has been shown to cooperate with RecQ DNA helicase in the catenation of circular dsDNA (Harmon et al. 1999). These results support the suggestion that eukaryotic Top3 may act at paused or converging replication forks to unlink the parental duplex (Wang 1991).
The exact mechanism by which these proteins act to restore replication forks is not known. One line of thought involves the regression of stalled forks into Holliday junctions (HJs). There is good evidence that this occurs in E. coli cells deficient in replicative DNA helicase activity (Seigneur et al. 1998). In support of this idea, Sgs1 is known to bind branched DNA and its DNA helicase activity can be assayed on a number of branched substrates including HJs (Bennett et al. 1999). In the case of BLM, it has been suggested that the role of the helicase is to act on these regressed forks and return the HJs to their original form (Karow et al. 2000). On the other hand, mounting evidence suggests that the activities of these RecQ helicases and Top3 are codependent. Sgs1 appears to be constitutively bound to Top3 in yeast cell extracts (Fricke et al. 2001) and most, if not all, sgs1 phenotypes are mimicked or exacerbated in Top3 mutants (Gangloff et al. 1994). It is also revealing that sgs1Δ phenotypes are more efficiently complemented by the Top3 interaction domain of Sgs1 than by its helicase domain (Mullen et al. 2000).
To further characterize the in vivo function of Sgs1, we previously isolated several slx mutants that require SGS1 for viability (Mullen et al. 2001). The mutations identified in this screen appear to define a pathway for bypassing Sgs1–Top3 function because they are lethal in combination with sgs1Δ, sgs1-hd (a helicase-defective allele), or top3Δ. Mutations in MMS4 and MUS81 (formerly SLX2 and SLX3), were found to generate identical phenotypes that parallel those of sgs1 mutants. These include UV and MMS sensitivity, complete loss of sporulation, and synthetic growth defects with mutations in TOP1 (Mullen et al. 2001). Homologs of Mms4 and Mus81 are found in numerous species, including humans, and they share amino acid sequence similarity with the Rad1–Rad10 (XPF-ERCC1) endonuclease that functions in nucleotide excision repair (NER; Bardwell et al. 1994). In this study, we tested the biochemical implications of these results. We show that Mms4 and Mus81 form a structure-specific endonuclease and that one of its preferred substrates is a replication fork. The data suggest that the Mms4–Mus81 endonuclease cleaves stalled replication forks, allowing replication to continue in the absence of Sgs1–Top3 function.
Results
Recombinant Mms4 and Mus81 form a heterodimeric complex
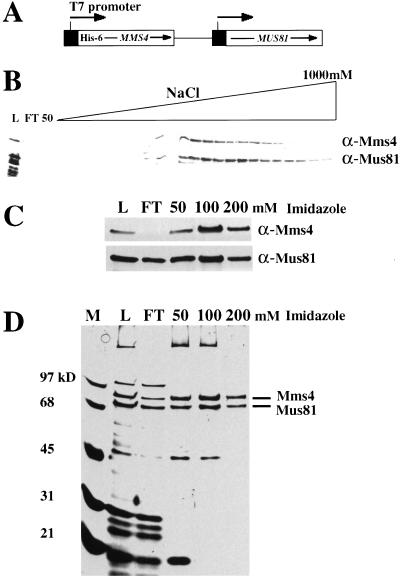
We had shown previously that the Mms4 and Mus81 proteins are associated in crude yeast extracts (Mullen et al. 2001). To determine whether these proteins form a simple heterodimeric complex, we expressed them together in E. coli. Each gene was subcloned downstream of an inducible promoter and the two expression cassettes were combined in a single plasmid (Fig. 1A). For purification purposes, the Mms4 protein was provided with a hexa-histidine tag at its N terminus. Extracts from induced cells were fractionated by phosphocellulose column chromatography and analyzed by immunoblot using antibodies raised against Mms4 or Mus81. Both proteins bound the resin and they eluted simultaneously at ∼650 mM NaCl (Fig. 1B). Peak fractions from this column were applied to a Ni-agarose column and eluted with increasing concentrations of imidazole. Immunoblotting these fractions indicated that both proteins bound the resin and could be eluted with imidazole (Fig. 1C). Silver staining revealed that the 50 and 100-mM imidazole fractions contained highly purified Mms4 and Mus81, whereas the 200-mM imidazole fraction consisted of a nearly homogeneous preparation of these proteins (Fig. 1D). Further analysis of this material revealed that Mms4 and Mus81 cofractionated under stringent immunoprecipitation (Mullen et al. 2001) and gel filtration conditions (500 mM NaCl; data not shown). We conclude that Mms4 and Mus81 form a tight complex. To determine the stoichiometry of the complex, additional preparations were subjected to gel filtration chromatography, SDS-PAGE, and densitometric scanning of Coomassie blue-stained gels. On the basis of size, a simple 1:1 molar ratio of Mms4–Mus81 predicts that the intensity of the stained bands will have a ratio of 1.13 (711/632 amino acids). Experimentally, we obtained a value of 1.15 ± 0.02 (data not shown). Thus, we conclude that Mms4 and Mus81 are present in a 1:1 molar ratio in this complex.
Figure 1.
Expression and purification of the Mms4–Mus81 complex. (A) MMS4 and MUS81 were subcloned downstream of the T7 promoters of pET28a and pET11a, respectively, and further combined to yield a single plasmid, pNJ6220, carrying the indicated expression cassettes. (B) An extract from induced E. coli cells carrying plasmid pNJ6220 was applied to a phosphocellulose column, washed, and eluted with the indicated salt gradient. Column fractions were then resolved by SDS-PAGE, Western blotted, and probed with antiserum against Mms4 or Mus81 as indicated at right. (L) Load; (FT) flow through. (C) Fractions containing immunoreactive material in B were pooled, applied to a Ni-agarose column, washed, and eluted with buffer containing the indicated concentrations of imidazole. Fractions were Western blotted and probed with the indicated anti-serum. (D) Fractions in C were resolved by SDS-PAGE and stained with silver. (M) Molecular weight markers.
Structure-specific endonuclease activity of the Mms4–Mus81 complex
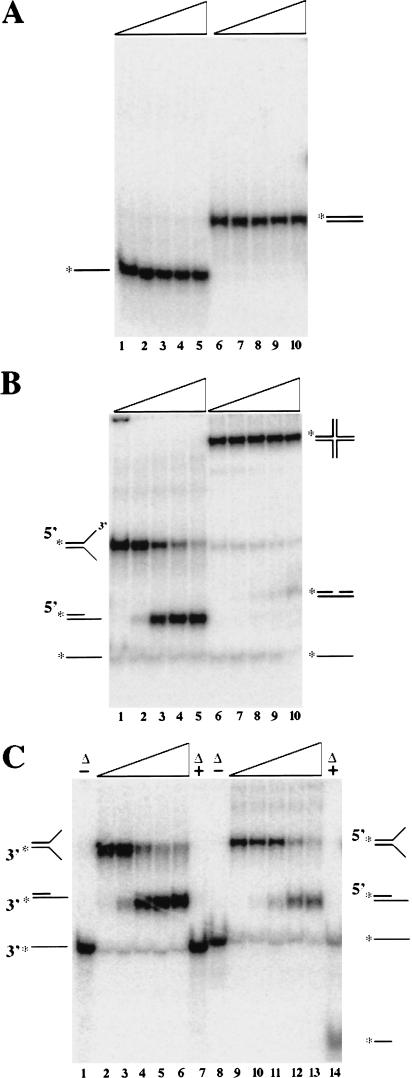
The amino acid sequence similarity of Mms4–Mus81 to the yeast Rad1–Rad10 proteins suggested that it might possess a nuclease activity. To test this hypothesis, we incubated the Mms4–Mus81 complex with a variety of 32P-labeled DNA substrates in the presence of Mg2+ and analyzed the products by polyacrylamide gel electrophoresis. Neither single-stranded nor double-stranded DNA was affected by incubation with Mms4–Mus81 (Fig. 2A). In contrast, when increasing amounts of Mms4–Mus81 were incubated with a branched substrate consisting of duplex DNA with noncomplementary tails (Y-form), nearly complete cleavage was obtained (Fig. 2B, lanes 1–5). The product migrated to a position that was intermediate to that of the substrate and linear ssDNA, indicating that a portion of the molecule had been removed. Nuclease activity was dependent on Mg2+ and was completely inhibited in the presence of excess EDTA (data not shown). Cleavage is relatively specific for the Y-form as another branched substrate, a HJ, was only partially cleaved by the highest concentration of the complex (Fig. 2B, lanes 6–10). When the cleaved products of fixed HJs were analyzed by denaturating gel electrophoresis, we observed cutting at numerous positions that were not localized to the crossover site (data not shown). We conclude that this weak cleavage is nonspecific and may be analogous to the random cleavage of HJs that has been observed with the yeast Rad1 protein (Habraken et al. 1994).
Figure 2.
The Mms4–Mus81 complex is a structure-specific endonuclease. A variety of 32P-labeled model substrates were incubated with increasing amounts of Mms4–Mus81 and the products were resolved on a native polyacrylamide gel prior to autoradiography. (A) Unbranched substrates. (Lanes 1–5) A 49-nucleotide single-stranded DNA (oligonucleotide 892*); (lanes 6–10) double-stranded DNA (892*/897). (B) Branched substrates. (Lanes 1–5) A Y-shaped substrate (892*/895); (lanes 6–10) a Holliday junction (892*/893/894/895). (C) Cleavage specificity. (Lanes 1–7) A Y-shaped substrate (892/895*), in which oligo 895 is 3′ end labeled; (lanes 8–14) the same substrate in which oligo 892 is 5′ end labeled. Titrations illustrated by a triangle represent 0, 2, 4, 8, and 20 ng of Mms4–Mus81. (*) The presence of a 32P-phosphate label at the 5′ position unless indicated otherwise. (Δ) The reaction mixture was boiled prior to loading.
To determine which of the strands had been cleaved from the Y-form substrate, a portion of the cleavage product was denatured by boiling prior to gel electrophoresis. By use of a substrate in which the 3′ end of the duplex portion was labeled, the digested and boiled product migrated at the position of unit-length ssDNA (Fig. 2C, lane 7). This indicates that the unlabeled strand of this substrate had been cleaved. To confirm this interpretation, we labeled the Y-form substrate at the 5′ end of the duplex and treated it as above. As expected, a band migrating faster than unit length linear was obtained (Fig. 2C, lane 14). We conclude that Mms4–Mus81 cleaves the 3′ noncomplementary strand of the Y-form substrate. With respect to this substrate, Mms4–Mus81 has the same cleavage specificity as the Rad1–Rad10 (XPF–ERCC1) endonuclease (Bardwell et al. 1994; de Laat et al. 1998a).
To further characterize the Mms4–Mus81 endonuclease, we tested a variety of branched DNA substrates ranging from the simple Y-form to a completely duplex form that resembles a replication fork. A titration of Mms4–Mus81 revealed that less than half of the Y-form substrate was cleaved with 10 ng of the complex (Fig. 3A). The cleaved product migrated with a marker consisting of the same labeled oligo annealed to a 24-nucleotide complementary strand (Fig. 3A, lane X). This product is consistent with cleavage of the noncomplementary 3′ tail, at or near the branch point. When the nuclease was incubated with duplex DNA containing a protruding 3′ ssDNA branch, complete cleavage was observed with just 0.6 ng of Mms4–Mus81 (Fig. 3B). Migration of the product relative to a specific marker (Fig. 3B, lane Y) again indicates that cleavage occurred at or near the branch point. By use of duplex DNA with a 5′ ssDNA protrusion, we observed very little digestion with as much as 10 ng of Mms4–Mus81 (Fig. 3C). The lack of cleavage on this substrate, relative to the simple Y-form, suggests that duplex 3′ branches are not substrates for the nuclease. However, when the nuclease was incubated with a completely duplex Y-form, a replication fork (RF) substrate, complete digestion was again obtained with 0.6 ng of Mms4–Mus81 (Fig. 3D). Densitometric quantitation of these gels indicates a 25-fold preference for this substrate over the simple Y-form. We conclude that the optimal substrates for the Mms4–Mus81 nuclease are duplex DNA with a 3′ ssDNA branch and the RF structure. This activity contrasts with that of Rad1–Rad10 and XPF–ERCC1. Relative to the simple Y-form, the NER complexes are less active on duplex DNA with a protruding 3′ branch (Rodriguez et al. 1996; de Laat et al. 1998a).
Figure 3.
Mms4–Mus81 endonuclease preferentially cleaves a replication fork substrate. A variety of 32P-labeled Y-shaped substrates were incubated with the indicated amounts of Mms4–Mus81 complex and the products were resolved on a native polyacrylamide gel prior to autoradiography. (A) Simple Y-form substrate (888*/891). (B) Duplex DNA with a protruding 3′ single-stranded branch (888*/891/994). (C) Duplex DNA with a protruding 5′ single-stranded branch (888*/891/992). (D) Replication fork substrate (888*/891/992/994). Markers were loaded as follows: (lanes X) dsDNA with a 3′ overhang (888*/940); (lanes Y) dsDNA containing a nick (888*/992/940); (lanes Z) Y-form substrate (888*/891). (*) The presence of a 32P-phosphate label at the 5′ position. (Δ) The reaction mixture was boiled prior to loading.
Binding and cleavage by the Mms4–Mus81 dimer
To characterize the roles of Mms4 and Mus81 in the cleavage reaction, we separately purified recombinant epitope-tagged versions of the two subunits and tested them for nuclease activity. Although immunoblot analysis revealed that the individual subunits were expressed poorly relative to their simultaneous expression, phosphocellulose and Ni-agarose chromatography yielded highly purified preparations of the two proteins (Fig. 4A). The full-length protein was the major component of each preparation, although smaller polypeptides were detected that are likely to be breakdown products, as they reacted with antibodies raised against the respective protein. Incubation of Mms4 or Mus81 alone with the RF substrate resulted in little or no nuclease activity. As shown in Figure 4B, only minor digestion products were obtained with 10 ng of Mms4, whereas 10 ng of Mus81 yielded no detectable product. In contrast, only 1 ng of the heterodimer was required for complete cutting of the RF substrate. Densitometric quantitation of this assay revealed that the heterodimer was 2300-fold more active than the Mms4 subunit alone. Attempts to reconstitute endonuclease activity from the individual subunits were unsuccessful. On the basis of these results, we conclude that the individual subunits have little or no nuclease activity on their own, and that the heterodimer is the active form of these proteins.
Figure 4.
Monomeric Mms4 and Mus81 lack endonuclease activity. (A) Purified dimeric Mms4–Mus81 and monomeric forms of Mms4 and Mus81 were resolved by SDS-PAGE and stained with Coomassie blue (Mms4–Mus81) or silver (Mms4 and Mus81). Molecular weight standards are shown in kilodaltons. (B) A 32P-labeled replication fork substrate was incubated with the indicated amounts of Mms4, Mus81, or Mms4–Mus81 complex as indicated. The products were then resolved on a native polyacrylamide gel prior to autoradiography. (Δ) The reaction mixture was boiled prior to loading.
The individual subunits were then tested for the ability to bind substrate DNA by use of a UV cross-linking assay in the absence of Mg2+. Mms4, Mus81, or the Mms4–Mus81 dimer was incubated with labeled RF substrate in the presence of EDTA. The mixture was then UV cross-linked and the proteins were immunoprecipitated and subjected to SDS-PAGE to identify protein–DNA cross-links. As shown in Figure 5A, neither Mms4 nor Mus81 bound the RF substrate under these conditions. However, the heterodimer resulted in a strong signal, indicating that it bound the RF substrate. This complex could be immunoprecipitated with antisera raised against either Mms4 or Mus81, but not an unrelated antiserum. We conclude that both subunits are required for substrate binding activity and that both subunits are present on the substrate DNA.
Figure 5.
The Mms4–Mus81 complex requires both subunits for substrate binding and cleavage activities. (A) A total of 50 ng of Mms4 (4), Mus81 (81), or Mms4–Mus81 dimer (4/81) was incubated with 24 fmole of 32P-labeled RF substrate and cross-linked with UV light. The products were then immunoprecipitated with anti-Mms4, anti-Mus81, or anti-Hmo1 serum as indicated, resolved on a 15% SDS–polyacrylamide gel, and visualized with a PhosphorImager. (B) 32P-labeled RF substrate was incubated in the presence or absence of 1 ng of Mms4–Mus81 complex, as indicated at bottom. The Mms4–Mus81 had been preincubated with the indicated amount of specific antiserum shown at top. The products were then resolved on a native polyacrylamide gel prior to autoradiography. (Δ) The reaction mixture was boiled prior to loading.
Finally, we tested the role of the individual subunits in the endonuclease activity of the complex by assaying Mms4–Mus81 activity in the presence of specific antisera. Mms4–Mus81 complex was incubated with increasing amounts of antiserum raised against Mms4 or Mus81, and the mixtures were used in a standard nuclease assay with the RF substrate. Incubation with anti-serum raised against Mms4 reduced the activity of the complex although substantial cleavage was observed even at the highest antibody concentration (Fig. 5B). Treatment with anti-serum raised against Mus81 inhibited Mms4–Mus81 nuclease activity almost entirely, whereas incubation with preimmune serum had no effect (Fig. 5B). The failure of the anti-Mms4 serum to completely inhibit nuclease activity may be due to the fact that it was raised against a portion of the protein (residues 1–471). We therefore conclude that optimal endonuclease activity requires both subunits.
MMS4 and MUS81 are required for meiosis in response to recombination
Among the phenotypes shared by yeast strains with mutations in MMS4 or MUS81 is a defect in meiosis. Although homozygous mms4 or mus81 diploids are unable to produce four-spored asci when grown under sporulation conditions, microscopic analysis of these sporulating diploids revealed that both produced two-spored asci at a very low frequency. This phenotype contrasts with that of the slx4 diploid, which sporulated normally (Mullen et al. 2001). Quantitation of this phenotype indicated that mms4 and mus81 diploids produced two-spored asci at nearly identical rates (∼0.5%) and microdissection revealed that spores from either diploid had very low viability (∼10%; Table 1). This phenotype is reminiscent of the reduced sporulation and viability seen in sgs1 and top3 homozygous diploids (Gangloff et al. 1999). To determine whether this meiotic defect was dependent on recombination, we constructed mms4 and mus81 mutants containing deletions of the SPO11 and SPO13 genes. Cells lacking SPO11 are unable to initiate recombination due to the lack of DSBs, whereas spo13 mutants bypass meiosis I. The spo11 spo13 double mutants allow one to recover viable meiotic products in the absence of recombination (Malone and Esposito 1981). As shown in Table 1, ∼20% of the spo11 spo13 double mutants, in an otherwise wild-type background, are able to sporulate. Although this rate is reduced relative to wild type, the resulting spores show near wild-type levels of viability (77%). Loss of either SPO11 or SPO13 in an mms4 background weakly suppressed the sporulation defect, whereas loss of both SPO11 and SPO13 resulted in complete suppression; 22% of mms4 spo11 spo13 cells sporulated compared with 20% for spo11 spo13 cells (Table 1). Further, the viability of spores derived from mms4 spo11 spo13 cells was improved to near wild-type levels (Table 1). The meiotic defects associated with mus81 were suppressed by the spo11 spo13 mutations in a similar manner. We conclude that the essential role of Mms4 and Mus81 in meiosis depends on recombination and may involve the processing of recombination intermediates. Consistent with previous observations (Mullen et al. 2001), the mms4 and mus81 phenotypes are identical in every assay we have tested.
Table 1.
Suppression of mms4 and mus81 sporulation defects
| Genotype
|
Sporulation efficiencya (%)
|
Spore viabilityb (%)
|
|---|---|---|
| wild type | 35 | 83 |
| spo11 | 4.9 | ND |
| spo13 | 19 | ND |
| spo11 spo13 | 20 | 77 |
| mms4 | 0.43 | 6 |
| mms4 spo11 | 3.1 | ND |
| mms4 spo13 | 1.5 | ND |
| mms4 spo11 spo13 | 22 | 73 |
| mus81 | 0.52 | 12 |
| mus81 spo11 | 4.8 | ND |
| mus81 spo13 | 1.2 | ND |
| mus81 spo11 spo13 | 21 | 83 |
For wild-type cells, sporulation refers to four-spored asci, and for all others it refers to two-spored asci. Sporulation efficiency was determined following 6 d in sporulation medium at 30°C. The value presented for each genotype is an average of two strains (at least 500 cells were counted per strain).
Viability was determined by the ability of micro-dissected spores to form colonies. At least 50 spores were examined for each genotype.
Discussion
The results presented here confirm that Mms4 and Mus81 comprise a complex with a distinct biochemical activity as suggested by earlier genetic studies. Multiple alleles of MMS4 and MUS81 were isolated in an sgs1 synthetic-lethal screen and subsequent epistasis experiments indicated that they were required in the same pathway for DNA damage resistance (Mullen et al. 2001). Additional genetic evidence presented here indicates that the meiotic phenotypes of these mutants are quantitatively similar and suppressible by eliminating recombination. Mms4 and Mus81 were suspected to function together in vivo as they coimmunoprecipitated from yeast cell extracts (Mullen et al. 2001), however, it was not clear whether this interaction required additional components. We found that recombinant Mms4 and Mus81 are poorly expressed individually, but are synthesized in good yield with relatively few degradation products when expressed together in the same cell. This behavior is often observed when multisubunit complexes are reconstituted in recombinant form (Bochkareva et al. 1998). Mms4–Mus81 bound phosphocellulose at high salt, as expected for a DNA-binding protein, and was stable in the presence of 0.5 M NaCl. Thus, Mms4 and Mus81 are capable of forming a complex in the absence of other yeast proteins. The 1:1 ratio of subunits suggests that the complex may be a simple heterodimer in solution. Mms4 and Mus81 are codependent for structure-specific DNA binding and endonuclease activity, as neither subunit displays these activities on their own.
The Mms4–Mus81 complex was tested for endonuclease activity on the basis of its amino acid sequence similarity to the Rad1–Rad10 (XPF–ERCC1) complex involved in NER (Mullen et al. 2001). The C terminus of Mus81 contains two regions of similarity to Rad1 (amino acids 823–1047), including a helix–hairpin–helix (HhH) domain, whereas the C terminus of Mms4 shows weak similarity to a region of Rad10 (amino acids 93–210) (Interthal and Heyer 2000; Mullen et al. 2001). These domains are known to mediate the Rad1–Rad10 and XPF–ERCC1 interactions (Bardwell et al. 1993; de Laat et al. 1998b), suggesting that the dimerization interface is conserved in Mms4–Mus81 (Mullen et al. 2001). However, amino acid sequence analysis has suggested that these C-terminal regions also contain residues important for metal ion coordination (Aravind et al. 1999). Thus, the conserved C-terminal domains may contain both catalytic and dimerization functions.
Mms4–Mus81 differs from the NER complexes in several ways. The most notable difference is the large size of Mms4 (691 amino acids) compared with Rad10 (210 amino acids) and ERCC1 (297 amino acids). These additional residues comprise a unique 300 amino acid N-terminal extension that is essential for activity, as the mms4-1 loss-of-function mutation (G173R) maps to this region (Xiao et al. 1998). This domain may be involved in a novel activity, such as substrate recognition, or mediation of specific protein–protein interactions. At the cellular level, there is no obvious overlap in the mutant phenotypes of mms4–mus81 and rad1–rad10. Although mms4 and mus81 mutants are sensitive to DNA-damaging agents, the level of sensitivity resembles that of sgs1 strains and is orders of magnitude less than that of rad1 or rad10 mutants. This level of sensitivity suggests that the Mms4–Mus81 complex plays an indirect role in DNA damage resistance, rather than a direct role in DNA repair. We also note that rad1–rad10 mutants have no reported defects in meiosis similar to those found in mms4–mus81 strains.
Enzymatically, both Mms4–Mus81 and the NER complexes are structure-specific endonucleases that cleave 3′ ssDNA extensions at duplex–ssDNA junctions (Bardwell et al. 1994). Although substrate specificity has not been analyzed exhaustively for either enzyme, we have identified a significant difference between them. Relative to the simple Y-form, both Rad1–Rad10 and XPF–ERCC1 are less active on duplex DNA with a 3′ ssDNA protrusion or branch (Rodriguez et al. 1996; de Laat et al. 1998a). In contrast, Mms4–Mus81 endonuclease activity is more active on this substrate relative to the simple Y-form. When the 3′ protrusion is duplex, as in duplex DNA with a 5′ ssDNA branch, Mms4–Mus81 fails to cleave, suggesting that the duplex 3′ arm is not recognized as a substrate. The fact that Mms4–Mus81 activity is restored on the RF substrate suggests that the enzyme recognizes the single- or double-stranded nature of the uncleaved 5′ arm with dsDNA acting as a positive effector. Taken together with the fact that mms4–mus81 phenotypes are distinct from rad1–rad10 phenotypes, we suggest that the RF substrate, and not the simple Y-form, is the in vivo substrate for Mms4–Mus81.
To explain the roles of Mms4–Mus81 and Sgs1–Top3 in vegetative growth, we propose the following model (Fig. 6A). Replication forks arrested in response to DNA damage can be re-established by one of two pathways. On the basis of RecQ function in E. coli, we suggest that Sgs1/Top3 acts to remove the nascent DNA strands from the parental duplex. This may occur in conjunction with a still unidentified exonuclease analogous to RecJ. The exposed DNA template is then bound by proteins (e.g., RPA, Pol α, etc.) to stabilize the fork and recruit DNA replication proteins. Simultaneously, excision repair proteins remove the damage so that fork movement can resume. The stalled fork is a reasonable substrate for RecQ helicases as both Sgs1 and BLM bind branched DNA structures (Bennett et al. 1999; Karow et al. 2000). Although Top3 may relax the topological stress induced by Sgs1 helicase activity, its function in this model may be analogous to the decatenation of strands by Top3 at the termination of pBR322 replication in vitro (DiGate and Marians 1988; Wang 1991). The demonstration that E. coli RecQ and Top3 cooperate to catenate dsDNA (Harmon et al. 1999) supports the combined role of these proteins in this pathway.
Figure 6.
Proposed role of Mms4–Mus81 in resolving stalled replication forks and meiotic recombination intermediates. (A) Replication fork arrest. A replication fork that encounters DNA damage (in this case, on the leading strand template) arrests and can be resolved by one of two pathways. In the left pathway, Sgs1 and Top3 (dark gray ovals) remove the nascent DNA (gray) from the parental duplex by virtue of its helicase and decatenation activities. The fate of these strands is not shown and they may be degraded. The ssDNA template formed in this step nucleates stabilization and replication proteins (gray circles), while the DNA repair machinery (white circles) fixes the damage. Newly loaded replication proteins (data not shown) re-establish fork movement. In the pathway at right, Mms4–Mus81 cleaves the leading strand template from the stalled fork. Following repair of the damage, ssDNA gaps are filled in. The DSB is repaired by a form of homologous recombination off the sister chromatid, which is initiated by the 3′ end of the cleaved arm invading the sister duplex. Extension of the invading strand creates a D-loop, lagging strand synthesis, and establishment of a replication fork. (B) Meiotic DSBs that occur in regions of heterogeneity initiate homologous recombination by pairing with target sequences on the homologous chromosome. The nonhomologous 3′ end is a substrate for cleavage by Mms4–Mus81. This creates the appropriate 3′ terminus, which allows elongation by DNA polymerase and subsequent formation of HJs.
The requirement for Sgs1–Top3 can be bypassed by use of the structure-specific endonuclease activity of Mms4–Mus81 (Fig. 6A, right). In this case, the leading strand template is cleaved from the fork while repair proteins correct the DNA duplex. Following gap repair, the DSB created by Mms4–Mus81 cleavage recombines with the repaired sister chromatid through an undefined mechanism. The process that re-establishes the fork may be a form of break-induced replication (BIR) (Paques and Haber 1999). We note that this process may be dependent on activities distinct from other forms of BIR, and obvious candidates for these activities include the four novel SLX genes that were isolated along with MMS4 and MUS81 (Mullen et al. 2001). This model makes several predictions including the possibility that Mms4–Mus81 is responsible for replication-dependent sister chromatid recombination that is induced by unrepaired UV damage (Kadyk and Hartwell 1993). In addition, human Mms4–Mus81 may play a role in the increased levels of SCE that are the hallmark of BLM−/− cells (Chaganti et al. 1974). The model also accounts for the moderate DNA damage sensitivity of cells lacking Sgs1–Top3 or Mms4–Mus81, as they retain the major DNA repair pathways (e.g., NER) and are only compromised in their ability to recover from fork arrest. We note that alternative models include the possibility that the substrate for Mms4–Mus81 is a recombination intermediate consisting of duplex DNA with a 3′ branch.
To explain the role of Mms4–Mus81 in meiosis, we propose that it is required to resolve recombination intermediates that consist of duplex DNA with a 3′ ssDNA protrusion. Such an intermediate might occur during the initiation of meiotic recombination (Fig. 6B). DSBs are normally substrates for recombinational repair. However, breaks that occur in regions of heterozygosity may generate 3′ ends lacking complete homology. Upon invasion of the homologous chromosome, this strand would be expected to pair with homologous sequences and generate an unpaired 3′ tail (Fig. 6B). This nonhomologous tail can be cleaved by Mms4–Mus81 to provide the proper terminus for extension by DNA polymerase and subsequent formation of HJs. Although the removal of the 3′ tail in this model appears to resemble that catalyzed by Rad1–Rad10 in single strand annealing (SSA) (Paques and Haber 1999), this substrate is devoid of ssDNA downstream of the 3′ branch. This is expected to be different from the substrate in SSA, in which long regions of the 5′ ends are resected prior to annealing. We note that additional roles for this cleavage activity are conceivable. For example, Mms4–Mus81 activity would be required subsequent to any strand annealing step that produces duplex DNA with a 3′ ssDNA protrusion.
The Mms4–Mus81 complex may interact physically with additional proteins to execute its function in vivo. In the case of NER, the damage-recognition protein XPA binds ERCC1 and is thought to target the endonuclease to sites of DNA damage (Park and Sancar 1994). It has not yet been tested whether Rad14 (XPA) binds Mms4. On the other hand, Mus81 was shown to interact with Rad54 (Interthal and Heyer 2000), a protein required for recombination, but whose function is not clearly defined. Rad54 stimulates the strand-pairing activity of Rad51 and displays DNA-dependent ATPase activity (Petukhova et al. 1998). A role for Mus81 in recombination is consistent with the meiotic phenotypes reported here, however, our preliminary efforts to identify a functional interaction between these proteins have failed to find an effect of Mms4–Mus81 on the ATPase activity of Rad54 (W.M. Fricke and S.J. Brill, unpubl.).
Finally, S. pombe mus81+ was isolated in a two-hybrid screen with the FHA1 domain of the Cds1 checkpoint kinase (Boddy et al. 2000). As in budding yeast, mus81+ was shown to play a role in meiosis and to be essential for viability in the absence of the RecQ homolog rqh1+ (Boddy et al. 2000). Epistasis experiments support an interaction with recombination proteins and suggest that spMus81 and Cds1 participate in the repair of DNA damage. Interestingly, MMS4 from S. cerevisiae was isolated recently in a two-hybrid screen with the meiotic checkpoint kinase Mek1, which contains a Rad53-like FHA domain (N. Hollingsworth, pers. comm.). Taken together, these results suggest that Mms4–Mus81 may interact functionally with checkpoint kinases. A regulatory role for Mms4–Mus81 is suggested by the fact that Mus81 is phosphorylated in S. pombe (Boddy et al. 2000) and Mms4 is phosphorylated in S. cerevisiae (S.J. Brill, unpubl.). Further experimentation will be required to test the possibility that Mms4–Mus81 plays a role in damage-dependent cell cycle regulation.
Materials and methods
Strains and plasmid construction
The yeast strains used in this study are derived from the wild-type strain W303-1a (MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100). Standard genetic techniques and reagents were used in the construction, transformation, and growth of yeast (Rose et al. 1990). Homozygous diploid strains were constructed by crossing haploids derived from JMY375 (W303-1a mms4-10∷KAN) and JMY380 (W303-1a mus81-10∷KAN) (Mullen et al. 2001). These derivatives contain the complete deletion alleles spo11-11∷KAN,loxP and/or spo13-20∷HGR, which were constructed using PCR amplification with gene-specific primers off plasmids pUG6 (kanamycin) (Guldener et al. 1996) or pAG32 (hygromycin B) (Goldstein and McCusker 1999), respectively. Proper integrative transplacement and segregation were determined by analytical PCR.
Recombinant proteins were expressed by use of the T7 RNA polymerase system and vectors pET11a (Studier et al. 1990) and pET28a (Novagen). Plasmid pNJ6217 expresses a 711 amino acid version of Mms4 (80,876 daltons) containing a hexa-histidine tag at its N terminus. Plasmid pJM6301 expresses the 632 amino acid wild-type version of Mus81 (72,263 daltons). The double expression plasmid pJM6220 was constructed by moving the 2.7-kb MluI/BamHI fragment from pJM6301 into the MluI/BglII sites of pNJ6217. Plasmid pNJ6313 expresses a 665 amino acid version of Mus81 (75,952 daltons) that contains a V5-epitope and a hexa-histidine tag at its C terminus.
Purification of recombinant proteins
E. coli BL21-RIL cells (Stratagene) containing plasmids pNJ6220 (His6-MMS4 MUS81), pNJ6217 (His6-MMS4), and pNJ6313 (MUS81-V5-His6) were grown at 37°C until the OD600 = 0.5, at which time the cells were induced with 0.2 (pNJ6313) or 0.4 mM (pNJ6217, pNJ6220) IPTG and were grown an additional 2.5 h at 37°C. The induced cells were pelleted and resuspended and frozen at −80°C in Buffer B (25 mM HEPES at pH 7.5, 10% glycerol, 1 mM EDTA, 0.01% NP-40, 1 mM DTT, and 0.1 mM PMSF) containing 50 mM NaCl and protease inhibitors. All subsequent steps took place at 0 or 4°C. Induced cells were thawed, sonicated three times for 1 min, centrifuged at 26,900g for 20 min, and the supernatant taken as extract.
For the Mms4–Mus81 complex, the NaCl concentration was adjusted to 150 mM and loaded onto a 40-mL phosphocellulose column. The column was washed with three column volumes of Buffer B plus 150 mM NaCl and eluted with an eight column-volume gradient from 150 to 1000 mM NaCl in Buffer B without DTT and EDTA. Peak Mms4–Mus81 fractions eluted at 650 mM NaCl, which were pooled and batch bound to 2 mL of Ni Probond resin (Invitrogen) in the presence of 10 mM imidazole for 3 h, after which the resin was poured into a column. The column was washed with 10 column volumes of Buffer N (25 mM Tris-HCl at pH 7.5, 500 mM NaCl, 10% glycerol, 0.01% NP-40, 1 mM PMSF) plus 10 mM imidazole, followed by seven column volumes of Buffer N plus 50 mM imidazole. The Mms4–Mus81 dimer was eluted in seven half-column volume fractions each of Buffer N plus 100 mM imidazole followed by Buffer N plus 200 mM imidazole. The Buffer N plus 200 mM imidazole fractions were pooled and dialyzed against Buffer A containing 100 mM NaCl. The Mms4 and Mus81 subunits were purified as above except that the extracts were loaded onto a phosphocellulose column at 75 mM KCl and washed with three column volumes of Buffer A plus 75 mM KCl. Proteins were eluted with an eight column-volume gradient from 100 to 1000 mM NaCl in Buffer B without DTT and EDTA. Peak fractions were pooled and batch bound to 1 mL Ni Probond resin for 2 h. The resin was poured into a column, washed consecutively with 10 column volumes of Buffer N containing 10 mM imidazole plus an additional 0.5 M NaCl, and three column volumes of Buffer N containing 50 mM imidazole. The bound proteins were then eluted with Buffer N plus 250 mM imidazole and dialyzed as above.
Immunological techniques
Rabbit antisera were raised against recombinant gel-isolated Mms4 (1–471) and amylose-purified MBP-Mus81 (1–632) (Cocalico). To immunoprecipitate proteins cross-linked to substrate DNA, 50 ng of purified Mms4, Mus81, or Mms4–Mus81 dimer was incubated with 24 fmole of 32P-labeled substrate DNA at 25°C for 15 min under the following conditions: 25 mM HEPES (pH 7.5), 0.5 mM DTT, 0.1% NP-40, 0.1 mg/mL BSA, and final volume of 15 μL. Binding reactions were cross-linked with UV light at a dose of 1000 J/m2 in a Stratalinker (Stratagene) and incubated with either 20 μL (anti-Mms4) or 2 μL (anti-Mus81) anti-serum, on ice for 1 h in 200 μL of RIPA buffer (50 mM Tris at pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, and 0.1% SDS). Protein A sepharose resin was added to each reaction, mixed at 4°C for 1 h, and washed three times with 1 mL of RIPA buffer. The bound products were resolved on a 15% SDS–polyacrylamide gel and visualized with a PhosphorImager.
Preparation of DNA substrates for nuclease assays
All DNA structures were prepared from labeled oligonucleotides that were annealed and purified essentially as described (White and Lilley 1996). The oligonucleotides (IDT) used in this study are: 888 (49 nucleotides), GACGCTGCCGAATTCTG GCGTTAGGAGATACCGATAAGCTTCGGCTTAA; 891 (49 nucleotides), ATCGATGTCTCTAGACAGCACGAGCCCTA ACGCCAGAATTCGGCAGCGT; 892 (49 nucleotides), GAC GCTGCCGAATTCTGGCTTGCTAGGACATCTTTGCCCA CGTTGACCC; 893 (50 nucleotides), TGGGTCAACGTGGGC AAAGATGTCCTAGCAATGTAATCGTCTATGACGTT; 894 (51 nucleotides), CAACGTCATAGACGATTACATTGCTAG GACATGCTGTCTAGAGACTATCGA; 895 (50 nucleotides), ATCGATAGTCTCTAGACAGCATGTCCTAGCAAGCCAG AATTCGGCAGCGT; 897 (49 nucleotides), GGGTCAACGT GGGCAAAGATGTCCTAGCAAGCCAGAATTCGGCAGCG TC; 940 (24 nucleotides), TAGCAAGCCAGAATTCGGCAG CGT; 992 (24 nucleotides), TTAAGCCGAAGCTTATCGGTA TCT; 994 (25 nucleotides), GCTCGTGCTGTCAGAGACATC GAT.
Nuclease assays
Assays were performed essentially as described (Habraken et al. 1994). Protein was incubated with 10,000 cpm 32P-labeled DNA substrate (2 fmole) in a final volume of 10 μL containing 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 10 mM MgCl2, 0.2 mM dithiothreitol, 0.1% NP-40, 0.1 mg/mL BSA, and 5% glycerol at 37°C for 30 min. After deproteinization by incubation with 0.1% SDS and 10 μg of proteinase K at 37°C for 10 min, the reaction products were electrophoresed through a 10% polyacrylamide gel. The gel was then dried and visualized by a PhosphorImager. For antibody inhibition, the Mms4–Mus81 protein was incubated with the antibodies at 4°C for 30 min prior to adding the cutting assay buffer and substrate.
Acknowledgments
We thank Justin Courcelle, Abram Gabriel, Nancy Hollingsworth, and Kim McKim for helpful discussions. We also thank Nancy Hollingsworth for communicating results prior to publication, and Patrick Sung and Orlando Scharer for purified proteins. This research was supported by NIH grant AG16637.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL brill@mbcl.rutgers.edu; FAX (732) 235-4880.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.932201.
References
- Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell AJ, Bardwell L, Johnson DK, Friedberg EC. Yeast DNA recombination and repair proteins Rad1 and Rad10 constitute a complex in vivo mediated by localized hydrophobic domains. Mol Microbiol. 1993;8:1177–1188. doi: 10.1111/j.1365-2958.1993.tb01662.x. [DOI] [PubMed] [Google Scholar]
- Bardwell AJ, Bardwell L, Tomkinson AE, Friedberg EC. Specific cleavage of model recombination and repair intermediates by the yeast Rad1–Rad10 DNA endonuclease. Science. 1994;265:2082–2085. doi: 10.1126/science.8091230. [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Keck JL, Wang JC. Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S. cerevisiae. J Mol Biol. 1999;289:235–248. doi: 10.1006/jmbi.1999.2739. [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Noirot-Gros MF, Wang JC. Interaction between yeast Sgs1 helicase and DNA topoisomerase III. J Biol Chem. 2000;275:26898–26905. doi: 10.1074/jbc.M003137200. [DOI] [PubMed] [Google Scholar]
- Bochkareva E, Frappier L, Edwards AM, Bochkarev A. The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain. J Biol Chem. 1998;273:3932–3936. doi: 10.1074/jbc.273.7.3932. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, Russell P. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol Cell Biol. 2000;20:8758–8766. doi: 10.1128/mcb.20.23.8758-8766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti RS, Schonberg S, German J. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc Natl Acad Sci. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraverty RK, Hickson ID. Defending genome integrity during DNA replication: A proposed role for RecQ family helicases. BioEssays. 1999;21:286–294. doi: 10.1002/(SICI)1521-1878(199904)21:4<286::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Courcelle J, Hanawalt PC. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol Gen Genet. 1999;262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- ————— Participation of recombination proteins in rescue of arrested replication forks in UV-irradiated Escherichia coli need not involve recombination. Proc Natl Acad Sci. 2001;98:8196–8202. doi: 10.1073/pnas.121008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Carswell-Crumpton C, Hanawalt PC. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc Natl Acad Sci. 1997;94:3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM. Historical overview: Searching for replication help in all of the rec places. Proc Natl Acad Sci. 2001;98:8173–8180. doi: 10.1073/pnas.131004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat WL, Appeldoorn E, Jaspers NG, Hoeijmakers JH. DNA structural elements required for ERCC1-XPF endonuclease activity. J Biol Chem. 1998a;273:7835–7842. doi: 10.1074/jbc.273.14.7835. [DOI] [PubMed] [Google Scholar]
- de Laat WL, Sijbers AM, Odijk H, Jaspers NG, Hoeijmakers JH. Mapping of interaction domains between human repair proteins ERCC1 and XPF. Nucleic Acids Res. 1998b;26:4146–4152. doi: 10.1093/nar/26.18.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGate RJ, Marians KJ. Identification of a potent decatenating enzyme from Escherichia coli. J Biol Chem. 1988;263:13366–13373. [PubMed] [Google Scholar]
- Fricke WM, Kaliraman V, Brill SJ. Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J Biol Chem. 2001;276:8848–8855. doi: 10.1074/jbc.M009719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, de Massy B, Arthur L, Rothstein R, Fabre F. The essential role of yeast topoisomerase III in meiosis depends on recombination. EMBO J. 1999;18:1701–1711. doi: 10.1093/emboj/18.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Goodwin A, Wang SW, Toda T, Norbury C, Hickson ID. Topoisomerase III is essential for accurate nuclear division in Schizosaccharomyces pombe. Nucleic Acids Res. 1999;27:4050–4058. doi: 10.1093/nar/27.20.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habraken Y, Sung P, Prakash L, Prakash S. Holliday junction cleavage by yeast Rad1 protein. Nature. 1994;371:531–534. doi: 10.1038/371531a0. [DOI] [PubMed] [Google Scholar]
- Harmon FG, DiGate RJ, Kowalczykowski SC. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: A conserved mechanism for control of DNA recombination. Mol Cell. 1999;3:611–620. doi: 10.1016/s1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- Heo SJ, Tatebayashi K, Ohsugi I, Shimamoto A, Furuichi Y, Ikeda H. Bloom's syndrome gene suppresses premature ageing caused by Sgs1 deficiency in yeast. Genes Cells. 1999;4:619–625. doi: 10.1046/j.1365-2443.1999.00288.x. [DOI] [PubMed] [Google Scholar]
- Hu P, Beresten SF, van Brabant AJ, Ye TZ, Pandolfi PP, Johnson FB, Guarente L, Ellis NA. Evidence for BLM and topoisomerase III alpha interaction in genomic stability. Hum Mol Genet. 2001;10:1287–1298. doi: 10.1093/hmg/10.12.1287. [DOI] [PubMed] [Google Scholar]
- Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3′→5′ exonuclease. Nat Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Interthal H, Heyer WD. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol Gen Genet. 2000;263:812–827. doi: 10.1007/s004380000241. [DOI] [PubMed] [Google Scholar]
- Johnson FB, Lombard DB, Neff NF, Mastrangelo MA, Dewolf W, Ellis NA, Marciniak RA, Yin Y, Jaenisch R, Guarente L. Association of the Bloom syndrome protein with topoisomerase IIIα in somatic and meiotic cells. Cancer Res. 2000;60:1162–1167. [PubMed] [Google Scholar]
- Kadyk LC, Hartwell LH. Replication-dependent sister chromatid recombination in rad1 mutants of Saccharomyces cerevisiae. Genetics. 1993;133:469–487. doi: 10.1093/genetics/133.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow JK, Constantinou A, Li JL, West SC, Hickson ID. The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc Natl Acad Sci. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RA, Wang JC. Identification of the yeast TOP3 gene product as a single strand-specific DNA topoisomerase. J Biol Chem. 1992;267:17178–17185. [PubMed] [Google Scholar]
- Lebel M, Spillare EA, Harris CC, Leder P. The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J Biol Chem. 1999;274:37795–37799. doi: 10.1074/jbc.274.53.37795. [DOI] [PubMed] [Google Scholar]
- Malone RE, Esposito RE. Recombinationless meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1981;1:891–901. doi: 10.1128/mcb.1.10.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Brill SJ. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2000;154:1101–1114. doi: 10.1093/genetics/154.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CH, Sancar A. Formation of a ternary complex by human XPA, ERCC1, and ERCC4(XPF) excision repair proteins. Proc Natl Acad Sci. 1994;91:5017–5021. doi: 10.1073/pnas.91.11.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- Rodriguez K, Wang Z, Friedberg EC, Tomkinson AE. Identification of functional domains within the RAD1–RAD10 repair and recombination endonuclease of Saccharomyces cerevisiae. J Biol Chem. 1996;271:20551–20558. doi: 10.1074/jbc.271.34.20551. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Seigneur M, Bidnenko V, Ehrlich SD, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- Stewart E, Chapman CR, Al-Khodairy F, Carr AM, Enoch T. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Meth Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Wang JC. DNA topoisomerases: Why so many? J Biol Chem. 1991;266:6659–6662. [PubMed] [Google Scholar]
- Wang JC, Caron PR, Kim RA. The role of DNA topoisomerases in recombination and genome stability: A double-edged sword? Cell. 1990;62:403–406. doi: 10.1016/0092-8674(90)90002-v. [DOI] [PubMed] [Google Scholar]
- Watt PM, Hickson ID, Borts RH, Louis EJ. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MF, Lilley DM. The structure-selectivity and sequence preference of the junction-resolving enzyme CCE1 of Saccharomyces cerevisiae. J Mol Biol. 1996;257:330–341. doi: 10.1006/jmbi.1996.0166. [DOI] [PubMed] [Google Scholar]
- Wu L, Davies SL, North PS, Goulaouic H, Riou JF, Turley H, Gatter KC, Hickson ID. The Bloom's syndrome gene product interacts with topoisomerase III. J Biol Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- Xiao W, Chow BL, Milo CN. Mms4, a putative transcriptional (co)activator, protects Saccharomyces cerevisiae cells from endogenous and environmental DNA damage. Mol Gen Genet. 1998;257:614–623. doi: 10.1007/s004380050689. [DOI] [PubMed] [Google Scholar]
- Zhong S, Hu P, Ye TZ, Stan R, Ellis NA, Pandolfi PP. A role for PML and the nuclear body in genomic stability. Oncogene. 1999;18:7941–7947. doi: 10.1038/sj.onc.1203367. [DOI] [PubMed] [Google Scholar]