Abstract
Double-stranded RNAs can suppress expression of homologous genes through an evolutionarily conserved process named RNA interference (RNAi) or post-transcriptional gene silencing (PTGS). One mechanism underlying silencing is degradation of target mRNAs by an RNP complex, which contains ∼22 nt of siRNAs as guides to substrate selection. A bidentate nuclease called Dicer has been implicated as the protein responsible for siRNA production. Here we characterize the Caenorhabditis elegans ortholog of Dicer (K12H4.8; dcr-1) in vivo and in vitro. dcr-1 mutants show a defect in RNAi. Furthermore, a combination of phenotypic abnormalities and RNA analysis suggests a role for dcr-1 in a regulatory pathway comprised of small temporal RNA (let-7) and its target (e.g., lin-41).
Keywords: RNAi, siRNA, PTGS, Dicer, development
RNA interference (RNAi) is a process through which exposure to double-stranded RNA (dsRNA) leads to the silencing of homologous genes, most often post-transcriptionally (Fire et al. 1998). RNAi is a widespread phenomenon, being present in organisms ranging from fungi to mice (for reviews, see Bass 2000; Hammond et al. 2001; Plasterk and Ketting 2000; Sharp 2001). However, RNAi was first discovered as cosuppression, a response in plants, which leads to simultaneous silencing of exogenous transgenes and homologous endogenous genes (Napoli et al. 1990; van der Krol et al. 1990). In retrospect, it is clear that cosuppression is most likely provoked by dsRNA derived from the multiple transgene copies (Kooter et al. 1999; Chuang and Meyerowitz 2000).
One of the natural functions of post-transcriptional gene silencing (PTGS) phenomena appears to be protection against molecular parasites (Ketting et al. 1999; Tabara et al. 1999; Mourrain et al. 2000; Dalmay et al. 2001). Somehow, viruses and transposons activate the RNAi machinery, ultimately causing the silencing of these elements. The fact that many plant viruses encode proteins that counteract the PTGS response (Anandalakshmi et al. 1998; Kasschau and Carrington 1998; Voinnet et al. 1999; Lucy et al. 2000) indicates the critical nature of this host response.
A characteristic feature of PTGS and RNAi is the presence of low molecular weight RNA derived from either the PTGS-inducing transgene or the exogenous dsRNA, respectively (Hamilton and Baulcombe 1999; Hammond et al. 2000; Parrish et al. 2000; Zamore et al. 2000). These small RNA molecules have been named guide RNA, or short interfering RNA (siRNA), and are present in a protein complex that can target homologous mRNA molecules for destruction (Hammond et al. 2000) by cleaving the mRNA molecule within the region spanned by the siRNA molecule (Elbashir et al. 2001).
In Drosophila, an RNAse III-family nuclease called Dicer has been implicated in the production of siRNAs (Bernstein et al. 2001). To study Dicer in a genetically accessible system we set out to develop an in vitro system that recapitulates individual steps of RNAi in Caenorhabditis elegans extracts. This has allowed us to verify that the predicted C. elegans Dicer protein, K14H4.8 (dcr-1), is indeed a functional ortholog of the Drosophila protein. Furthermore, we isolated a dcr-1 deletion mutant from a C. elegans deletion library, and show that this protein is required for RNAi, at least in the germ line.
Interestingly, the dcr-1 deletion mutant displays developmental abnormalities, reminiscent of phenotypes caused by loss of function of let-7, a gene encoding a small RNA essential for the proper timing of C. elegans development (Reinhart et al. 2000). This RNA, known more generically as a member of a class of small temporal RNAs (stRNAs; Pasquinelli et al. 2000), suppresses the expression of target genes at the translational level by base-pairing to complementary sequences in the 3′ UTR (Slack et al. 2000). The mature let-7 RNA is predicted to derive from a precursor RNA, which has the potential to form a stem-loop structure, creating a dsRNA molecule (Pasquinelli et al. 2000). We show that in vitro Dicer can process the let-7 precursor to its mature form, and that in vivo, in dcr-1 mutant animals, the levels of processed let-7 RNA are reduced, but the let-7 precursor RNA accumulates.
Considered together, our results show that the C. elegans DCR-1 and the Drosophila Dicer proteins are indeed functional orthologs; that DCR-1 is required for functional RNAi, at least in the germ line of C. elegans; and that Dicer is involved in the maturation of small RNA molecules that regulate developmental timing.
Results and Discussion
A cell-free system for C. elegans Dicer
A signature feature of RNA interference and related gene silencing phenomena is the production of small, ∼22-nt RNAs termed guide RNAs or siRNAs. These have been observed in plants undergoing cosuppression or virus-induced gene silencing (Hamilton and Baulcombe 1999) and in C. elegans and Drosophila during RNA interference (Hammond et al. 2000; Parrish et al. 2000; Yang et al. 2000; Zamore et al. 2000). In Drosophila, biochemical studies have indicated that siRNAs are produced by nucleolytic digestion of the dsRNA silencing trigger (Zamore et al. 2000; Bernstein et al. 2001). To test whether siRNAs are produced by an evolutionarily conserved mechanism, we have taken a combination of biochemical and genetic approaches.
We prepared extracts from C. elegans embryos and tested these for Dicer activity, as evidenced by the ability to process long dsRNA into siRNAs. Such an activity is clearly detectable (Fig. 1a). A comparison of reactions performed in parallel shows that Drosophila and C. elegans extracts produce siRNA species of different sizes (Fig. 1c). In C. elegans, siRNAs produced in vitro are 23 bp in length, consistent with the size of siRNAs found in vivo (M. Tijsterman, R.F. Ketting, T. Sijen, R.H.A. Plasterk, and K. Okihara, unpubl.). Drosophila siRNAs produced in vitro are predominantly 21 bp in length and comigrate with siRNAs that are associated with the RISC enzyme in S2 cells (Hammond et al. 2000). As was observed with Drosophila Dicer, longer dsRNAs are processed more efficiently, which correlates with the observation that long dsRNAs are more effective inducers of RNAi than are short dsRNAs (Fig. 1d; Fire et al. 1998; Parrish et al. 2000; Yang et al. 2000).
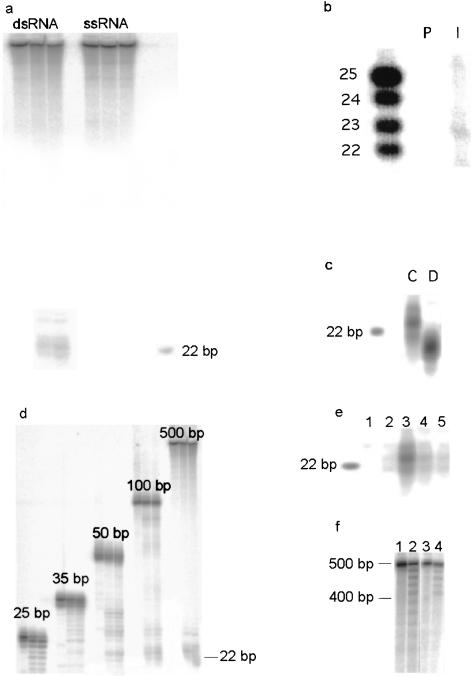
Figure 1.
(a) Double-stranded RNA (ds) and single-stranded RNA (ss), both 500 bp long, were incubated with extract from C. elegans embryos. siRNAs are produced in a time-dependent fashion from the dsRNA but not from the ssRNA (time points: 0, 30, and 60 min). (b) siRNA-producing activity can be immunoprecipitated with antisera (I) raised against DCR-1. The preimmune serum (P) is inactive. The left lane contains an RNA marker. (c) siRNA produced by C. elegans extract (C) is ∼2 nt bigger than siRNA produced by Drosophila extract (D). In both reactions a 500-bp dsRNA was used as substrate. (d) Time courses (0, 30, 60 min) with dsRNA of different lengths, using C. elegans extract. siRNA is produced from dsRNA of 35 bp and longer, but the larger dsRNA molecules are clearly better substrates (equal counts and equal mass of RNA were added to each reaction). (e) siRNA production is ATP-dependent. (Lane 1) The substrate (500 bp) was not incubated with extract; (lane 2) a reaction with ATP-depleted extract; (lane 3) ATP is added back to the depleted extract; and (lanes 4,5) nonhydrolyzable ATP analogs are added (ATPγS and ADPNP). (f) Incubation of 500-bp dsRNA with C. elegans embryo extract results in the formation of a ladder, with a spacing characteristic of siRNAs. (Lane 1) Untreated RNA; (lane 2) the same RNA after treatment with extract; (lanes 3,4) identical reactions to lanes 1 and 2, but with a 100-bp dsRNA substrate with a 400-bp single-stranded tail. The laddering stops at the position where the RNA substrate is no longer double stranded.
We have previously proposed an RNAse III-family nuclease, Dicer, as a candidate for catalyzing the production of siRNAs (Bernstein et al. 2001). The genome of C. elegans encodes a possible ortholog of the Drosophila Dicer protein, K12H4.8, which shares with Dicer a predicted domain structure comprising (from the N terminus to the C terminus) a helicase domain, a PAZ domain (Cerutti et al. 2000), dual RNAse III domains, and a double-stranded RNA-binding domain. We raised a polyclonal antiserum to the C terminus of K12H4.8. This antiserum specifically immunoprecipitated from C. elegans embryo extracts an activity that digests dsRNA into siRNAs (Fig. 1b). These results suggest that K12H4.8 is indeed the functional ortholog of Drosophila Dicer, and we, therefore, refer to this gene as dcr-1.
DCR-1, like Drosophila Dicer (Bernstein et al. 2001), requires ATP for efficient cleavage, and ATP hydrolysis further enhances siRNA production (Fig. 1e). We previously hypothesized that ATP hydrolysis by the helicase domain might drive a processive cleavage of dsRNA substrates by Dicer (Bernstein et al. 2001). A prediction of this model is that examination of reaction intermediates might reveal a ladder of products. Indeed, we find that 500-bp dsRNA is shortened by DCR-1 in increments of ∼23 nt (Fig. 1f). Reactions performed in extracts depleted of ATP produce only the first decrement of the ladder (data not shown). Furthermore, when a partial dsRNA is offered as substrate, the reaction terminates at the point where the RNA becomes single-stranded. Our results therefore suggest that DCR-1 converts dsRNA into siRNAs through a processive processing reaction, extracting energy for translocation along the dsRNA from ATP hydrolysis. This proposed mechanism is consistent with the observation that in vitro RNAi in Drosophila embryo extracts leads to cleavage of mRNA at characteristic ∼22-nt intervals (Zamore et al. 2000).
dcr-1 is involved in RNAi
To test the involvement of C. elegans Dicer in RNAi in vivo, we isolated a deletion mutant of dcr-1 (Jansen et al. 1997). Thus far, screens for RNAi-resistant mutants have yielded viable and fertile mutants (Tabara et al. 1999). Animals homozygous for the dcr-1 deletion are, however, sterile. Oocytes are abnormal, and no fertilized eggs can be detected (Fig. 2a,b). These defects can be partially rescued by introduction of a transgene containing a wild-type copy of dcr-1 (not shown). Fertilized eggs are formed in the presence of this transgene, but the eggs do not hatch, presumably because of loss of the transgene or limited expression of DCR-1 from the transgene in the germ line.
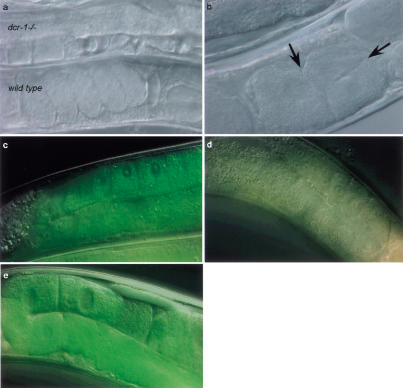
Figure 2.
(a) A wild-type animal next to a dcr-1 homozygous deletion animal. Fertilization of oocytes does not occur, or occurs sporadically. Lack of fertilization is not caused by a sperm defect as mating to wild-type males does not rescue this phenotype (data not shown). This phenotype can be rescued by introducing a wild-type copy of dcr-1. However, the fertilized eggs do not hatch, and the mother displays an egg-laying-defective phenotype (data not shown). (b) Oocytes are abnormal. Oocytes normally do not divide in the gonad, but in dcr-1(pk1531) mutant animals this is frequently observed (arrows indicate division planes). (c–e) Animals carrying a GFP transgene expressing in the germ line. Wild-type animals (n = 25) display clear fluorescence in the gonad (c), which is silenced by dsRNA against GFP (d) (n = 29). Animals homozygous for the dcr-1 deletion displayed the fluorescence both before (n = 18) and after RNAi (e) (n = 22), indicating that the RNAi response is defective.
To test whether dcr-1 animals are resistant to RNAi, we fed these worms dsRNA homologous to the unc-22 gene. Surprisingly, we observed a wild-type RNAi response. This might indicate that RNAi can proceed independently of Dicer activity. Alternatively, maternally contributed DCR-1 protein might be sufficient to generate an RNAi response. In fact, we have found that at least one other gene required for RNAi (mut-15; R.F. Ketting and R.H.A. Plasterk, unpubl.) displays such a maternal effect. It was expected that maternally contributed Dicer protein would be absent from the germ line of adult dcr-1 homozygotes. We therefore tested whether RNAi could be used to silence a GFP transgene that is expressed specifically in that tissue. Although RNAi is quite effective at silencing germ-line GFP in wild-type worms, dcr-1 homozygotes are RNAi-resistant (Fig. 2c–e), and we conclude that Dicer is essential for RNAi in at least some tissues. We cannot exclude that there is a second pathway that induces RNAi in the absence of Dicer; however, we prefer the most parsimonious explanation: that the residual RNAi effects result from persistence of maternal Dicer protein.
dcr-1 is involved in developmental timing
In addition to being sterile, dcr-1 homozygotes show a number of additional phenotypic abnormalities. Among these is a defect of the seam cells in the L4-to-adult transition. These cells fail to fuse, and in some cases undergo an additional round of cell division (Fig. 3a,b). As a result, the alae are absent in ∼60% (38/62) of the dcr-1(pk1531) homozygous animals. Interestingly, this phenotype is also characteristic of loss-of-function mutations in the let-7 gene (Reinhart et al. 2000).
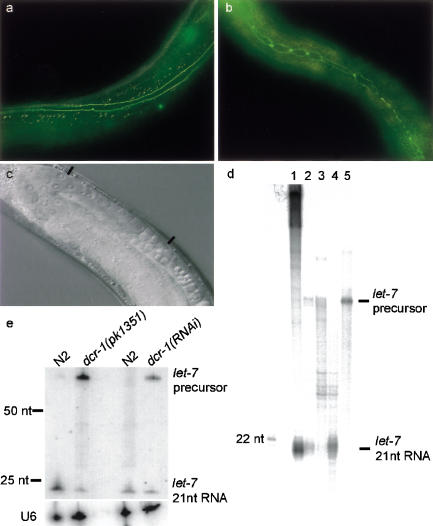
Figure 3.
(a) The wild-type pattern of jam-1::gfp staining in seam cells in adult animals. The seam cells have fused, and no individual cells can be recognized. (b) In the dcr-1 deletion mutant, the seam cells of the adult animal have failed to fuse and have undergone an additional round of cell division. (c) An animal is presented that contains a high copy number hsp::dcr-1 transgene (see Materials and Methods). The animal is sterile owing to the absence of oocytes, similar to a lin-41 knockout phenotype. The region where the oocytes should be is located between the two black bars. (d) Dicer converts the let-7 double-stranded precursor into 21-nt RNA molecules in vitro. Radiolabeled let-7 precursor RNA (Drosophila sequence; Pasquinelli et al. 2000; shown in lane 5) was incubated with Drosophila embryo extract (lane 2), dicer preimmune precipitate (lane 3), and dicer immune precipitate (lane 4). (Lane 1) A control reaction with 500-bp dsRNA. (e) A Northern blot on which RNA from wild-type and mutant animals is probed for let-7; U6 was used for a loading control. (Lanes 1,2) RNA from L4-stage wild-type and dcr-1(pk1351) homozygous mutant animals, respectively. The processed let-7 signal drops 10-fold (corrected for loading) in the dcr-1 mutant population, and the longer let-7 precursor RNA accumulates. We find a similar result (twofold reduction of the processed let-7) when we inhibit dcr-1 by RNAi, as shown in lanes 3 and 4; the RNA in these two lanes was isolated from adult animals.
The let-7 gene product is a small, noncoding RNA that regulates the timing of developmental events in C. elegans (therefore named small temporal RNA or stRNA; Pasquinelli et al. 2000). Of interest, the let-7 RNA is 21 nt in length, and it has been hypothesized that the let-7 RNA is produced by post-transcriptional processing of a longer precursor that is predicted to form an extended hairpin structure, which may be a substrate for DCR-1 (Pasquinelli et al. 2000; Sharp 2001). Regulation by let-7 occurs at the translational level and presumably is mediated by complementary base-pairing between let-7 and the 3′-untranslated regions of target genes (Slack et al. 2000).
One of the in vivo targets of let-7 is lin-41 (Slack et al. 2000), and the increased expression of this protein in let-7 mutants leads to the burst vulva phenotype. Interestingly, dcr-1 homozygous mutants also display a burst vulva phenotype, up to 80% (17/21), which can be rescued by introducing the wild-type dcr-1 gene. We tested if this phenotype can be partially suppressed by down-regulating LIN-41 protein through RNAi; and indeed it can: after RNAi of lin-41 we find only 25% burst vulva (5/20). This suggests that the burst vulva phenotype in dcr-1 mutant animals is at least partially caused by an up-regulation of LIN-41, and the epistatic effect is an indication that dcr-1 and lin-41 indeed act in the same pathway. Conversely, hypomorphic alleles of lin-41 have an Egl phenotype (an egg-laying defect), whereas null alleles of lin-41 are sterile owing to absence of oocytes (Slack et al. 2000). Accordingly, different levels of ectopic expression of DCR-1 might, via down-regulation of lin-41, induce an Egl phenotype or sterility. This is indeed what we find (Fig. 3c; results not shown). Although the phenotypes described above are not specific enough to directly imply dcr-1 as an actor in the let-7/lin-41 pathway, the phenotypic relationship between animals with altered DCR-1 levels and animals with alterations in the let-7/lin-41 pathway, are suggestive.
To test this more directly we took two approaches. Using Drosophila embryo extracts and immunoprecipitates as a source of Dicer, we tested whether Dicer could process Drosophila let-7 precursor RNA into its mature form in vitro. Indeed, the ∼75-nt hairpin was processed into an ∼21-nt mature RNA with a disproportionately high efficiency as compared to perfect duplexes of similar size (Fig. 3d; data not shown). Second, we asked whether the dcr-1 mutation had an effect on the levels of let-7 RNA in vivo. We find that levels of mature let-7 RNA are reduced in dcr-1 mutant animals, and that this reduction is accompanied by an accumulation of the longer let-7 RNA precursor (Fig. 3e). Together this shows that dcr-1 is directly involved in the conversion of the double-stranded let-7 precursor RNA into the active, 21-nt species.
RNAi and development
RNAi and PTGS can clearly function to protect the genome against viruses and transposons (Kasschau and Carrington 1998; Baulcombe 1999; Ketting et al. 1999; Tabara et al. 1999; Mourrain et al. 2000; Wu-Scharf et al. 2000; Dalmay et al. 2001). In addition, there is some evidence that factors involved in RNAi or PTGS also play a role in proper germ-line development (Bohmert et al. 1998; Jacobsen et al. 1999; Smardon et al. 2000). We here show that at least one component of the RNAi machinery in animals, Dicer, also plays a role in generating small RNAs involved in developmental timing.
The mechanisms by which RNAi and stRNAs regulate the expression of target genes are quite distinct. In the former case, mRNAs are destroyed, whereas in the latter, expression is inhibited at the translational level. This raises the possibility that 22-nt RNAs produced by Dicer might act in multiple, distinct regulatory pathways that are not otherwise mechanistically related (Fig. 4). Alternatively, the effector machinery may be shared by both processes, with an altered outcome of target recognition. The let-7 RNA is not perfectly homologous to its target substrates, and such a mismatch may inhibit the ability of RISC to cleave its substrates, effectively switching the mode of regulation from degradation to translational repression. It should be noted that let-7 is, most likely, not the only substrate for Dicer that is required for normal development. There may be many other endogenously encoded dsRNAs that are processed by Dicer to produce stRNA molecules, for example, lin-4 (Olsen and Ambros 1999). For this gene it has been shown that the mismatch between lin-4 and its target is critically required for proper regulation (Ha et al. 1996).
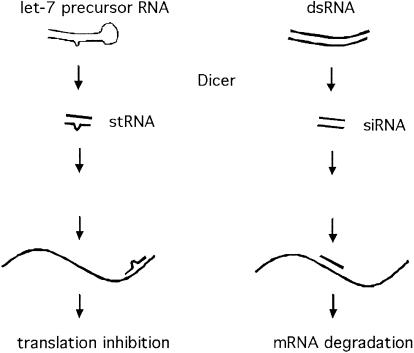
Figure 4.
Dicer may be involved in two distinct processes, both of which require the processing of dsRNA into small RNA (stRNA, small temporal, or regulatory; or siRNA, short interfering). One of these pathways (dsRNA-induced) leads to RNA degradation via the RISC complex and RNAi. The second pathway (let-7-dependent) leads to translational suppression via interaction between endogenous, small temporal RNAs and their mRNA targets. In the stRNA branch the double-stranded precursor and the match between stRNA and target RNA are not perfect (at least for let-7 and lin-4). We indicate this by including a bulge in the dsRNA molecules. This is not intended to specify size or number of mismatched regions.
The 22-nt siRNAs that act in RNAi/PTGS have been found in multiple species (Hamilton and Baulcombe 1999; Hammond et al. 2000; Parrish et al. 2000; Yang et al. 2000; Zamore et al. 2000). We have recently detected Dicer activity in extracts of plants and fungi (Nicotiana benthamiana and Neurospora crassa, A. Denli and G.J. Hannon, unpubl.; N. crassa and Phytophthora infestans, R.F. Ketting and R.H.A. Plasterk, unpubl.). Thus far, stRNA genes (like let-7) have been identified in animals ranging from C. elegans to humans (Pasquinelli et al. 2000). Considered together, these observations suggest conserved roles for Dicer proteins in both dsRNA-induced silencing and in regulating developmental timing. Ironically, small temporal RNAs have yet to be identified in plants, in which developmental defects have been associated with mutants in the RNAi machinery (Bohmert et al. 1998; Jacobsen et al. 1999). It will therefore be of interest to determine whether small temporal RNAs also regulate developmental timing in plants and to investigate whether this mode of gene regulation might also extend to nondevelopmental programs of gene expression.
Materials and methods
In vitro processing reactions
C. elegans embryonic extracts were made from eggs isolated from pos-1 dsRNA-fed mothers. They were resuspended in 0.5 volume of lysis buffer (10 mM Hepes at pH 7.0, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, 10% glycerol, 1 mM DTT, protease inhibitors; Roche). The eggs were ground in a dounce tissue grinder. The lysate was cleared by spinning 10 min at 12,000g. The supernatant was frozen at −80°C. Drosophila embryos and S2 cell extracts were prepared and all processing reactions were performed as described (Bernstein et al. 2001).
Antibody production and immunoprecipitation
Polyclonal antisera were raised against a protein fragment encoded by the two C-terminal exons of the K12H4.8 gene. The protein fragment was isolated from inclusion bodies and further purified using a C-terminal 6×His tag. Both rabbits produced antibodies that are capable of precipitating a fraction of the Dicer. Immunoprecipitations were performed as described (Bernstein et al. 2001) except that 100 μL of C. elegans extract was incubated with 10 μL of preimmune or immune serum at 4°C for 1 h.
K12H4.8 deletion mutant
The deletion allele pk1351 of K12H4.8 was isolated from a C. elegans deletion library (Jansen et al. 1997). pk1351 deletes 1629 nt, starting at base pair 2132 in K12H4.8 (in an intron), removing part of the helicase domain and part of the PAZ domain. Splicing to exons further downstream leads to frameshifts and early stops. Most likely this is a null allele of K12H4.8. The deletion allele was followed by PCR, using primers K12F21 and K12R21. The wild-type allele was visualized using primers K12F21 and K12R22. The mutant was outcrossed seven times with Bristol N2. The sterility and burst vulva phenotype cosegregated perfectly in these crosses. The experiments described here were all done with the outcrossed deletion mutant.
Transgenics
A PCR fragment was made using AccuTaq (Sigma) that contains the complete K12H4.8 plus 500 bp of the upstream region. A transgene (pkEx1415) was made from a mix of linearized pRF4 plasmid DNA at 5 ng/μL, PCR fragment at 1 ng/μL, and PvuII-digested genomic DNA at 100 ng/μL. pkEx1415 results in an Egl phenotype (>80%) and partially rescues dcr-1(pk1351). A heat shock promoter fusion (hsp16_2::dcr-1) was made by PCR: primers hspA and hspB were used to make the promoter fragment and hsp-dcr and K12H4.8 R1 to make the dcr-1 fragment. The two PCR products were fused using primers hspC and K12H4.8 R2. This product was injected at 50 ng/μL, together with pRF4 at 50 ng/μL. Among the F1 transgenics grown at 15°C, 7 out of 70 displayed the phenotype shown in Figure 3c. One line was obtained (pkEx1416), reproducing the sterility phenotype when the animals are heat shocked at the L3 stage, at 30°C for 2 h. Sequences are available on request.
Seam cell analysis
The seam cells were visualized using the jcIs1 transgene. Animals containing this transgene were grown at 15°C. Of the dcr-1(pk1351) mutant animals, 60% (38/62) displayed no alae, and showed the seam cell defect depicted in Figure 3b. This phenotype and the burst vulva phenotype become less penetrant at higher temperatures. This temperature effect may be explained by a better activity of maternally contributed protein at higher temperatures, as also found in the in vitro assay (data not shown).
RNAi assays
RNAi against unc-22, dcr-1, and gfp was performed by feeding Eschericha coli that expresses dsRNA (Fraser et al. 2000). Weak dcr-1 phenotypes became apparent after culturing the animals on food expressing dcr-1 dsRNA for three generations: animals started to develop protruding vulvas, a low level of sterility was detected, and an accumulation of let-7 precursor RNA was detected. Apparently it is possible to lower the activity of a gene required for RNAi by performing RNAi against this gene. Similar results were obtained in Drosophila (Bernstein et al. 2001). The GFP dsRNA-expressing clone (L4417) was obtained from A. Fire (Carnegie Institution of Washington, Baltimore, MD). The GFP-expressing transgene that was used to assay RNAi in the germ line is ruIs48[pAZ147: pie-1/tubulin/GFP fusion], from strain AZ235.
RNA analysis
RNA for the Northern blots was isolated from 800–2000 young adult animals displaying the dcr-1(pk1351) phenotype, or from young adult N2 animals, fed with normal food, or with dcr-1 food. RNA was run on a 15% denaturing acrylamide gel. Radiolabeled DNA complementary to the let-7 mature RNA was used as probe.
Acknowledgments
We thank Amy Caudy and Karen Thijssen for technical assistance, and Marcel Tijsterman and Rik Korswagen for helpful discussions. We thank A. Fire for reagents. Some of the strains used in this work were obtained from the Caenorhabditis Genetics Center (CGC). R.F.K. is supported by the Centre for Biomedical Genetics. S.E.J. is supported by the Dutch Science Foundation (NWO). This work was supported in part by a grant from the NIH (RO1-GM62534, GJH). G.J.H. is a Rita Allen Foundation Scholar.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
After submission of this work several other labs independently reported on the role of Dicer in RNAi and development: Grishok et al. (2001), Hutvagner et al. (2001), and Knight and Bass (2001).
Footnotes
E-MAIL hannon@cshl.org; FAX (516) 367-8874.
E-MAIL plasterk@niob.knaw.nl; FAX 31-302516464.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.927801.
References
- Anandalakshmi R, Pruss GJ, Ge X, Marathe R, Mallory AC, Smith TH, Vance VB. A viral suppressor of gene silencing in plants. Proc Natl Acad Sci. 1998;95:13079–13084. doi: 10.1073/pnas.95.22.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL. Double-stranded RNA as a template for gene silencing. Cell. 2000;101:235–238. doi: 10.1016/s0092-8674(02)71133-1. [DOI] [PubMed] [Google Scholar]
- Baulcombe DC. Gene silencing: RNA makes RNA makes no protein. Curr Biol. 1999;9:R599–R601. doi: 10.1016/s0960-9822(99)80383-2. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: The novel PAZ domain and redefinition of the Piwi domain. Trends Biochem Sci. 2000;25:481–482. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- Chuang CF, Meyerowitz EM. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci. 2000;97:4985–4990. doi: 10.1073/pnas.060034297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T, Horsefield R, Hartig Braunstein T, Baulcombe DC. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 2001;20:2069–2077. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie D, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Ha I, Wightman B, Ruvkun G. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes & Dev. 1996;10:3041–3050. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Caudy AA, Hannon GJ. Post-transcriptional gene silencing by double-stranded RNA. Nat Rev Genet. 2001;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Running MP, Meyerowitz EM. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development. 1999;126:5231–5243. doi: 10.1242/dev.126.23.5231. [DOI] [PubMed] [Google Scholar]
- Jansen G, Hazendonk E, Thijssen KL, Plasterk RH. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat Genet. 1997;17:119–121. doi: 10.1038/ng0997-119. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Carrington JC. A counterdefensive strategy of plant viruses: Suppression of posttranscriptional gene silencing. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- Knight, S.W. and Bass, B.L. 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in C. elegans. [Published online 2 August/10.1126/Science 1062039] [DOI] [PMC free article] [PubMed]
- Kooter JM, Matzke MA, Meyer P. Listening to the silent genes: Transgene silencing, gene regulation and pathogen control. Trends Plant Sci. 1999;4:340–347. doi: 10.1016/s1360-1385(99)01467-3. [DOI] [PubMed] [Google Scholar]
- Lucy AP, Guo HS, Li WX, Ding SW. Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J. 2000;19:1672–1680. doi: 10.1093/emboj/19.7.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel J-B, Jouette D, Lacombe A-M, Nikic S, Picault N, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen RA. Introduction of a chimeric chalcone synthase gene into Petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Parrish S, Fleenor J, Xu S, Mello C, Fire A. Functional anatomy of a dsRNA trigger. Differential requirement for the two trigger strands in RNA interference. Mol Cell. 2000;6:1077–1087. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Plasterk RH, Ketting RF. The silence of the genes. Curr Opin Genet Dev. 2000;10:562–567. doi: 10.1016/s0959-437x(00)00128-3. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Sharp PA. RNA interference 2001. Genes & Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- van der Krol AR, Mur LA, Beld M, Mol JN, Stuitje AR. Flavonoid genes in Petunia: Addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell. 1990;2:291–299. doi: 10.1105/tpc.2.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Pinto YM, Baulcombe DC. Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci. 1999;96:14147–14152. doi: 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu-Scharf D, Jeong B, Zhang C, Cerutti H. Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science. 2000;290:1159–1162. doi: 10.1126/science.290.5494.1159. [DOI] [PubMed] [Google Scholar]
- Yang D, Lu H, Erickson JW. Evidence that processed small dsRNAs may mediate sequence-specific mRNA degradation during RNAi in Drosophila embryos. Curr Biol. 2000;10:1191–1200. doi: 10.1016/s0960-9822(00)00732-6. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]