Abstract
Bacterial tmRNA mediates a trans-translation reaction, which permits the recycling of stalled ribosomes and probably also contributes to the regulated expression of a subset of genes. Its action results in the addition of a small number of C-terminal amino acids to protein whose synthesis had stalled and these constitute a proteolytic recognition tag for the degradation of these incompletely synthesized proteins. Previous work has identified pseudoknots and stem–loops that are widely conserved in divergent bacteria. In the present work an alignment of tmRNA gene sequences within 13 β-proteobacteria reveals an additional sub-structure specific for this bacterial group. This sub-structure is in pseudoknot Pk2, and consists of one to two additional stem–loop(s) capped by stable GNRA tetraloop(s). Three-dimensional models of tmRNA pseudoknot 2 (Pk2) containing various topological versions of the additional sub-structure suggest that the sub-structures likely point away from the core of the RNA, containing both the tRNA and the mRNA domains. A putative tertiary interaction has also been identified.
INTRODUCTION
Even the finding of ribosomes bypassing 50 nt present in the coding sequence of an mRNA did not prepare us for the surprise that ribosomes can switch from one mRNA to another while synthesizing one protein. This feat is accomplished when bacterial or plastid ribosomes come to the 3′-end of a mRNA that lacks a translation termination codon (1–4). The favored model is that aminoacylated tmRNA enters the ribosomal A site, probably in a complex with elongation factor Tu (5,6), with the help of a specific protein, SmpB (7), thereby providing an escape valve for ‘stuck’ ribosomes. tmRNA is so named because in this process it functions both as tRNA and mRNA. In Escherichia coli, tmRNA is 363 nt long and the two ends pair to form the partial tRNA analog that is aminoacylated with alanine (8–10). After donation of this alanine to the stalled peptide, the ribosomes resume translation of an internal sequence in tmRNA itself (11), allowing ribosomes to reach a terminator and then recycle. The amino acids added to the stalled polypeptide due to tmRNA function constitute a tag recognized by a specific protease, leading to degradation of the aberrant protein product (12). Recent work has focused on a role for tmRNA in the control of gene expression (13), a requirement for tmRNA for the growth of stressed Bacillus subtilis (14), a demonstration that functioning tmRNA is essential for Neisseria gonorrhoeae though not for its role in proteolysis (15), the finding that tmRNA is important for Salmonella enterica pathogenesis (16) and that tmRNA is present even in the α-proteobacteria, where it is encoded in two pieces (17).
Structural analyses based on phylogenetic (18,19) and probing data (20,21) have led to a compact secondary structure model of E.coli tmRNA, encompassing six helices and four pseudoknots. One of the four pseudoknots, Pk1, is essential for protein tagging in vitro (22) and its structure has been confirmed in solution by a systematic mutational analysis combined with NMR spectroscopy (23). The rapid completion of several eubacterial DNA sequencing projects has recently made available tmRNA gene sequences from 89 species. These sequences are available at the tmRNA Web site (24; http://www.indiana.edu/~tmrna/) and 61 of these sequences are also shown at the tmRDB website of (25; http://psyche.uthct.edu/dbs/tmRDB/rna/tmrnaphylolist.html). Moreover, the sequence of 50 additional tmRNA genes, including members of eight additional phyla, were recently determined by PCR amplification (B.Felden, J.F.Atkins and R.F.Gesteland, unpublished results), bringing the total tmRNA gene sequences to 139 species from 21 bacterial phyla. These sequences permit an overall phylogenetic survey of tmRNA structure as refinement of sequence alignments within each bacterial phylum or within a specific subgroup is only achievable when a sufficiently large set of tmRNA sequences is available.
The present study was initiated to perform an extensive phylogenetic survey of tmRNA sequences within a single bacterial subdivision (β) of the proteobacteria (purple bacteria or purple non-sulfur bacteria). Within a subset of closely related sequences, tmRNA may possess peculiar structural signatures specific to the bacterial subdivision. Also, the high homogeneity between the subset of tmRNA sequences should be sufficient to obtain an unambiguous sequence alignment, especially around the coding sequence as well as for all the unpaired nucleotide stretches. A sequence consensus specific to a single bacterial subgroup will be proposed and compared to the one derived from all known tmRNA sequences. Such an alignment within a selected bacterial subgroup should also help the search for putative tertiary interactions in tmRNAs, a task that has been difficult to achieve. In the course of a recent study (26), we noticed that the tmRNA sequences from the β-proteobacteria may possess an additional structural domain within pseudoknot 2, Pk2, one of the four pseudoknots that is generally conserved throughout the known tmRNAs sequences. Thus, the β-proteobacteria constitute an interesting bacterial subgroup for extensive sequence analysis.
MATERIALS AND METHODS
Bacterial genomic DNAs were prepared as described (26). Sequences of tmRNA genes were obtained by PCR using primer sets A (5′-GGG GCT GAT TCT GGA TTC GAC-3′ and 5′-TGG AGC TGG CGG GAG TTG AAC-3′ based on E.coli tmRNA termini) or B (5′-GGG GGC GGA AAG GAT TCG ACG –3′ and 5′-TGG AGG CGG CGG GAA TCG AAC-3′, based on Thermotoga neapolitana tmRNA termini), combined to dye terminator sequencing with the PCR primers. Some β-proteobacteria tmRNA gene sequences already known have been verified in this study, e.g. Bordetella pertussis tmRNA, and some sequence mistakes, or possibly strain variations, have been found.
An initial computer alignment was performed by using the algorithm COSEQ (27), and the alignment was subsequently refined manually, considering both primary and secondary structure, attempting to avoid gaps. Three-dimensional models were constructed as described (28).
RESULTS AND DISCUSSION
Nine previously uncharacterized tmRNA genes from the β-proteobacteria have been amplified, sequenced and aligned. The genes are from six families (Alcaligenaceae, Ammonia-oxidizing bacteria, Burkholderiaceae, Comamonadaceae, Methylophilaceae and Neisseriaceae) and nine genera (Alcaligenes, Bordetella, Chromobacterium, Comamonas, Hydrogenophaga, Methylobacillus, Nitrosomonas, Ralstonia and Variovorax) (Table 1) that were not determined in a previous study (26). The organisms utilized were chosen with the aim of covering a broad range of families and genera within a single bacterial subdivision. Three additional tmRNA sequences (B.pertussis, Neisseria meningitidis and N.gonorrhoeae), available at both the tmRNA and tmRDB Web sites (24,25), are also included in Table 1. All predicted proteolysis tags of the 13 tmRNA gene sequences presumably start with a non-coded alanine residue (from the chargeable tmRNA 3′-end) followed by 10 internally encoded amino acids, eight of which are strictly conserved among the subgroup (Table 1, bold letters). The two encoded amino acids that are not strictly conserved are centered in the tag at positions 5 and 6 (three different amino acids are found at both positions), suggesting a stronger selective pressure at both ends of the tag. Hydrophobic amino acids (ALAA) at the C-terminus of the tag are required for efficient proteolysis of the tagged proteins (2), likely accounting for the sequence conservation at the C-terminus of the tag. The reason for the strict conservation of the first four amino acids in the tag (ANDE) within this bacterial subgroup is, however, unknown.
Table 1. List of β-proteobacteria tmRNA sequences used in this study.
| Complete name |
Abbreviation |
Putative proteolysis tag |
Strain |
Reference |
| N.gonorrhoeae | Nis.gonorr | (A)ANDETYALAA* | 24 | |
| N.meningitidis | Nis.meninS | (A)ANDETYALAA* | 24 | |
| Methylobacillus glycogenes | Mtb.glycog | (A)ANDETYALAA* | ATCC no. 29475 | This work |
| Chromobacterium violaceum | Crb.violac | (A)ANDETYALAA* | ATCC no. 07461 | This work |
| Nitrosomonas cryotolerans | Nms.cryoto | (A)ANDENYALAA* | ATCC no. 49181 | This work |
| Nitrosomonas europaea | Nms.europa | (A)ANDENYALAA* | 24 | |
| Comamonas testosteronia | Com.testos | (A)ANDERFALAA* | ATCC no. 11996 | This work |
| V.paradoxus | Vrv.parado | (A)ANDERFALAA* | ATCC no. 25301 | This work |
| Hydrogenophaga pallerii | Hph.paller | (A)ANDERFALAA* | ATCC no. 17728 | This work |
| Ralstonia eutrophab | Ral.eutrop | (A)ANDERYALAA* | DSM no. 531 | This work |
| Alcaligenes faecalis | Alc.faecal | (A)ANDERFALAA* | This work | |
| B.pertussisc | Brd.pertus | (A)ANDERLALAA* | 24 |
aPreviously misnamed Pseudomonas testosteroni.
bPreviously known as Alcaligenes eutrophus. This tmRNA was resequenced since the previous sequence (38) lacks 12 nt at positions that are conserved in all the other sequences from β-proteobacteria.
cAlso sequenced in the present work.
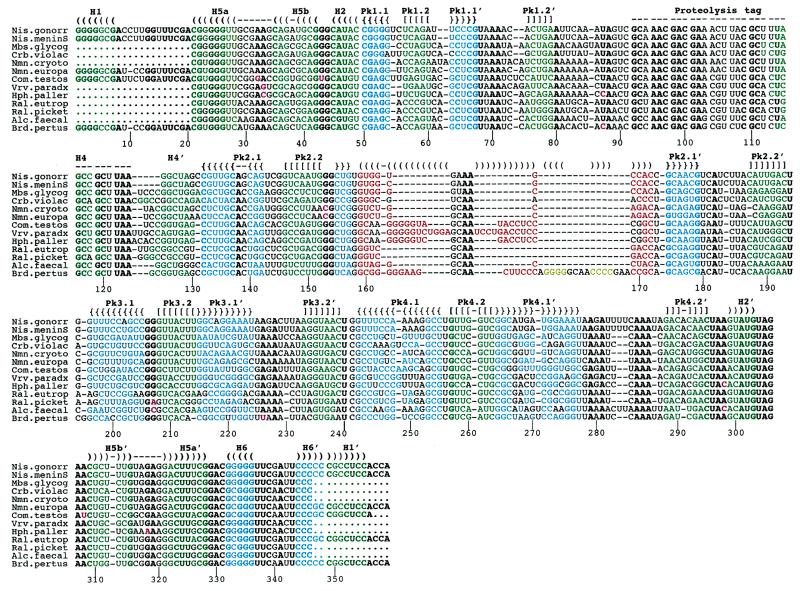
A sequence alignment of these 13 tmRNA genes from the β-purple subgroup is shown in Figure 1. The primary sequences are homogeneous and can be aligned with accuracy. As anticipated, the usual tmRNA secondary structural domains including four pseudoknots (Pk1–Pk4) and six helices (H1–H6) are apparent in all the new tmRNA sequences. The nomenclature of helices and pseudoknots in tmRNAs is from Felden et al. (19). The equivalence to another nomenclature (18) is as follows: Pk1–Pk4 are ψ1–ψ4, H1 is P1, H2 is P2c, H4 is P5, H5 is P2a-b and H6 is P12. For consistency, stems H1–H6 (green) and pseudoknots Pk1–Pk4 (blue-green) are color-coded as in Figure 2 (top). Unexpectedly, there is a region of strict sequence conservation in stems H1, H2, H5a and Pk1.1, suggesting the importance of the nucleotide sequence of these RNA helices for tmRNA structure and/or function. On the contrary, the primary sequences in helices Pk1.2 and H5b and all the helices of pseudoknots Pk2, Pk3 and Pk4, although maintaining canonical or wobble base pairings, are not conserved within this bacterial subgroup. This suggests that the architecture, but not the specific sequences, of these paired regions is involved in tmRNA function.
Figure 1.
Alignment of 13 tmRNA gene sequences from the β-proteobacteria. Conserved nucleotides within this bacterial subgroup are in bold. The color code is as follows: green and blue, canonical base pairs within stems and pseudoknots conserved in all known bacterial tmRNAs; red and yellow, insertions present in tmRNAs from the β-proteobacteria only; purple, nucleotides from the β-proteobacteria that do not follow the consensus of tmRNA sequences. For clarity, identical brackets and dashes cap the two RNA strands located apart within the primary sequence that form a helix. The sequence numbering follows the consensus sequence of β-proteobacteria tmRNAs.
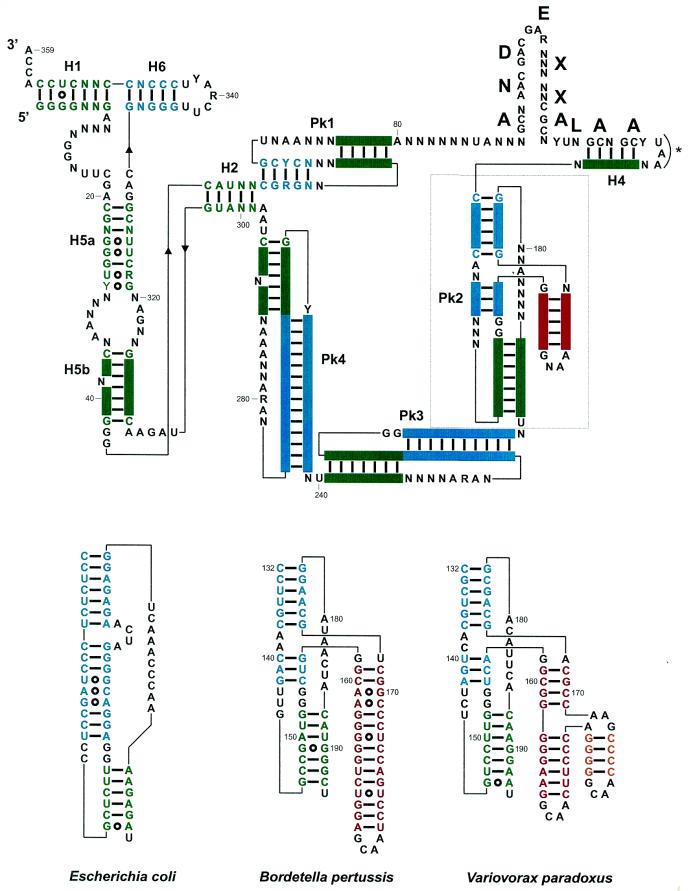
Figure 2.
(Top) Consensus secondary structure for tmRNAs from the β-proteobacteria, with the color code used in Figure 1. Structural domains are H1–H6 for the RNA helices, and Pk1–Pk4 for the pseudoknots. A predicted amino acid sequence consensus is shown above the coding sequence. The bold letters correspond to the encoded amino acids, with X corresponding to non-conserved amino acids. Pk2 is boxed, and its structural insertion, specific of tmRNAs from the β-proteobacteria, is in red. The stop codon is marked by an asterisk. (Bottom) Phylogenetically supported secondary structures of the additional domain (in red and yellow) within Pk2 from representative β-proteobacteria, V.paradoxus and B.pertussis compared to E.coli. Notice the incremental intricacy of the structural insertion. Solid bars between bases represent the Watson–Crick base pairs and open circles the non-Watson–Crick pairs. The two invariant G residues (positions 153 and 154) at the junction between the helices forming Pk2 require some discussion. They could form pairs with positions 143 and 144 (as positions 151–152 and 163–164 do in E.coli) and the drawings shown in Figure 3 would tend to support that view. However, two points prevent a definite conclusion; first, the bases vary on only one strand and, secondly, the relative lengths of the helices forming Pk2 vary so that one cannot derive a unique alignment between the β-proteobacteria and E.coli. Thus, they are shown as unpaired.
All the conserved nucleotides in the tmRNA sequences from β-proteobacteria are shown bold in Figure 1. Although they can be used only with difficulty in covariation analysis, some conserved nucleotides maintain Watson–Crick G=C pairs at the ends of helices H5a, H5b, Pk1.1, Pk2.1 and Pk4.2 (Fig. 2, top), which would tend to suggest that they do exist as such. They probably either restrict the overall folding of tmRNA in that bacterial subgroup by locking in the ends of helices or, alternatively, they might serve as recognition elements. Conserved nucleotides are also found upstream and downstream of Watson–Crick paired regions. In some of those cases, they may well form non-Watson–Crick pairs, extending the nearby stems by one to two additional base pairs. Specific examples are a GA tandem (G19–A330/A20–G329) upstream of H5a and two putative GA pairs (G43–A308 and G44–A307) downstream of H5b. Conserved nucleotides are also located in the four single strands that are predicted to cross the shallow grooves of all four pseudoknots: U68NAA71 in Pk1, A182 in Pk2, A226RA228 in Pk3 and A279RANNAAA286 in Pk4, with R being a purine and N being any of the four nucleotides (Fig 1; Fig 2, top). These conserved nucleotides, especially the adenines, may be required for pseudoknot stability by forming non-Watson–Crick pairs in the shallow grooves of helices, or might also interact with other parts of the molecule.
Residues U194, U240 and U296 are also strictly conserved. Interestingly, they are all located 3′ (downstream) of pseudoknots Pk2, Pk3 and Pk4, respectively. Uridine residues have a propensity for inducing sharp turns in a phosphodiester backbone, and they may be required to assemble the three pseudoknots in tmRNA into a compact RNA structure. As a consequence, pseudoknots in tmRNA may not be packed linearly as proposed in the tmRDB database (http://psyche.uthct.edu/dbs/tmRDB/rna/3d-models/pdb/esccoltm3D-model-12.pdb), but rather folded together in a compact array, head to tail and in an anti-parallel fashion, as in the structured group I introns (29) and RNase P RNA (27).
As a first indication about how tmRNA pseudoknots might interact with each other, we noticed a covariation between positions 227 and 280 from Pk3 and Pk4 that might reflect a long-range interaction. Nucleotide 227 (A or G in Fig. 1; R in Fig. 2, top) is located in the single strand crossing the shallow groove in Pk3 and nucleotide 280 (A or G in Fig. 1; R in Fig. 2, top) in the single strand crossing the shallow groove in Pk4. These two nucleotides are both surrounded by two conserved adenines (Fig. 1; Fig. 2, top), forming two 5′-ARA-3′ motifs. Interestingly, when R227 is a guanine, R280 is also a guanine in five cases (sequences 1–2 and 7–9, from top to bottom in Fig. 1). On the other hand, when R227 is an adenine, R280 is also an adenine in eight instances (sequences 3–6 and 10–13, from top to bottom in Fig. 1). The phylogenetic tree based on 16S rRNA sequences suggests that covariations of this type result from at least two independent events (in one instance, the G227/G280 converts into A227/A280 and, in the other case, there is the reverse conversion). These two independent covariation events provide evidence that these two nucleotides interact via non-Watson–Crick A-A or G-G pairings. These two purine-rich motives in both Pk3 and Pk4 are found at similar locations in all known tmRNA sequences. If one assumes that Pk3 and Pk4 adopt a standard pseudoknot with some co-axiality of their constitutive helices (Pk3.1/Pk3.2 and Pk4.1/Pk4.2), the segments containing the conserved 5′-ARA-3′ motifs would be located in the shallow groove of the first helix (Pk3.1 and Pk4.1) leading to potential contacts with base pairs of Pk3.1 and Pk4.1 as, for example, in the structure of the HDV RNA where conserved purines (in J4/2) interact with the shallow groove of helix P3 of a pseudoknot (30). In order for positions 227 and 280 to interact, the two pseudoknots Pk3 and Pk4 would have to be packed in a roughly antiparallel arrangement. At present, we cannot rule out that these nucleotide changes are just a coincidence or have occurred for reasons other than tmRNA structure, such as being part of a binding site for a tmRNA-binding protein. A similar example of exchange between trans Watson–Crick A.A (N1–N6 and N6–N1) and G.G (N1–O6 and O6–N1) pairs, which are isosteric, has been observed for the RRE RNA (31–33).
This proposed interaction needs further experimental testing, e.g. by UV cross-links and mutational analysis. If confirmed, it suggests that at least two pseudoknots in tmRNA, Pk3 and Pk4, are packed together via a tertiary interaction involving specific sequences located in the single strands crossing their shallow grooves. Again, this tertiary interaction conflicts with previous reports about the tertiary structure of tmRNAs (tmRDB database, http://psyche.uthct.edu/dbs/tmRDB/rna/3d-models/pdb/esccoltm3D-model-12.pdb) and favors the idea that tmRNA pseudoknots may well be folded together in a compact array.
In tmRNA sequences from all known γ-proteobacteria (the lineage that eventually gave rise to the β subdivision), a 4–10 nt bulge is found within the first stem (Pk2.1) of pseudoknot Pk2 (see E.coli, Fig. 2). This internal bulge is experimentally supported by probing data, at least for E.coli tmRNA (20,21). An additional structural domain, shown in red and yellow in Figures 1–3, replaces this internal bulge in all 13 tmRNA gene sequences surveyed here, and probably also in all β-proteobacteria. In most cases, this additional domain consists of a phylogenetically supported stem–loop of 3–13 bp (Fig. 1). With one exception (Chromobacterium violaceum), these additional helices are capped by a stable ‘GNRA tetraloop’ (34). In B.pertussis, a second additional stem–loop is predicted to insert into the first additional stem (in yellow in Figs 1–3). Interestingly, two stable GNRA tetraloops, including a ‘nicked tetraloop’ (35), cap the two ends of this second additional stem (for details see B.pertussis, Fig. 2). Thus, the 5′- and 3′-ends of this second insertion are brought into very close proximity, avoiding perturbation of the helical context of the first additional stem (Fig. 2, B.pertussis; Fig. 3).
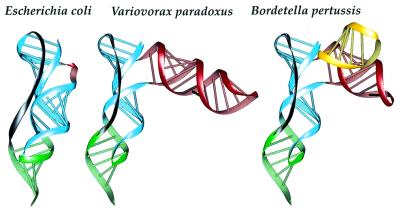
Figure 3.
Ribbon representation following the ribose–phosphate backbone of pseudoknot Pk2 for tmRNAs from E.coli, V.paradoxus and B.pertussis. RNA strands are color-coded as follows: blue, Pk2.1; green, Pk2.2; red, first additional stem–loop; yellow, second additional stem–loop; dark gray, single-strand junction.
With only one exception out of 13 sequences, the conservation of GNRA tetraloops within the additional domain is striking, raising the possibility of involvement in long-range tertiary interactions with other structural domains in tmRNA (36,37). However, the observed conservation of the ‘GNAA consensus’ (no ‘GNGA loop’ is observed) and the absence of the 11 nt receptor of GAAA tetraloops (37) do not support this hypothesis. Moreover, the large variation in the length of the additional stem within Pk2 is hardly compatible with the conservation of a tertiary contact. It should also be noted that the observed tetraloops mostly follow the sequence ‘GAAA’ or ‘GCAA’, i.e. the two most stable GNRA tetraloops. Therefore, the observed conservation of these GNRA tetraloops most likely reflects thermodynamic stability constraints and/or recent divergence.
Predictive three-dimensional models of pseudoknot Pk2 from E.coli, V.paradoxus and B.pertussis have been constructed, and are presented in Figure 3. These three bacterial species have been selected according to the various lengths of their structural insertions within Pk2, from simple to intricate (Figs 1 and 2). The predicted configuration of the three-way junction, between the two parts of stem Pk2.1 and the structural insertion, is rather unusual and provides no evidence for specific interactions between unpaired nucleotides. For this reason, the orientation of the additional stem(s) to both stems of pseudoknot Pk2, respectively, is somewhat arbitrary and a parallel arrangement of the new substructure with the other helices cannot be excluded. However, the lack of joining nucleotides at the three-way junction of the helical stems themselves suggests that a significant bend of stem Pk2.1 is required to accommodate the structural insertion (see Fig. 3, where it can be seen that the two parts of Pk2.1, in blue, make opposite bends of ~20°). On the other hand, the pseudoknotted fold is strongly constrained by the length of the single strand that crosses the shallow groove of stem Pk2.1 (7–8 nt in the known tmRNA sequences from β-proteobacteria versus 10 in E.coli). This single strand is relatively short when compared to the length of Pk2.1, which forms about a complete helical turn long. In that respect, the bend introduced by the structural insertion permits accommodation of a short junction. The presence of the additional sub-structures in only some subgroups of sequences suggests that, in the overall architecture of tmRNA, the sub-structures should be pointing away from the core structure.
Acknowledgments
ACKNOWLEDGEMENTS
Genomic DNAs from Ralstonia eutropha and B.pertussis were kindly provided by Drs J.W.Brown (North Carolina State University, USA) and L.Kirsebom (Uppsala University, Sweden), respectively. This work was supported by a Human Frontier Science Program research grant (RG0291/2000-M 100) to B.F. and J.F.A., and by a grant from the Department of Energy DEFG03-99ER62732 to R.F.G. and NIH grant GM48152 to J.F.A. C.M. was supported by a Human Frontier Science Program research grant (CRG0291/19997-M) to E.W.
References
- 1.Tu G.F., Reid,G.E., Zhang,J.G., Moritz,R.L. and Simpson,R.J. (1995) C-terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide. J. Biol. Chem., 270, 9322–9326. [DOI] [PubMed] [Google Scholar]
- 2.Keiler K.C., Waller,P.R.H. and Sauer,R.T. (1996) Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science, 271, 990–993. [DOI] [PubMed] [Google Scholar]
- 3.Himeno H., Sato,M., Tadaki,T., Fukushima,M., Ushida,C. and Muto,A. (1997) In vitro trans translation mediated by alanine charged 10Sa RNA. J. Mol. Biol., 268, 803–808. [DOI] [PubMed] [Google Scholar]
- 4.Karzai A.W., Roche,E.D. and Sauer,R.T. (2000) The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nature Struct. Biol., 7, 449–455. [DOI] [PubMed] [Google Scholar]
- 5.Rudinger-Thirion J., Giegé,R. and Felden,B. (1999) Aminoacylated tmRNA from Escherichia coli interacts with prokaryotic elongation factor Tu. RNA, 5, 989–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barends S., Wower,J. and Kraal,B. (2000) Kinetic parameters for tmRNA binding to alanyl-tRNA synthetase and elongation factor Tu from Escherichia coli. Biochemistry, 39, 2652–2658. [DOI] [PubMed] [Google Scholar]
- 7.Karzai A.W., Susskind,M.M. and Sauer,R.T. (1999) SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA. EMBO J., 18, 3793–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ushida C., Himeno,H., Watanabe,T. and Muto,A. (1994) tRNA-like structures in 10Sa RNAs of Mycoplasma capricolum and Bacillus subtilis. Nucleic Acids Res., 22, 3392–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komine Y., Kitabatake,M., Yokogawa,T. and Nishikawa,K. (1994) A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl Acad. Sci. USA, 91, 9223–9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nameki N., Tadaki,T., Muto,A. and Himeno,H. (1999) Amino acid acceptor identity switch of Escherichia coli tmRNA from alanine to histidine in vitro. J. Mol. Biol., 289, 1–7. [DOI] [PubMed] [Google Scholar]
- 11.Williams K.P., Martindale,K.A. and Bartel,D.P. (1999) Resuming translation on tmRNA: a unique mode of determining a reading frame. EMBO J., 18, 5423–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottesman S., Roche,E., Zhou,Y. and Sauer,R.T. (1998) The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev., 12, 1338–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abo T., Inada,T., Ogawa,K. and Aiba,H. (2000) SsrA-mediated tagging and proteolysis of LacI and its role in the regulation of the lac operon. EMBO J., 19, 3762–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muto A., Fujihara,A., Ito,K., Matsuno,J., Ushida,C. and Himeno,H. (2000) Requirement of transfer-messenger RNA for the growth of Bacillus subtilis under stress. Genes Cells, 5, 627–635. [DOI] [PubMed] [Google Scholar]
- 15.Huang C., Wolfgang,M.C., Withey,J., Koomey,M. and Friedman,D.I. (2000) Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO J., 19, 1098–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Julio S.M., Heithoff,D.M. and Mahan,M.J. (2000) ssrA (tmRNA) plays a role in Salmonella enterica Serovar typhimurium pathogenesis. J. Bacteriol., 182, 1558–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keiler K.C., Shapiro,L. and Wiliams,K.P. (2000) tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: a two-piece tmRNA functions in Caulobacter. Proc. Natl Acad. Sci. USA, 97, 7778–7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams K.P. and Bartel,D.P. (1996) Phylogenetic analysis of tmRNA secondary structure. RNA, 2, 1306–1310. [PMC free article] [PubMed] [Google Scholar]
- 19.Felden B., Himeno,H., Muto,A., Atkins,J.F. and Gesteland,R.F. (1996) Structural organization of Escherichia coli tmRNA. Biochimie, 78, 979–983. [DOI] [PubMed] [Google Scholar]
- 20.Felden B., Himeno,H., Muto,A., McCutcheon,J.P., Atkins,J.F. and Gesteland,R.F. (1997) Probing the structure of the Escherichia coli 10Sa RNA (tmRNA). RNA, 3, 89–103. [PMC free article] [PubMed] [Google Scholar]
- 21.Hickerson R.P., Watkins-Sims,C.D., Burrows,C.J., Atkins,J.F., Gesteland,R.F. and Felden,B. (1998) A nickel complex cleaves uridines in folded RNA structures: application to E.coli tmRNA and related engineered molecules. J. Mol. Biol., 279, 577–587. [DOI] [PubMed] [Google Scholar]
- 22.Nameki N., Felden,B., Atkins,J.F., Gesteland,R.F., Himeno,H. and Muto,A. (1999) Functional and structural analysis of a pseudoknot upstream of the tag-encoded sequence in E.coli tmRNA. J. Mol. Biol., 286, 733–744. [DOI] [PubMed] [Google Scholar]
- 23.Nameki N., Chattopadhyay,P., Himeno,H., Muto,A. and Kawai,G. (1999) An NMR and mutational analysis of an RNA pseudoknot of Escherichia coli tmRNA involved in trans-translation. Nucleic Acids Res., 27, 3667–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams K.P. (2000) The tmRNA website. Nucleic Acids Res., 28, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zwieb C. and Wower,J. (2000) tmRDB (tmRNA database). Nucleic Acids Res., 28, 169–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Felden B., Gesteland,R.F. and Atkins,J.F. (1999) Eubacterial tmRNAs: everywhere except the α-proteobacteria? Biochim. Biophys. Acta, 1446, 145–148. [DOI] [PubMed] [Google Scholar]
- 27.Massire C., Jaeger,L. and Westhof,E. (1998) Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol., 279, 773–793. [DOI] [PubMed] [Google Scholar]
- 28.Massire C. and Westhof,E. (1998) MANIP: an interactive tool for modelling RNA. J. Mol. Graph. Model, 16, 197–205. [DOI] [PubMed] [Google Scholar]
- 29.Lehnert V., Jaeger,L., Michel,F. and Westhof,E. (1996) New loop–loop tertiary interactions in self-splicing introns of subgroup IC and ID: a complete 3D model of the Tetrahymena thermophila ribozyme. Chem. Biol., 3, 993–1009. [DOI] [PubMed] [Google Scholar]
- 30.Ferre-D’Amare A.R., Zhou,K. and Doudna,J.A. (1998) Crystal structure of a hepatitis δ virus ribozyme. Nature, 395, 567–574. [DOI] [PubMed] [Google Scholar]
- 31.Battiste J.L., Mao,H., Rao,N.S., Tan,R., Muhandiram,D.R., Kay,L.E., Frankel,A.D. and Williamson,J.R. (1996) Helix-RNA major groove recognition in an HIV-1 rev peptide–RRE RNA complex. Science, 273, 1547–1551. [DOI] [PubMed] [Google Scholar]
- 32.Ye X., Gorin,A., Ellington,A.D. and Patel,D.J. (1996) Deep penetration of an α-helix into a widened RNA major groove in the HIV-1 rev peptide–RNA aptamer complex. Nat. Struct. Biol., 3, 1026. [DOI] [PubMed] [Google Scholar]
- 33.Hermann T. and Westhof,E. (1999) Non-Watson–Crick base pairs in RNA–protein recognition. Chem. Biol., 6, R335–R343. [DOI] [PubMed] [Google Scholar]
- 34.Woese C.R., Winker,S. and Gutell,R.R. (1990). Architecture of ribosomal RNA: constraints of the sequences of ‘tetra-loops’. Proc. Natl Acad. Sci. USA, 87, 8467–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abramovitz D.L. and Pyle,A.M. (1997) Remarkable morphological variability of a common RNA folding motif: the GNRA tetraloop-receptor interaction. J. Mol. Biol., 266, 493–506. [DOI] [PubMed] [Google Scholar]
- 36.Jaeger L., Michel,F. and Westhof,E. (1994) Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J. Mol. Biol., 236, 1271–1276. [DOI] [PubMed] [Google Scholar]
- 37.Costa M. and Michel,F. (1995) Frequent use of the same tertiary motif by self-folding RNAs. EMBO J., 14, 1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown J.W., Hunt,D.A. and Pace,N.R. (1990) Nucleotide sequence of the 10Sa RNA gene of the β-purple eubacterium Alcaligenes eutrophus. Nucleic Acids Res., 18, 2820. [DOI] [PMC free article] [PubMed] [Google Scholar]