Abstract
Nonsense-mediated mRNA decay (NMD) prevents the accumulation of transcripts bearing premature termination codons. Here we show that Saccharomyces cerevisiae NMD mutants accumulate 5′–extended RNAs (CD-CUTs) of many subtelomeric genes. Using the subtelomeric ZRT1 and FIT3 genes activated in response to zinc and iron deficiency, respectively, we show that transcription of these CD-CUTs mediates repression at the bona fide promoters, by preventing premature binding of RNA polymerase II in conditions of metal repletion. Expression of the main ZRT1 CD-CUT is controlled by the histone deacetylase Rpd3p, showing that histone deacetylases can regulate expression of genes through modulation of the level of CD-CUTs. Analysis of binding of the transcriptional activator Zap1p and insertion of transcriptional terminators upstream from the Zap1p binding sites show that CD-CUT transcription or accumulation also interferes with binding of the transcriptional activator Zap1p. Consistent with this model, overexpressing Zap1p or using a constitutively active version of the Aft1p transcriptional activator rescues the induction defect of ZRT1 and FIT3 in NMD mutants. These results show that cryptic upstream sense transcription resulting in unstable transcripts degraded by NMD controls repression of a large number of genes located in subtelomeric regions, and in particular of many metal homeostasis genes.
Author Summary
Precise expression of genes relies on their accurate and timely transcription and on turning off these genes when production of the proteins is not required. Our study describes that in the baker's yeast, S. cerevisiae, repression of many genes relies on transcription of long extended RNAs upstream from where transcription normally initiates. These long extended RNAs are degraded by a machinery that recognizes that these RNAs do not encode any functional proteins. Using genes involved in controlling the uptake of the essential elements zinc and iron, we find that transcription of these long extended non-coding RNAs represses transcription by preventing the binding of the transcriptional machinery to the normal transcriptional control elements. Our findings show that repression of transcription of many genes relies on the transcription of unstable long RNAs and that this mechanism of control is particularly prevalent for genes involved in controlling the level of metals inside cells.
Introduction
A large fraction of eukaryotic genomes is transcribed, even in the non-coding regions (reviewed in [1]). One of the major questions that arise from these observations is to understand whether the RNAs expressed from these regions serve any functional purpose or whether they correspond to genomic noise that is ultimately routed for degradation. Transcription of non-coding (nc) RNAs nearby protein-coding genes has emerged as a means of transcriptional control (reviewed in [1], [2]). In the yeast S.cerevisiae, the first and best-documented example is the SRG1 ncRNA, which regulates transcription of the SER3 gene involved in serine metabolism through transcriptional interference [3]. While the SRG1 ncRNA is transcribed in the sense direction upstream of its target gene, most ncRNAs found to regulate S.cerevisiae transcription are antisense transcripts, such as the ones described for the PHO84, Ty-1 and GAL10 loci [4]–[7], which control chromatin modification marks at these genes. However, upstream sense transcription resulting in the ICR1 ncRNA has been found to regulate the FLO11 gene [8].
Cryptic transcripts can also be generated through the transcription of elements that control the expression of bona fide protein coding genes. There is ample evidence that promoter regions are associated with bidirectional transcription in S.cerevisiae [2], [9]. In addition, it has been shown that enhancers are transcribed by RNA polymerase II [10]. In S.cerevisiae, many transcripts associated with intergenic or promoter transcription are unstable under normal conditions. They are hardly detectable in wild-type strains because of their rapid degradation by nuclear RNA turnover. Indeed, many transcripts associated with cryptic transcription are detectable only when the activity of the nuclear exosome, or that of the TRAMP-complex, which stimulates exosome activity, is inhibited [2], [11], [12]. Therefore, these transcripts have been labeled “CUTs” for Cryptic Unstable Transcripts. Most of the degradative activities targeting CUTs seem to be concentrated in the nucleus. However, the cytoplasmic exonuclease Xrn1p can degrade the antisense RNAs that regulate the Ty-1 gene [5]. In addition, several CUTs can be degraded by the cytoplasmic degradation machinery [12]–[13], showing that the degradation of RNAs arising from transcription in non-coding regions can result from both nuclear and cytoplasmic RNA degradation pathways.
Nonsense mediated decay is an RNA surveillance mechanism that recognizes transcripts containing premature translation termination codons (PTCs; [14], [15]). This degradation system is used to prevent accumulation of aberrant mRNAs that would encode potentially toxic proteins [16], [17], but is also used for gene expression control. For example, NMD degrades alternatively or inefficiently spliced mRNAs that contain PTCs [18]–[21], and transcripts containing long 3′-UTRs [22]. In S.cerevisiae, NMD also controls Mg++ uptake by degrading the transcript encoding the major Mg++ transporter [23]. Previous studies using microarray analysis of S.cerevisiae NMD mutants have shown that NMD influences directly or indirectly the expression of many genes, including some that do not exhibit PTCs [24]–[26]. The identification of RNAs that associate with the NMD factor Upf1p by RNA pull-down has proved to be an efficient way to discriminate direct and indirect NMD targets [27]. However, many NMD targets do not accumulate to high levels in the presence of a functional NMD system, which renders this approach difficult for low expression transcripts. In this study, using tiling microarrays, we show that NMD degrades 5′-extended transcripts generated by cryptic transcription upstream from bona fide promoters, and that upstream transcription is responsible for the repression of metals homeostasis genes in conditions of metal repletion. These results show that NMD controls the expression of a large number of genes by modulating the expression of 5′-extended RNAs that control transcription.
Results
Accumulation of 5′-extended unstable RNAs of subtelomeric genes in NMD mutants
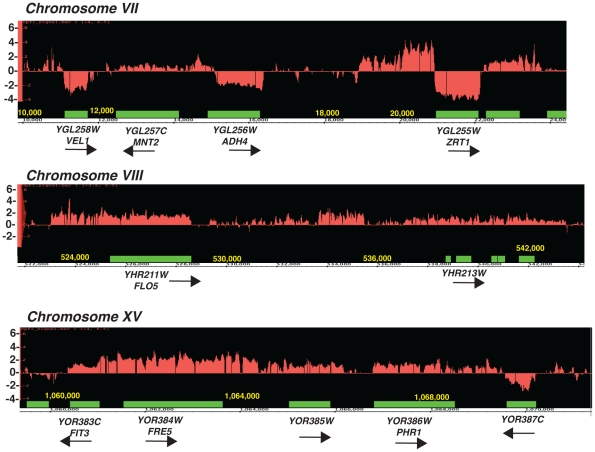
We previously used tiling microarrays to show that NMD controls the degradation of inefficiently spliced S.cerevisiae pre-mRNAs [21]. Further analysis of these microarrays in intergenic areas revealed that many subtelomeric regions accumulated RNA signal outside and upstream from the open-reading frames (ORF) in the upf1Δ and xrn1Δ mutants. Examples shown in Figure 1 include the subtelomeric ZRT1 gene encoding the primary zinc transporter [28], the FIT3 gene involved in siderophore-iron transport facilitation [29], and the FLO5 gene encoding a cell wall protein (Figure 1). Many genes located in subtelomeric regions are involved in the response to adverse growth conditions [30]. In the case of the FIT3 and FLO5 genes, the array profiles suggested that these species originate 5′-to the normal transcriptional start site and extend in the open-reading frame (which was confirmed by northern analysis, see below). In the case of ADH4 and ZRT1, increase of signal in the upstream regions correlated with a decrease of signal in the downstream regions (Figure 1). Northern blot analysis using antisense riboprobes confirmed that these species correspond to 5′-extended transcripts originating upstream from the bona fide promoters and partially or completely overlapping the open-reading frame (Figure 2 and see below). Based on the array profiles, some of these species were very long, containing 5′-extensions up to several kb for ZRT1 (Figure 1, Figure S2). In the case of FIT3, a previous study had mapped in detail the 5′-extended species that extends into the ORF [31]. Thus, these species are different from the short antisense CUTs transcribed divergently from the promoters [2], [9]. Accumulation of 5′-extended transcripts in NMD mutants has been reported for a few transcripts, but was interpreted as the result of transcriptional noise [27]. We hypothesized that some of these 5′-extended unstable transcripts might be used for regulatory purposes, as shown in other systems [3], [32], [33]. Because the accumulation of 5′-extended transcripts relied on the inactivation of cytoplasmic degradation pathways, we named these 5′-extended species CD-CUTs (Cytoplasmically Degraded Cryptic Unstable Transcripts).
Figure 1. Tiling arrays profiles of the upf1Δ mutant relative to the wild-type strain.
Green boxes represent the boundaries of open-reading frames, red the log2 ratio of the RNA signals detected in the upf1 Δ mutant relative to wild-type. Arrows indicate the direction of transcription of the ORFs. Shown are three segments of three chromosomes.
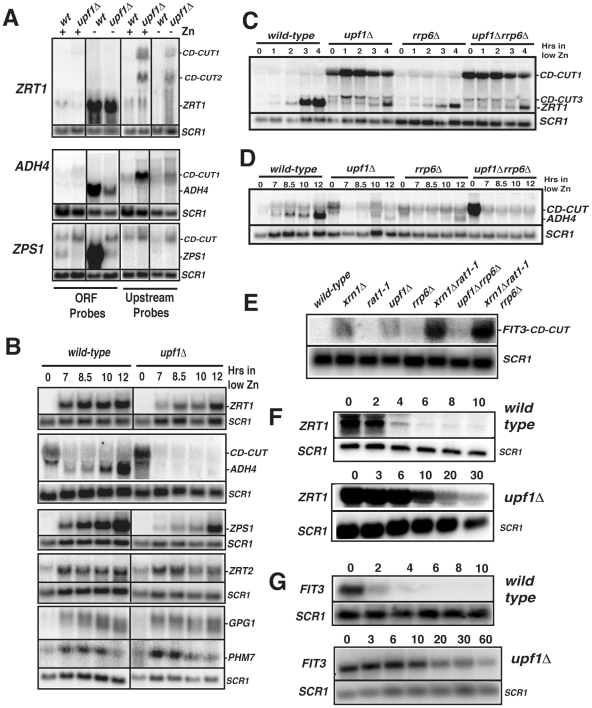
Figure 2. Characterization of CD-CUT of zinc and iron responsive genes.
A. Northern blots of indicated genes in wild-type and upf1Δ strains grown in the presence (+) or absence (−) of zinc. Extended species are marked as CD-CUT1 and 2. SCR1 was used as a loading control. B. Kinetics of induction of zinc regulon genes in wild-type and upf1Δ strains after a shift to a medium lacking zinc. C–E. Analysis of ZRT1, ADH4 and FIT3 CD-CUT and mRNA expression in ribonuclease mutants. C. Northern analysis of ZRT1 induction in wild-type, upf1Δ, rrp6Δ and upf1Δrrp6Δ prior to and after a shift into low zinc medium. D, as in A for ADH4. E, northern analysis of the FIT3 CD-CUT in iron-replete conditions in the indicated strains. F–G. Analysis of ZRT1 (F) and FIT3 (G) CD-CUT turnover in wild-type and upf1Δ strains. The GAL promoter was inserted upstream from the major site of transcription initiation of the ZRT1 or FIT3 CD-CUTs. After growth in galactose containing medium, cells were switched to glucose-containing medium and cell aliquots were harvested at the indicated times after the switch. CD-CUT levels were analyzed by northern blots using upstream riboprobes.
NMD mutants show a delay in the induction of subtelomeric metal homeostasis genes
In the case of subtelomeric genes involved in zinc metabolism (ZRT1, ADH4, VEL1, YOR387C, Figure 1; ZPS1, Figure S1), the accumulation of CD-CUTs was correlated with a decreased signal in the ORF regions, suggesting that they might play a negative role in the expression of the bona fide mRNAs. Interestingly, zinc regulon genes not located in subtelomeric areas, such as the one encoding the low affinity zinc transporter ZRT2 [34] did not exhibit CD-CUTs (data not shown). We found that the growth conditions used for the microarrays (synthetic complete minimal medium) corresponded to mild zinc deficiency conditions, in which some of the zinc regulon genes were partially induced (data not shown). Therefore the decrease of signal observed in the ORF regions for ZRT1 and ADH4 (Figure 1) might be due to a defect in the partial induction of these genes in minimal medium. We further analyzed the expression of zinc responsive mRNAs and their corresponding CD-CUTs in wild-type and upf1Δ strains grown in the presence or absence of zinc, using northern blots and antisense riboprobes covering the open-reading frames (ORFs) or the upstream (UP) regions (Figure 2A). This analysis confirmed the accumulation of CD-CUTs of ZRT1, ADH4 and ZPS1 in the upf1Δ mutant strain. For ZPS1, CD-CUTs were readily detectable in the wild-type as well as in the upf1Δ strain, explaining why only a modest increase in upstream signal was observed for this gene on the arrays, which compare the transcripts levels of the upf1Δ mutant to the wild-type (Figure S1). Some CD-CUTs were detected only with the upstream riboprobes, demonstrating that they are independent from the ORFs (Figure 2A). However, most CD-CUTs were also detected using the ORF riboprobes (Figure 2A and see below), indicating that they extend through the ORFs. A detailed characterization of the ZRT1 and ADH4 CD-CUTs using various upstream probes and ORF probes is shown in Figure S2 and described below. ORF riboprobes detected the induction of the normal ZPS1, ADH4, and ZRT1 mRNAs in wild-type cells grown in a medium lacking zinc (Figure 2A). However, these mRNAs were less abundant in the upf1Δ mutant (Figure 2A). This observation suggested that the accumulation of CD-CUTs due to NMD inactivation was deleterious to the expression of the bona fide mRNAs. We further characterized the expression of these genes in the upf1Δ mutant by monitoring their kinetics of induction (Figure 2B). This experiment showed that the upf1Δ mutant exhibited a delay in the induction of the subtelomeric ZPS1, ZRT1, ADH4 (Figure 2B) and VEL1/YOR387C genes (Figure S3A). The induction defect was also observed in the upf2Δ and upf3Δ strains (Figure S3A), indicating that it is a general feature of NMD mutants. In contrast, the induction of other zinc regulon genes such as ZRT2, GPG1 (YGL121C) and PHM7 (YOL084W), which are not localized in subtelomeric regions and do not exhibit CD-CUTs (data not shown) was unaffected by Upf1p absence (Figure 2B). A previous study found that NMD can influence gene expression by controlling the level of transcriptional activators [35]. We found that the mRNA levels of the Zap1p activator (which activates zinc regulon genes) were similar in wild-type and upf1Δ mutants (Figure S3B), suggesting that the delay of induction in the absence of Upf1p was not due to reduced Zap1p levels. Overall these results show that the defective induction of subtelomeric zinc-responsive genes in the upf1Δ mutant is not due to a global deficiency in zinc sensing in this mutant, but is specific to genes that exhibit CD-CUTs. Defective induction was also observed for the FIT3 iron depletion-responsive gene in a strain lacking Xrn1p or Upf1p activity (Figure S4). Overall these observations suggest that the presence or transcription of CD-CUTs represses the expression of many subtelomeric genes, some of which are involved in zinc or iron homeostasis.
Long 5′-extended transcripts of subtelomeric genes are primarily targeted by NMD, while short 5′-extended transcripts are degraded by NMD and nuclear RNA degradation pathways
To further investigate the mechanisms of turnover of CD-CUTs by the different RNA degradation machineries, we analyzed the accumulation of CD-CUTs of ZRT1 and ADH4 in strains lacking Upf1p, the nuclear exosome component Rrp6p, or both. These two genes were chosen because the 5′-extensions found in the CD-CUTs of ZRT1 and ADH4 are very different in size (extension of 2 kb for ZRT1 vs. a few hundred nucleotides for ADH4). This analysis showed that the major ZRT1 CD-CUT (CD-CUT1, Figure 2C) accumulated dramatically in the upf1Δ strain, and to a much lesser extent in the rrp6Δ mutant. Accumulation of the ZRT1 main CD-CUT-1 was not increased in the upf1Δrrp6Δ double mutant compared to the upf1Δ single mutant (Figure 2C). These observations suggest that the degradation of the longer CD-CUT of ZRT1 relies mostly on NMD, consistent with the long upstream region lacking any extended ORF. In contrast, the short ADH4 CD-CUT accumulated to similar levels in the upf1Δ and rrp6Δ strains (Figure 2D). Strikingly, the accumulation of this CD-CUT was dramatically increased in the upf1Δrrp6Δ double mutant (Figure 2D, time zero), suggesting that this CD-CUT is degraded by the cooperative action of the nuclear exosome and of NMD. Interestingly the induction of the bona fide ADH4 mRNA was completely defective in this double mutant strain, while the single mutants were only partially delayed. The correlation between the strong accumulation of the ADH4 CD-CUT in the upf1Δrrp6Δ double mutant and the severe induction defect of the bona fide ADH4 mRNA provides further evidence for the repression of this subtelomeric gene by its CD-CUT.
The CD-CUT of the subtelomeric iron responsive gene FIT3 was previously shown to accumulate in the xrn1Δrat1-1 double mutant [31], raising the question of which exonuclease was primarily responsible for its degradation. We analyzed the expression of this CD-CUT in strains lacking Xrn1p, Upf1p, Rrp6p, in the xrn1Δrat1-1 mutant strain and in other double mutant strains. This analysis revealed that the long CD-CUT of FIT3 accumulated to the highest level in the strain lacking Xrn1p and to a lesser extent Upf1p (Figure 2E), and its accumulation was not dramatically increased by Rrp6p inactivation, in contrast to what was found for ADH4. Thus, based on this steady-state analysis, the long CD-CUT of FIT3 is also primarily targeted by cytoplasmic turnover pathways that include Xrn1p and Upf1p.
The turnover of the ZRT1 and FIT3 CD-CUTs is dependent on NMD
Because CD-CUTs exhibit a lack of extended ORFs, we interpreted their accumulation in the upf1Δ and xrn1Δ mutant strains as the result of a lack of degradation by NMD. Alternatively, we could not rule out that transcription of these upstream regions might be indirectly up-regulated in these mutants. To test this hypothesis, GFP-HIS3 cassettes [36] were inserted upstream from the normal ZRT1 or ADH4 promoters in wild-type and upf1Δ strains, such that the expression of the GFP mRNA was under the control of the upstream regions (Figure S5A). Northern blot analysis of GFP inserted upstream from ZRT1 or ADH4 showed that this reporter transcript was expressed at similar levels in the wild-type and upf1Δ strains (Figure S5B). These results suggested that the accumulation of CD-CUTs in NMD mutants is not due to an increased transcription of these upstream regions, but to the absence of ORF in the 5′- extension of the CD-CUTs.
To gain further evidence that the turnover of CD-CUTs is directly dependent on NMD, we replaced the region upstream from the site of transcription initiation of the ZRT1 and FIT3 CD-CUTs with a galactose inducible promoter. This allowed us to measure the rate of decay of these CD-CUTs in the presence or absence of functional NMD. The kinetics of turnover of the ZRT1 CD-CUT (Figure 2F) or of the FIT3 CD-CUT (Figure 2G) showed that these species are much more unstable in the presence of functional NMD (t1/2 = 2–3 min.) than in the absence of Upf1p (t1/2 = 20–30 min.). Because the turnover rate of these species is strongly decreased when NMD is inactivated, we conclude that these CD-CUTs are directly targeted by NMD for their degradation.
The main CD-CUT of ZRT1 is activated by the histone deacetylase Rpd3p
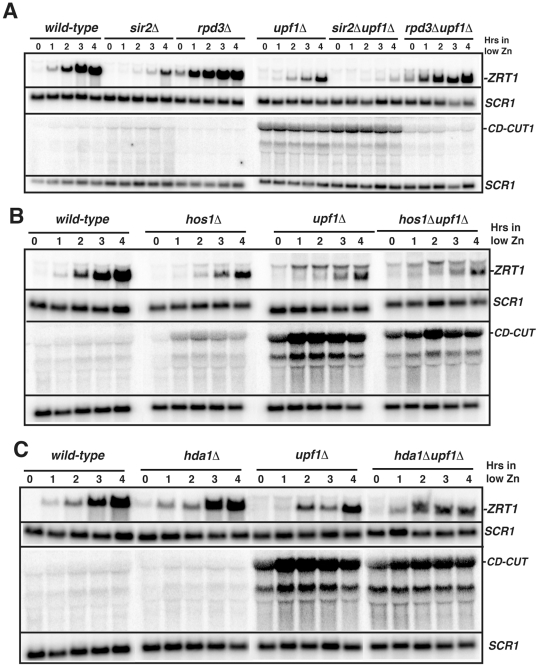
Previous microarray analysis of a strain inactivated for the histone deacetylase Rpd3p showed that ZRT1 is derepressed in the rpd3Δ strain and that Sir2p played a role antagonistic to Rpd3p in ZRT1 expression [37]. To investigate whether Rpd3p or Sir2p control ZRT1 by modulating the expression of its CD-CUT, we inactivated Upf1p in rpd3Δ or sir2Δ backgrounds and studied the induction of ZRT1 in these strains. ZRT1 was strongly derepressed in the rpd3Δ strain (Figure 3A, time zero), in agreement with previous data [37]. Strikingly, inactivation of Rpd3p in the upf1Δ strain completely rescued the induction defect of this NMD mutant, and resulted in a strong derepression of ZRT1 in normal zinc conditions (Figure 3A). Inactivation of Rpd3p also resulted in the almost complete disappearance of the ZRT1 CD-CUT observed in the upf1Δ strain. These results show that Rpd3p positively controls the expression of CD-CUT of ZRT1, and suggest that the derepression of ZRT1 in the rpd3Δ mutant [37] is due to the absence of the CD-CUT. In contrast, Sir2p inactivation reduced ZRT1 levels (Figure 3A), in agreement with the previous results [37]. Combining the sir2Δ deletion to the upf1Δ deletion exacerbated the ZRT1 induction delay when compared to the upf1Δ mutant (Figure 4A), but the sir2Δupf1Δ mutant did not exhibit higher levels of CD-CUT than the upf1Δ single mutant (Figure 4A). Therefore, the negative effects of Sir2p inactivation on ZRT1 expression are unlikely to be directly linked to its effect on the ZRT1 CD-CUT.
Figure 3. The main ZRT1 CD-CUT is controlled by the histone deacetylase Rpd3p.
A. Northern blot analysis of ZRT1 mRNA and CD-CUTs in wild-type, sir2Δ, rpd3Δ, upf1Δ and sir2Δupf1Δ and rpd3Δupf1Δ deletion strains. B. Northern blot analysis of ZRT1 mRNA and CD-CUTs in wild-type, hos1Δ, upf1Δ and hos1Δupf1Δ strains. C. Northern blot analysis of ZRT1 mRNA and CD-CUTs in wild-type, hda1Δ, upf1Δ and hda1Δupf1Δ strains.
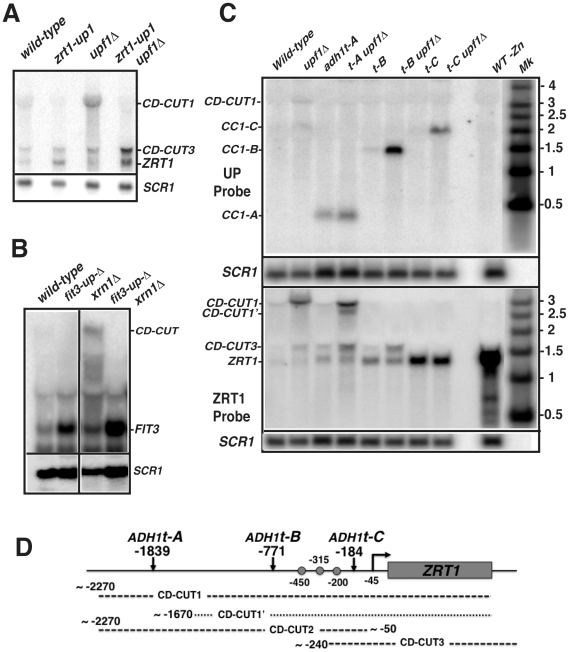
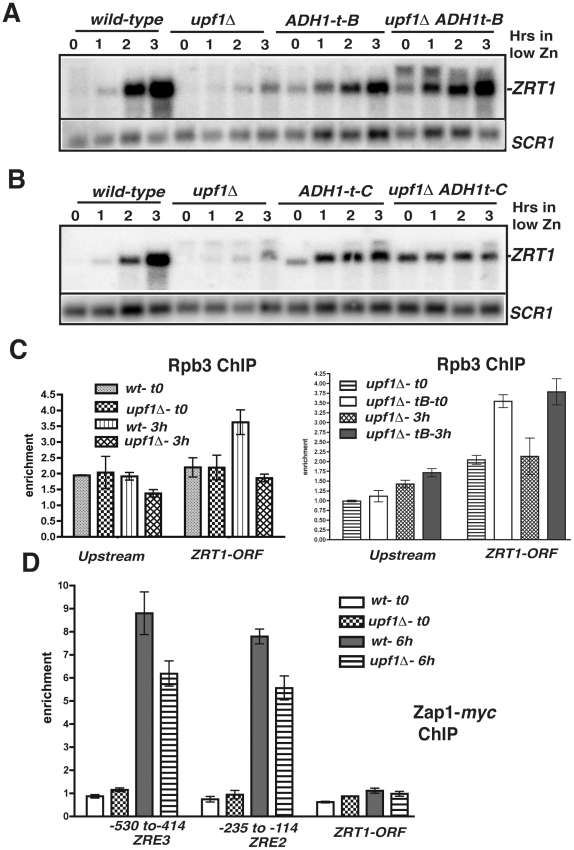
Figure 4. Mutations of the upstream regions and insertion of transcription terminators result in a derepression of ZRT1 and FIT3.
A. Effects of the insertion of a GFP-HIS3 cassette upstream ZRT1 (zrt1-up1) on ZRT1 mRNA levels in zinc repletion conditions in wild-type and upf1Δ strains. ZRT1 RNAs were assessed by northern blot using an ORF probe. B. Effects of the deletion of the region upstream FIT3 (fit3-upΔ) on FIT3 levels in iron repletion conditions in wild-type and xrn1Δ strains. FIT3 RNAs were assessed by northern blot using an ORF probe. C. Effects of the insertion of ADH1t transcriptional terminators at various sites upstream ZRT1 on ZRT1 mRNA and CD-CUT levels in zinc repletion conditions in wild-type and upf1Δ strains. Mk is a size marker. The upper membrane was hybridized with a probe upstream from the ZRT1 ORF, while the lower membrane was hybridized with a probe corresponding to the ZRT1 ORF. D. ADH1t terminator insertion sites and schematic maps of ZRT1 CD-CUT species. Dots represent the three major Zap1p binding sites.
To investigate the specificity of the effect observed with Rpd3p on the ZRT1 CD-CUT levels, we performed the same genetic analysis with the Hos1p, Hda1p, Hda2p and Hda3p deacetylases. The hos1Δ strain showed a slight delay in the induction of ZRT1, correlated with an increase of CD-CUT levels, but the hos1Δupf1Δ double mutant showed no additive effect when combined with the upf1Δ deletion (Figure 3B). Neither Hda1p (Figure 3C), nor Hda2p or Hda3p (Figure S6) were found to affect ZRT1 induction or repression. These results show that the major effect observed with Rpd3p on the ZRT1 CD-CUT is specific to this deacetylase. We tried to corroborate these results by monitoring the presence of Rpd3p in the region 5′ to ZRT1 but could not obtain reproducible evidence for enrichment by ChIP (data not shown). However the genetic data shown above strongly suggest that Rpd3p mediates the repression of ZRT1 through the modulation of the transcription of the CD-CUT.
Inactivation of the transcription of the CD-CUTs by deletion or transcriptional termination relieves ZRT1 and FIT3 repression and can rescue the induction defect of the upf1Δ mutant
If CD-CUTs are involved in the repression of the ZRT1 gene, we predicted that the replacement of its upstream region by the GFP-HIS3 coding cassette (Figure S5A) might alleviate its repression. Northern blot analysis of the strain carrying one of the insertions upstream from ZRT1 (zrt1-up1; inserted 1978 to 578 nucleotides upstream from the ZRT1 ATG; Figure S5A) showed a four-fold derepression of ZRT1 in zinc repletion conditions, both in the wild-type and upf1Δ backgrounds (Figure 4A). Insertion of this cassette eliminated the detection of the main CD-CUT of ZRT1, with the exception of the short CD-CUT3 (Figure 4A). This insertion also partially suppressed the induction defect of the upf1Δ strain during zinc deficiency (Figure S5C). The kinetics of disappearance of ZRT1 upon shifting back to zinc-containing medium was also monitored in these strains after 4 hours of induction, but we found no difference in the rate of ZRT1 shutoff in the presence or absence of its main CD-CUT (Figure S5C). A similar derepression was observed for FIT3 in a strain carrying a 3 Kb deletion of the region upstream of the FIT3 gene (from −4 kb to −1 kb upstream FIT3; fit3-upΔ, Figure 4B). Analysis of the fit3-upΔxrn1Δ double mutant strain showed that the CD-CUTs of FIT3 were eliminated in this double mutant, which confirmed that the derepression was due to the absence of the CD-CUT. Thus, deleting the regions encoding the CD-CUTs is sufficient to trigger derepression of the bona fide mRNAs, even in a wild-type context.
To provide more direct evidence that transcription of CD-CUTs is responsible for repression of the downstream promoters, we inserted the ADH1 transcription terminator (ADH1t) at 3 positions upstream from ZRT1 (tA: −1839, tB: −771 and tC: −184 bp; Figure 4C, 4D). If CD-CUT transcription or accumulation prevents the binding of RNA polymerase or of the transcriptional activator Zap1p, we hypothesized that terminating transcription of the CD-CUTs prior to the ZRT1 transcriptional control elements could derepress ZRT1 and/or rescue of the induction defect of NMD mutants. We first assessed ZRT1 mRNA and CD-CUTs levels in these strains in normal zinc conditions (Figure 4C). A sample from a strain grown in low zinc conditions was included as a control for the ZRT1 mRNA. Insertion of ADH1t at position A did not result in major ZRT1 derepression, probably because terminating transcription at this site results in activation of an alternative CD-CUT downstream from that site (labeled CD-CUT1′. Figure 4C). However insertion of this terminator resulted in a much shorter transcript that was now insensitive to a upf1 deletion, further showing that the sensitivity of CD-CUTs to NMD is dependent on their long size, and potentially on the lack of extended ORF in the 5′-extension. Strikingly, insertion of the ADH1t at positions −771 (B) or −184 (C) resulted in a derepression of ZRT1 (Figure 4C). The strongest effect was observed for ADH1t-C, possibly because this terminator stops all CD-CUT transcription immediately before the ZRT1 TATA box. In the ADH1t-B strain, an increased accumulation of the CD-CUT3 is observed, while this species disappears in the ADH1t-C strain. These results show that terminating transcription of the CD-CUTs upstream from the ZRT1 promoter is sufficient to derepress ZRT1 in conditions of non-induction. Additionally, insertion of these terminators allowed us to map in further detail the CD-CUTs upstream from ZRT1. Based on the hybridization pattern with the different probes (Figure S2A), the effect of the various terminators on their mobility in northern blots (Figure 4C), the approximate architecture of these CD-CUTs is shown in Figure 4D.
Transcription of the zinc regulon genes is activated by binding of the transcriptional activator Zap1p to their promoters during zinc deficiency [28], [38]. The terminator sequence inserted at position 771 is located upstream from the three major Zap1 binding sites (ZRE; [38]), while the terminator sequence inserted at position 184 is inserted downstream from them (Figure 4D). Based on this, we hypothesized that if transcription of the CD-CUTs prevents binding of Zap1p, the two strains containing terminators at the two positions might behave differently during a shift into low zinc conditions. Indeed, insertion of the ADH1t at position B derepressed ZRT1, and also fully rescued the induction defect of the upf1Δ strain (Figure 5A). However strains carrying an insertion of the ADH1t at position C failed to induce ZRT1 in conditions of induction, even in a context of active NMD. This result suggests that the region located between positions B and C, which contains most of the Zap1p binding sites must be accessible for ZRT1 induction. It is unclear why the strain containing the ADH1t at position C failed to induce ZRT1, even when NMD is active. It is possible that the higher levels of expression of the ZRT1 transporter in non-induction conditions resulted in higher cellular zinc levels prior to induction, thus delaying the response. Additionally we cannot rule out that inserting the ADH1t at site C might have changed the chromatin structure, and thus perturbed the induction of ZRT1.
Figure 5. Effects of transcription terminators upstream ZRT1 on ZRT1 induction and analysis of RNA Polymerase II and Zap1p occupancies by ChIP.
A and B. Effects of the insertion of ADH1 transcriptional terminators at various sites upstream ZRT1 on ZRT1 mRNA during zinc depletion in wild-type and upf1Δ strains. C. Analysis of RNA polymerase II occupancy in wild-type, upf1Δ and upf1Δ ADH1t-B strains grown in normal zinc medium (t0) or after a 3 hours shift in low zinc. Shown are normalized qPCR analysis of the different regions from ChIP samples obtained using anti-Rpb3p antibodies normalized to input DNA and to a qPCR product for a control non-transcribed region. D. Analysis of Zap1p occupancy at different sites of the ZRT1 gene. Legends as in 5C except that a myc-tagged version of Zap1p was used.
Binding of RNA Polymerase II and of the transcriptional activator Zap1p is defective in the NMD mutant upf1Δ
To corroborate these results, we studied RNA Polymerase II occupancy in two regions, upstream from ZRT1 (−1223 to −1123), and within the ZRT1 ORF (+905 to +1021), using chromatin immunoprecipitation (ChIP) of the Rpb3p subunit. We found slightly above background levels of occupancy of the polymerase in conditions of zinc repletion in both regions in wild-type and upf1Δ strains (Figure 5C). Interestingly, Rpb3p occupancy was similar for both strains in the upstream region, indicative of a similar level of transcription of the CD-CUTs. This result further indicates that accumulation of the CD-CUTs in the upf1Δ strain is due to a lack of degradation rather than increased transcription. Upon a shift to low zinc medium, Rpb3p occupancy increased for the wild-type strain in the ZRT1 ORF, but not in the upstream region, reflecting the induction of the ZRT1 gene. However this increase was not observed in the upf1Δ strain, corroborating the results observed by northern analysis. To show that the insertion of the terminator upstream from ZRT1 rescues the induction defect of the upf1Δ strain by allowing polymerase binding, we performed the same analysis by comparing Rpb3p occupancy in the upf1Δ and upf1Δ-ADH1t-B strains. Strikingly, Rpb3p levels were increased upon insertion of ADH1t at position B in the upf1Δ strain, both in normal zinc medium and after 3 hrs of induction, showing that the derepression of ZRT1 and the rescue of the induction defects are due to increased RNA Polymerase occupancy.
We also analyzed binding of the transcriptional activator Zap1p by ChIP in wild-type and upf1Δ strains using a myc-tagged version of Zap1p inserted at the chromosomal locus. We found background levels of Zap1p occupancy to its binding sites (ZREs) in conditions of repression in both strains (Figure 5D). However increased occupancy was observed in the wild-type strain upon a shift to low zinc (Figure 5D). Binding was reduced in the upf1Δ mutant, further showing that the accumulation of CD-CUTs perturbs Zap1p binding during ZRT1 induction. Zap1p enrichment was highly specific, as it was not observed in the ZRT1 coding region (Figure 5D). Overall the differences in RNA polymerase II and Zap1p occupancies in the ZRT1 gene are consistent with the results described above by Northern blot, showing that the effects observed in NMD mutants upon accumulation of CD-CUTs are indicative of transcriptional defects of the ZRT1 gene.
Overexpression or constitutive activation of transcriptional activators of subtelomeric genes suppresses their induction defect in the upf1Δ and xrn1Δ strains
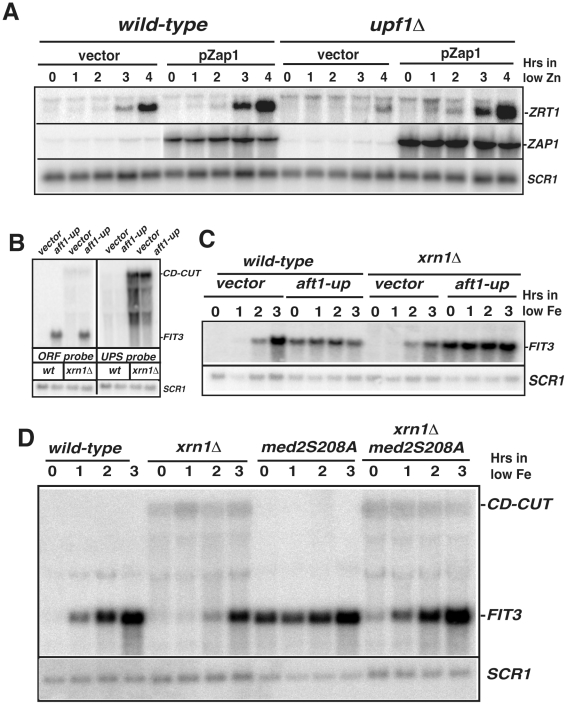
The previous result showed that binding of the Zap1p activator is deficient in the NMD mutant upf1Δ during the low zinc response. If so, we predicted that overexpressing Zap1p might suppress the induction delay in this strain. Indeed, overexpressing Zap1p in the upf1Δ mutant was sufficient to rescue ZRT1 induction to levels comparable to those observed in the wild-type strain (Figure 6A). This result shows that defective binding of Zap1p to the ZREs in the upf1Δ mutant can be overcome by overexpressing this activator. To extend these results to another gene induced in different conditions and controlled by a different activator, we monitored the expression of FIT3 in wild-type and xrn1Δ strains expressing aft1-up, a constitutively active version of Aft1p (kindly provided by J.Kaplan; [39], [40]. Aft1p is one of the two major transcriptional activators involved in the low iron response [39]. As expected, expression of the aft1-up allele resulted in derepression of the FIT3 mRNA in normal iron conditions in the wild-type strain (Figure 6B). However, similar levels of the mature FIT3 transcript were observed in an xrn1Δ background when the aft1-up allele was expressed, showing that the presence of a constitutively activated form of Aft1p can overcome CD-CUT-mediated repression. The accumulation of the FIT3 CD-CUT was not affected by expression of the aft1-up construct (Figure 6B), showing that this effect was not due to a decrease of expression of CD-CUT. We also monitored FIT3 induction in these strains upon a shift to low iron conditions (Figure 6C). Interestingly, FIT3 levels did not increase in the aft1-up strain upon a shift to low iron conditions (Figure 6C). However FIT3 accumulation was higher in the xrn1Δ aft1-up double mutant, possibly because of a reduced degradation of the FIT3 mRNA in the absence of Xrn1p.
Figure 6. CD-CUT mediated transcriptional repression can be rescued by overexpression or constitutive activation of transcriptional activators.
A. Overexpression of Zap1 rescues the ZRT1 induction defect observed in the upf1Δ mutant. Induction of ZRT1 in wild-type and upf1Δ mutant containing an empty vector or a vector overexpressing Zap1p (pZap1) under the control of a MET25 promoter. The WT or upf1Δ stains containing the vector pUG35 or pIT31 were grown in normal zinc conditions (time zero) or after overexpression of Zap1 and shift into low zinc medium. B and C. Expression of the aft1-up allele induces FIT3 expression even in the presence of the FIT3 CD-CUT. A plasmid expressing the aft1-up allele or a control vector were transformed into wild-type and xrn1Δ strain, and the levels of FIT3 mature mRNA or those of the CD-CUT were assessed by northern analysis using an ORF or an upstream (UP) probe, respectively. In panel C, the level of the FIT3 mRNA was assessed in these strains prior to and after a shift to low iron conditions. D. Analysis of FIT3 induction in wild-type, xrn1Δ, med2S208A and double mutant strains.
Accumulation of CD-CUTs abolishes the derepression of FIT3 induced by a Mediator component mutation
To investigate the specificity of the effects described above, we searched for conditions in which the induction of ZRT1 or FIT3 was uncoupled from activation by their transcriptional activators. Mutation of the Med2p tail component of the Mediator complex into a non-phosphorylated isoform (med2-S208A) was shown to result in a constitutive expression of FIT3 [41]. This observation led us to investigate the effect of the accumulation of the FIT3 CD-CUT on the derepression of FIT3 induced by this Mediator component mutation. We inactivated Xrn1p in a strain carrying the med2-S208A mutation (kind gift of F.Holstege) and analyzed the expression of FIT3. FIT3 derepression was observed in the med2-S208A mutant grown in normal medium (Figure 6D; time zero), in agreement with previous findings [41], but this mutant did not exhibit any further induction in low iron until 3 hours after the shift. Strikingly, inactivating Xrn1p in the med2-S208A strain abolished the derepression of FIT3 observed in the med2-S208A strain (Figure 6D). However the xrn1Δmed2-S208A mutant strain was not as defective for induction as the xrn1Δ strain, since the double mutant showed kinetics of FIT3 induction comparable to the wild-type strain. The result obtained in normal iron conditions (time zero, Figure 6D) shows that the accumulation of the FIT3 CD-CUT can inhibit the activation of FIT3 that results from a mediator component mutation. These results contrast with the result observed previously with the aft1-up mutation, which can activate expression of FIT3 even when CD-CUTs accumulate due to the inactivation of Xrn1p. Taken together, these results suggest that the function of the FIT3 and ZRT1 CD-CUTs is to prevent the premature binding of the RNA polymerase or of transcriptional activators such as Zap1 and Aft1p when these genes are transcriptionally repressed.
Discussion
Stable non-coding RNAs that can be detected without perturbations of the RNA degradation machinery have been shown to accumulate near many genes [42]–[44]. Here we show that strains defective for NMD accumulate 5′-extended forms (CD-CUTs) of many subtelomeric genes, some of which are involved in zinc and iron uptake and homeostasis. The accumulation of CD-CUTs is observed for a large number of genes located in subtelomeric regions in NMD mutants (Figure 1; Table S1). NMD also degrades the upstream sense ncRNA ICR1 that regulates the FLO11 gene ([8]; Figure S1B). Therefore CD-CUTs degraded by NMD are not exclusively involved in regulating metal homeostasis genes. Given the number of transcripts for which we detected a potential accumulation of CD-CUTs in NMD mutants by tiling arrays (Table S1), and the fact that extended forms of SRG1, which regulate SER3 transcription can be degraded by NMD [13], we speculate that cryptic upstream sense transcription might be used more widely than previously thought. However most of these transcripts are normally undetectable or present at very low levels because of active NMD.
CD-CUTs are clearly degraded by NMD, as shown by their extended half-life in the absence of Upf1p (Figure 2F, 2G) and because they are no longer stabilized by a upf1 deletion when a long ORF is inserted in their place (Figure S5B). CD-CUTs are likely recognized as NMD substrates because of the lack of extended ORFs in the 5′-regions upstream from the natural ORF. This might result in random translation initiation in the 5′-region, followed shortly by a stop codon, resulting in recognition of a faux/extended 3′-UTR [45], thus targeting them to NMD. In support of this model, terminating transcription of the main ZRT1 CD-CUT in a manner that results in a shorter transcript renders it insensitive to a upf1 deletion (Figure 4C, adh1t-A strain). CD-CUTs accumulating in NMD-deficient strains are different from the CUTs accumulating in nuclear exosome or TRAMP complex mutants [11], [46], [47]. CD-CUTs are much larger than CUTs, and most of them extend within the open-reading frames to terminate at or near the site of normal 3′ processing of the ORF mRNAs (Figure 1 and Figure 2, Figure S2, Figure 4D). Thus, both the nuclear exosome and cytoplasmic NMD degradation machineries are used to regulate gene expression but act on different sets of promoter-associated unstable RNAs. However, the two pathways can sometimes intersect, for example, in the case of the ADH4 CD-CUT, which is efficiently degraded only when both NMD and the nuclear exosome is inactivated (Figure 2; Figure 7).
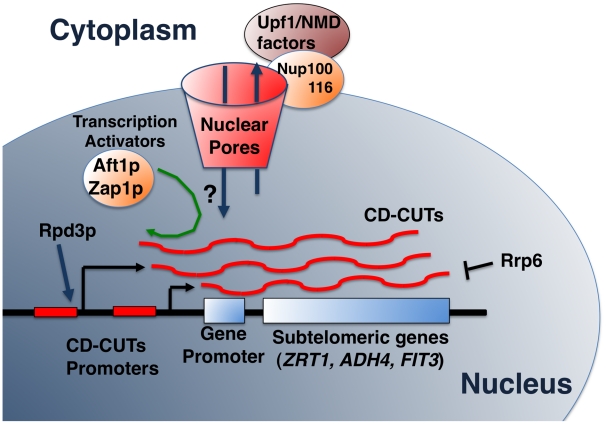
Figure 7. Model of biogenesis, action, and degradation of CD-CUTs.
The model shows the degradation of CD-CUTs by the NMD pathway, potentially localized at the vicinity of the nuclear pore due to the anchoring by the Nup100/116 nucleoporins [50]. Hypothetical retrograde transport of CD-CUTs is shown by a question mark but would explain the increased repression of subtelomeric genes in the absence of cytoplasmic degradation in NMD mutants.
How do CD-CUTs mediate transcriptional repression?
The precise mechanism by which CD-CUTs mediate transcriptional repression is not fully understood. Accumulation of CD-CUTs in NMD mutants negatively interferes with production of the normal transcripts and with RNA polymerase II and transcriptional activator binding (Figure 5, Figure 6, Figure 7). We do not know whether acting in cis is strictly required, which would be consistent with an SRG1-like transcriptional interference model [3]. We tried to express the ZRT1 CD-CUTs from a plasmid to test for the possibility of a trans effect, but could not detect any reproducible effect on ZRT1 induction (data not shown). A recent study showed that transcription of the sense upstream ncRNA ICR1 mediates transcriptional control of the subtelomeric FLO11 gene [8]. We found that the region upstream from the FLO11 gene encoding ICR1 shows elevated RNA levels in the upf1Δ strain (Figure S6). Like ICR1, expression of the ZRT1 CD-CUT is under the control of the histone deacetylase Rpd3p (Figure 4 and [8]). Based on these similarities, it is possible that the mechanisms of transcriptional control of the FLO11 gene mediated by ICR1 described in [8] may be applicable to the action of the CD-CUTs that control other subtelomeric regions.
The experiments in which we inserted transcription terminators upstream from ZRT1 do not allow us to differentiate between the cis and trans-acting models for CD-CUTs. Insertion of the terminator at position B relieves repression and allows the upf1Δ strain to induce ZRT1 in low zinc conditions (Figure 5). However in this strain, the CD-CUTs terminate before the Zap1 binding sites, so the results could be interpreted either way (transcriptional interference or trans-acting). Insertion of the terminator at position C relieves repression, but also inhibits ZRT1 induction even when NMD is active (Figure 5). Thus, we cannot conclude whether the CD-CUTs act in trans or are only the product of transcription that generates transcriptional interference. Because NMD mutants show higher CD-CUTs levels without a higher level of RNA Polymerase in the CD-CUT transcribed region (Figure 5C) and also result in stronger repression, we favor the hypothesis that these RNAs act in trans. However, further work is required to fully prove this point. Another unanswered question is to understand how the transcriptional machinery overcomes CD-CUT mediated repression in conditions of induction. It is possible that CD-CUT transcription is decreased in these conditions, but neither northern analysis nor the ChIP data seem to indicate that this is the case. Another alternative is that activation of the transcriptional activators is so potent during induction that it can overcome CD-CUTs mediated repression, even if the level of transcription of CD-CUTs does not change. The results obtained with the Zap1p overexpression or the constitutive Aft1p allele strains (Figure 6) are consistent with this model.
How does a cytoplasmic degradation pathway influence nuclear transcription?
One of the paradoxes raised by our observations is that 5′-extended species of subtelomeric genes are degraded by NMD, which is a cytoplasmic degradation pathway (Figure 7), yet, these CD-CUTs mediate transcriptional repression, and must therefore be localized to the nucleus if they mediate repression. Interestingly, many subtelomeric genes exhibit a perinuclear localization in S.cerevisiae [48]. In addition, connections have been made between nuclear activation of genes and localization at the periphery of the nuclear envelope near the nuclear pores (reviewed in [49]). Finally, Upf1p has been shown to interact with two nucleoporins localized on the outer side of the nuclear envelope, Nup100p and Nup116p ([50]; Figure 7), suggesting that at least part of the NMD process might occur at the vicinity of the nuclear envelope (Figure 7). Therefore if both subtelomeric genes and NMD components are localized close to the nuclear envelope but on opposite sides of the nuclear pores, the physical distance between the sites of transcription and action of CD-CUTs, and their site of degradation might be closer than thought from just considering the nuclear/cytoplasmic distribution (Figure 7). This would possibly allow retrograde transport of these CD-CUTs from their site of degradation to their site of action, and would allow a regulation of transcription by ncRNAs primarily degraded in the cytoplasm (Figure 7). It is also possible that CD-CUTs might be degraded when they emerge out of the nuclear pore complex, and that failure to degrade induces an increase of their nuclear localization, explaining a higher level of repression in NMD mutants. A better analysis of the mechanisms of nuclear/cytoplasmic trafficking of these CD-CUTs will ultimately allow a full understanding of their mechanisms of action.
Conserved functions for ncRNAs and NMD in the control of the expression of metal homeostasis genes
Despite these unanswered questions, our results have uncovered a novel function for NMD in controlling the accumulation of transcripts that negatively interfere with transcription of genes involved in zinc and iron homeostasis. Previous work has shown that upstream ncRNAs are involved in controlling gene expression related to metals homeostasis. It was shown that Zap1p activates the transcription of ncRNAs which mediate the repression of zinc-dependent alcohol dehydrogenases by transcriptional interference during zinc deficiency [51]. However it is unclear whether or not these ncRNAs are targeted by NMD, in a manner similar to CD-CUTs that regulate ZRT1, ADH4 and FIT3. A potential transcriptional interference mechanism involving long unstable upstream sense ncRNA has also been described in Chlamydomonas during copper deficiency [52], suggesting that this mechanism has been conserved during evolution to contribute generally to metal homeostasis genes regulation. Thus, there seems to be prevalent use of ncRNA transcription to control gene expression during metals homeostasis in different organisms. In addition to the potential crosstalk with transcription described here, NMD was also recently shown to regulate Mg++ cellular levels [23] by degrading the transcript encoding the main Mg++ transporter. The fact that this RNA surveillance system is so intimately implicated in the regulation of metals homeostasis in general might be linked to the prevalence of these metals in the ribosome and in their function of translation and in its fidelity [23], possibly revealing another layer of co-evolution between NMD and translation.
Materials and Methods
Yeast strains and media
Most strains were derived from BY4741 or 4742 (Open Biosystems). Strains in which the GFP-HIS cassette in the upstream region of ZRT1 or ADH4 gene were obtained by homologous recombination [36]. The strain carrying the deletion of the region upstream FIT3 and the strains containing the terminator insertions were obtained by delitto perfetto [53]. Double mutants in which the UPF1 or XRN1 genes were knocked out were obtained by direct disruption of these genes in other mutant strains, as described [21]. Insertion of the myc-tag for Zap1p was performed as described [36].
Strains were grown in conditions of non-induction in either YPD or Synthetic Complete medium (SC) supplemented with 2 mM ZnCl2. Growth in condition of low zinc gene induction was performed in either a Chelex-treated synthetic complete medium (CSC) or a SC medium containing 1 mM EDTA, pH adjusted to 4.4 with 20 mM citrate. CSC was prepared as described [28] except that all amino acid required were added and pH adjusted to 4.4. Growth in conditions of low iron gene induction was performed by adding BPS chelator as described [31]. Strains were grown in YPD (pre-low iron shifts) or SC+2 mM Zn (pre-low zinc shifts) until OD600 = 0.5, washed twice in sterile water, and shifted into YPD medium with BPS (low iron shift) or SC+EDTA medium (low zinc shift) for the indicated times. For the experiments including overexpression of Zap1p, WT or upf1Δ strains containing the vector pUG35 or pIT31 (see below) were grown in SC medium without Uracil (SC-URA) supplemented with 2 mM Zn. At OD600 = 0.45, cells were washed twice in sterile water and shifted in SC-URA without methionine (SC-URA-MET) supplemented with 2 mM Zn for 2 hours to overexpress Zap1p prior to zinc starvation. After 2 h, cells were washed twice in sterile water and maintained in log phase in SC-URA-MET medium containing 1 mM EDTA. Kinetics of induction were performed as described above.
Plasmids
A PCR product corresponding to the ZAP1 gene was generated from genomic DNA with primers containing the restriction sites ClaI and SalI and inserted in the vector pUG35 digested by the same restriction enzymes. After transformation and amplification in E. coli, the plasmid (pIT31) was confirmed by sequencing. The aft1-up expression plasmid was obtained from J.Kaplan (U.of Utah).
RNA analysis
Tiling Arrays used in this study were described previously [21], [31] and are accessible in the GEO database (accession number GSE11621). Northern blot hybridization analysis was performed as previously described [21], [31]. All riboprobes were synthesized with the T3 MAXIscript kit (Ambion). Riboprobes were hybridized at 67°C except for the ADH4 ORF probe (65°C).
Chromatin Immunoprecipitation (ChIP)
ChIPs using anti-Rpb3 RNA polymerase II subunit and a myc-tagged version of Zap1p inserted at the chromosomal locus were performed as described [54], [55].
Supporting Information
Tiling array profiles of ZPS1 and FLO11. A. Tiling array profile in the region of the ZPS1 gene. Shown is the log2 ratio of signal detected for the upf1Δ strain divided by the signal for the wild-type strain. B. Tiling Array profile of the upf1Δ mutant compared to the wild-type strain in the right subtelomeric region of Chr IX containing the FLO11 gene. The FLO11 gene is localized on the Crick strand (transcribed right to left) from positions 393,672 to 389,569. The increase of signal in the upf1Δ mutant upstream from the FLO11 gene is indicative of higher levels of the ICR1 ncRNA controlling FLO11. Note that the neighbor genes MRS1 and SEC11 located in region 397,000–399,000 are not affected by the upf1Δ deletion.
(TIF)
Mapping of the CD-CUTs of ZRT1 (A) and ADH4 (B) genes by northern blot analysis. The ZRT1 probes cover the following nucleotides: Probe 1: −2410 to −2089; Probe 2: −800 to −211; Probe 3: −257 to −84; Probe 4: +931 to +1131; Probe 5: +1141 to +1341 (downstream from the ORF).
(TIF)
Analysis of zinc regulon genes induction in NMD mutants. A. Kinetics of induction of zinc regulon genes in wild-type, upf2Δ and upf3Δ strains after a shift to a medium lacking zinc. Gene induction was monitored by northern blot using probes hybridizing to the corresponding genes. SCR1 was used as a loading control. B. Analysis of ZAP1 mRNAs levels in wild-type and upf1Δ strains. ZAP1 mRNA levels were analyzed by northern blot from cells grown in SC+2 mM Zn (time zero), or after a shift to SC+EDTA (low Zn). An RNA sample from a zap1Δ strain was included as a negative control for the detection of the ZAP1 mRNA.
(TIF)
Kinetics of FIT3 induction in strains lacking Xrn1p, Upf1p or in the double mutant xrn1Δrat1-1. Shown is a northern blot analysis of FIT3 expression in the indicated strains using a probe complementary to FIT3.
(TIF)
Insertion of GFP cassettes upstream from ZRT1 and ADH4. A. Schematic representation of the ZRT1 and ADH4 genes and location of the GFP-HIS3 cassettes deletions/insertions (UP1 and UP2, UP). The dashed lines represent the regions replaced by the cassettes. B. GFP mRNA levels analyzed by northern blot in the different insertion strains. Levels were normalized to SCR1. A strain without GFP insertion was included as negative control. C. Effects of the deletion of the upstream region and of the insertion of a GFP-HIS3 cassette on the kinetics of induction and shutoff of ZRT1. Insertion of the GFP-HIS3 cassette upstream from ZRT1 (zrt1-up1) results in a three-fold increase in the ZRT1 mRNA peak in the upf1Δ strain during zinc deficiency. ZRT1 induction was faster in the strain carrying this insertion compared to the wild-type, as shown by the amount of the ZRT1 mRNA expressed after 30 to 120 minutes. After 4 hours of induction, the kinetics of disappearance of ZRT1 upon shifting back to zinc-containing medium was also monitored in these strains (+zinc), and samples were harvested at the indicated times after addition of zinc to the medium. RNAs extracted from all four strains were loaded on the same gel and analyzed on the same membranes exposed to the same times, but each strain is shown as a separate panel for clarity and space purposes.
(TIF)
Kinetics of ZRT1 induction in wild-type, hda2Δ hda3Δ, upf1Δ, hda2Δupf1Δ and hda3Δupf1Δ strains.
(TIF)
List of S.cerevisiae ORFs for which 5′ extended species were observed by tiling microarrays in the upf1Δ and/or xrn1Δstrains.
(XLS)
Acknowledgments
We thank Cynthia Lee, Sabeeha Merchant, Michael Grunstein, and Siavash Kurdistani for helpful discussions; F. Holstege and S. Kurdistani for yeast strains; J.Kaplan for the aft1-up plasmid; Kevin Roy for critical reading of the manuscript; and one anonymous reviewer of a grant proposal for helpful suggestions.
Footnotes
The authors have declared that no competing interests exist.
This work is supported by grants GM61518 from NIH and RSG 06-040 from the American Cancer Society to GFC and by NRSA F31 GM084678 from the National Institute from General Medical Sciences to CRN. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 2.Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, et al. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 3.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 4.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 5.Berretta J, Pinskaya M, Morillon A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 2008;22:615–626. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camblong J, Beyrouthy N, Guffanti E, Schlaepfer G, Steinmetz LM, et al. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev. 2009;23:1534–1545. doi: 10.1101/gad.522509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bumgarner SL, Dowell RD, Grisafi P, Gifford DK, Fink GR. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc Natl Acad Sci U S A. 2009;106:18321–18326. doi: 10.1073/pnas.0909641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 12.Lee A, Hansen KD, Bullard J, Dudoit S, Sherlock G. Novel low abundance and transient RNAs in yeast revealed by tiling microarrays and ultra high-throughput sequencing are not conserved across closely related yeast species. PLoS Genet. 2008;4:e1000299. doi: 10.1371/journal.pgen.1000299. doi: 10.1371/journal.pgen.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson DM, Parker R. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:92–101. doi: 10.1128/MCB.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang YF, Imam JS, Wilkinson MF. The Nonsense-Mediated Decay RNA Surveillance Pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 15.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 16.Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 17.Kuzmiak HA, Maquat LE. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol Med. 2006;12:306–316. doi: 10.1016/j.molmed.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Mitrovich QM, Anderson P. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 2000;14:2173–2184. doi: 10.1101/gad.819900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 20.Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sayani S, Janis M, Lee CY, Toesca I, Chanfreau GF. Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol Cell. 2008;31:360–370. doi: 10.1016/j.molcel.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kebaara BW, Atkin AL. Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:2771–2778. doi: 10.1093/nar/gkp146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansson MJ, Jacobson A. Nonsense-mediated mRNA decay maintains translational fidelity by limiting magnesium uptake. Genes Dev. 2010;24:1491–1495. doi: 10.1101/gad.1930710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lelivelt MJ, Culbertson MR. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol. 1999;19:6710–6719. doi: 10.1128/mcb.19.10.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He F, Li X, Spatrick P, Casillo R, Dong S, et al. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell. 2003;12:1439–1452. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- 26.Guan Q, Zheng W, Tang S, Liu X, Zinkel RA, et al. Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast. PLoS Genet. 2006;2:e203. doi: 10.1371/journal.pgen.0020203. doi: 10.1371/journal.pgen.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansson MJ, He F, Spatrick P, Li C, Jacobson A. Association of yeast Upf1p with direct substrates of the NMD pathway. Proc Natl Acad Sci U S A. 2007;104:20872–20877. doi: 10.1073/pnas.0709257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyons TJ, Gasch AP, Gaither LA, Botstein D, Brown PO, et al. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc Natl Acad Sci U S A. 2000;97:7957–7962. doi: 10.1073/pnas.97.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Protchenko O, Ferea T, Rashford J, Tiedeman J, Brown PO, et al. Three cell wall mannoproteins facilitate the uptake of iron in Saccharomyces cerevisiae. J Biol Chem. 2001;276:49244–49250. doi: 10.1074/jbc.M109220200. [DOI] [PubMed] [Google Scholar]
- 30.Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, et al. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell. 2002;109:437–446. doi: 10.1016/s0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- 31.Lee A, Henras AK, Chanfreau G. Multiple RNA surveillance pathways limit aberrant expression of iron uptake mRNAs and prevent iron toxicity in S. cerevisiae. Mol Cell. 2005;19:39–51. doi: 10.1016/j.molcel.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 32.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Arai S, Song X, Reichart D, Du K, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008 doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao H, Eide D. The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J Biol Chem. 1996;271:23203–23210. doi: 10.1074/jbc.271.38.23203. [DOI] [PubMed] [Google Scholar]
- 35.Taylor R, Kebaara BW, Nazarenus T, Jones A, Yamanaka R, et al. Gene set coregulated by the Saccharomyces cerevisiae nonsense-mediated mRNA decay pathway. Eukaryot Cell. 2005;4:2066–2077. doi: 10.1128/EC.4.12.2066-2077.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 37.Bernstein BE, Tong JK, Schreiber SL. Genomewide studies of histone deacetylase function in yeast. Proc Natl Acad Sci U S A. 2000;97:13708–13713. doi: 10.1073/pnas.250477697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao H, Butler E, Rodgers J, Spizzo T, Duesterhoeft S, et al. Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J Biol Chem. 1998;273:28713–28720. doi: 10.1074/jbc.273.44.28713. [DOI] [PubMed] [Google Scholar]
- 39.Yamaguchi-Iwai Y, Dancis A, Klausner RD. AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 1995;14:1231–1239. doi: 10.1002/j.1460-2075.1995.tb07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Philpott CC, Rashford J, Yamaguchi-Iwai Y, Rouault TA, Dancis A, et al. Cell-cycle arrest and inhibition of G1 cyclin translation by iron in AFT1-1(up) yeast. EMBO J. 1998;17:5026–5036. doi: 10.1093/emboj/17.17.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van de Peppel J, Kettelarij N, van Bakel H, Kockelkorn TT, van Leenen D, et al. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol Cell. 2005;19:511–522. doi: 10.1016/j.molcel.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 42.Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 43.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 44.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amrani N, Dong S, He F, Ganesan R, Ghosh S, et al. Aberrant termination triggers nonsense-mediated mRNA decay. Biochem Soc Trans. 2006;34:39–42. doi: 10.1042/BST20060039. [DOI] [PubMed] [Google Scholar]
- 46.Davis CA, Ares M., Jr Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2006;103:3262–3267. doi: 10.1073/pnas.0507783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- 48.Grund SE, Fischer T, Cabal GG, Antunez O, Perez-Ortin JE, et al. The inner nuclear membrane protein Src1 associates with subtelomeric genes and alters their regulated gene expression. J Cell Biol. 2008;182:897–910. doi: 10.1083/jcb.200803098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dieppois G, Stutz F. Connecting the transcription site to the nuclear pore: a multi-tether process that regulates gene expression. J Cell Sci. 2010;123:1989–1999. doi: 10.1242/jcs.053694. [DOI] [PubMed] [Google Scholar]
- 50.Nazarenus T, Cedarberg R, Bell R, Cheatle J, Forch A, et al. Upf1p, a highly conserved protein required for nonsense-mediated mRNA decay, interacts with the nuclear pore proteins Nup100p and Nup116p. Gene. 2005;345:199–212. doi: 10.1016/j.gene.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 51.Bird AJ, Gordon M, Eide DJ, Winge DR. Repression of ADH1 and ADH3 during zinc deficiency by Zap1-induced intergenic RNA transcripts. EMBO J. 2006;25:5726–5734. doi: 10.1038/sj.emboj.7601453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moseley JL, Page MD, Alder NP, Eriksson M, Quinn J, et al. Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell. 2002;14:673–688. doi: 10.1105/tpc.010420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Storici F, Lewis LK, Resnick MA. In vivo site-directed mutagenesis using oligonucleotides. Nat Biotechnol. 2001;19:773–776. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- 54.Yang PK, Hoareau C, Froment C, Monsarrat B, Henry Y, et al. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol Cell Biol. 2005;25:3295–3304. doi: 10.1128/MCB.25.8.3295-3304.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, et al. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tiling array profiles of ZPS1 and FLO11. A. Tiling array profile in the region of the ZPS1 gene. Shown is the log2 ratio of signal detected for the upf1Δ strain divided by the signal for the wild-type strain. B. Tiling Array profile of the upf1Δ mutant compared to the wild-type strain in the right subtelomeric region of Chr IX containing the FLO11 gene. The FLO11 gene is localized on the Crick strand (transcribed right to left) from positions 393,672 to 389,569. The increase of signal in the upf1Δ mutant upstream from the FLO11 gene is indicative of higher levels of the ICR1 ncRNA controlling FLO11. Note that the neighbor genes MRS1 and SEC11 located in region 397,000–399,000 are not affected by the upf1Δ deletion.
(TIF)
Mapping of the CD-CUTs of ZRT1 (A) and ADH4 (B) genes by northern blot analysis. The ZRT1 probes cover the following nucleotides: Probe 1: −2410 to −2089; Probe 2: −800 to −211; Probe 3: −257 to −84; Probe 4: +931 to +1131; Probe 5: +1141 to +1341 (downstream from the ORF).
(TIF)
Analysis of zinc regulon genes induction in NMD mutants. A. Kinetics of induction of zinc regulon genes in wild-type, upf2Δ and upf3Δ strains after a shift to a medium lacking zinc. Gene induction was monitored by northern blot using probes hybridizing to the corresponding genes. SCR1 was used as a loading control. B. Analysis of ZAP1 mRNAs levels in wild-type and upf1Δ strains. ZAP1 mRNA levels were analyzed by northern blot from cells grown in SC+2 mM Zn (time zero), or after a shift to SC+EDTA (low Zn). An RNA sample from a zap1Δ strain was included as a negative control for the detection of the ZAP1 mRNA.
(TIF)
Kinetics of FIT3 induction in strains lacking Xrn1p, Upf1p or in the double mutant xrn1Δrat1-1. Shown is a northern blot analysis of FIT3 expression in the indicated strains using a probe complementary to FIT3.
(TIF)
Insertion of GFP cassettes upstream from ZRT1 and ADH4. A. Schematic representation of the ZRT1 and ADH4 genes and location of the GFP-HIS3 cassettes deletions/insertions (UP1 and UP2, UP). The dashed lines represent the regions replaced by the cassettes. B. GFP mRNA levels analyzed by northern blot in the different insertion strains. Levels were normalized to SCR1. A strain without GFP insertion was included as negative control. C. Effects of the deletion of the upstream region and of the insertion of a GFP-HIS3 cassette on the kinetics of induction and shutoff of ZRT1. Insertion of the GFP-HIS3 cassette upstream from ZRT1 (zrt1-up1) results in a three-fold increase in the ZRT1 mRNA peak in the upf1Δ strain during zinc deficiency. ZRT1 induction was faster in the strain carrying this insertion compared to the wild-type, as shown by the amount of the ZRT1 mRNA expressed after 30 to 120 minutes. After 4 hours of induction, the kinetics of disappearance of ZRT1 upon shifting back to zinc-containing medium was also monitored in these strains (+zinc), and samples were harvested at the indicated times after addition of zinc to the medium. RNAs extracted from all four strains were loaded on the same gel and analyzed on the same membranes exposed to the same times, but each strain is shown as a separate panel for clarity and space purposes.
(TIF)
Kinetics of ZRT1 induction in wild-type, hda2Δ hda3Δ, upf1Δ, hda2Δupf1Δ and hda3Δupf1Δ strains.
(TIF)
List of S.cerevisiae ORFs for which 5′ extended species were observed by tiling microarrays in the upf1Δ and/or xrn1Δstrains.
(XLS)