Abstract
In most instances, translation is regulated at the initiation phase, when a ribosome is recruited to the 5′ end of an mRNA. The eIF4E-binding proteins (4E-BPs) interdict translation initiation by binding to the translation factor eIF4E, and preventing recruitment of the translation machinery to mRNA. The 4E-BPs inhibit translation in a reversible manner. Hypophosphorylated 4E-BPs interact avidly with eIF4E, whereas 4E-BP hyperphosphorylation, elicited by stimulation of cells with hormones, cytokines, or growth factors, results in an abrogation of eIF4E-binding activity. We reported previously that phosphorylation of 4E-BP1 on Thr 37 and Thr 46 is relatively insensitive to serum deprivation and rapamycin treatment, and that phosphorylation of these residues is required for the subsequent phosphorylation of a set of unidentified serum-responsive sites. Here, using mass spectrometry, we identify the serum-responsive, rapamycin-sensitive sites as Ser 65 and Thr 70. Utilizing a novel combination of two-dimensional isoelectric focusing/SDS-PAGE and Western blotting with phosphospecific antibodies, we also establish the order of 4E-BP1 phosphorylation in vivo; phosphorylation of Thr 37/Thr 46 is followed by Thr 70 phosphorylation, and Ser 65 is phosphorylated last. Finally, we show that phosphorylation of Ser 65 and Thr 70 alone is insufficient to block binding to eIF4E, indicating that a combination of phosphorylation events is necessary to dissociate 4E-BP1 from eIF4E.
Keywords: 4E-BP1, translation initiation, eIF4E, phosphorylation, phosphospecific antibody, phosphopeptide mapping
The eukaryotic translation initiation factor 4E (eIF4E) recognizes the cap structure, m7GpppX (in which m is a methyl group and X is any nucleotide), present at the 5′ end of nuclear transcribed mRNAs (for reviews, see Hershey and Merrick 2000; Raught et al. 2000b). eIF4E directs the translation machinery to the 5′ end of mRNA via an interaction with the scaffolding protein eIF4G (for review, see Hershey and Merrick 2000). The eIF4E-binding proteins (4E-BPs) are a family of three small peptides that inhibit cap-dependent translation by binding to eIF4E and obstructing its interaction with eIF4G (Lin et al. 1994; Pause et al. 1994; Gingras et al. 1999b; Raught et al. 2000b). The 4E-BPs act as molecular mimics of the eIF4E-binding site in eIF4G, and thus effectively compete with eIF4G for eIF4E binding (Haghighat et al. 1995; Mader et al. 1995; Marcotrigiano et al. 1999).
Binding of the 4E-BPs to eIF4E is reversible. Whereas hypophosphorylated 4E-BPs bind avidly to eIF4E, 4E-BP hyperphosphorylation abrogates this interaction. Numerous types of extracellular stimuli, including serum, hormones, growth factors, mitogens, cytokines, and G-protein-coupled receptor agonists elicit 4E-BP1 hyperphosphorylation, and a concomitant loss of eIF4E-binding activity. Conversely, nutrient deprivation, environmental stress, or infection with certain picornaviruses induce 4E-BP1 dephosphorylation, accompanied by a dramatic increase in eIF4E-binding activity and an inhibition of translation (for reviews, see Gingras et al. 1999b; Raught et al. 2000b). Activation of phosphoinositide 3′-kinase (PI3K) or a downstream PI3K-effector kinase, Akt/PKB (protein kinase B), leads to 4E-BP1 hyperphosphorylation (e.g., von Manteuffel et al. 1996; Gingras et al. 1998; Kohn et al. 1998; Wu et al. 1998; Takata et al. 1999), and expression of an activated PI3K catalytic subunit or an activated Akt/PKB protein elicits 4E-BP1 phosphorylation on the same residues that are phosphorylated in response to serum stimulation (Gingras et al. 1998; Kohn et al. 1998; Dufner et al. 1999; Takata et al. 1999). Hyperphosphorylation of 4E-BP1 induced by extracellular stimuli is inhibited by the PI3K inhibitors wortmannin and LY294002. Hyperphosphorylation is also abrogated by rapamycin treatment (Lin et al. 1995; Beretta et al. 1996; von Manteuffel et al. 1996). Rapamycin inhibits downstream signaling from the kinase FRAP/mTOR (FKBP and rapamycin associated protein/mammalian target of rapamycin), and a rapamycin-resistant FRAP/mTOR protein confers rapamycin resistance to 4E-BP1 phosphorylation (e.g., Brunn et al. 1997a,b; Burnett et al. 1998; Gingras et al. 1998). Thus, a wealth of evidence indicates that PI3K and FRAP/mTOR signaling are required to effect the release of 4E-BP1 from eIF4E (for a recent review, see Gingras et al. 2001a).
Six phosphorylation sites have been identified in 4E-BP1: Thr 37, Thr 46, Ser 65, Thr 70, Ser 83, and Ser 112 (numbering according to human 4E-BP1; Fadden et al. 1997; Heesom et al. 1998). We showed previously that in HEK 293 cells, Thr 37 and Thr 46 are phosphorylated to a significant extent even in the absence of serum, and that serum stimulation only moderately increases the phosphorylation state of these residues (Gingras et al. 1999a). We also reported that phosphorylation of Thr 37/Thr 46 is required for the phosphorylation of additional, unknown serum-sensitive sites. Mutation of Thr 37 and/or Thr 46 to alanine residues prevents phosphorylation of the serum-sensitive sites, but substitution of Thr 37 and Thr 46 by glutamic acid residues partially restores the phosphorylation of the serum-sensitive sites (Gingras et al. 1999a). These data suggest that phosphorylation of Thr 37 and Thr 46 serves as a priming event, allowing for the succeeding serum-induced phosphorylation of the remaining 4E-BP1 sites. Phosphorylation of Ser 65 was also subsequently reported by other investigators to be dependent on prior phosphorylation of Thr 37, Thr 46, and Thr 70 (Mothe-Satney et al. 2000a,b). However, at variance with our data, these studies suggested that phosphorylation of Thr 70 does not depend on prior phosphorylation of Thr 37 and Thr 46 (Mothe-Satney et al. 2000a,b).
The number and identity of the phosphorylation sites required to effect the release of 4E-BP1 from eIF4E also remains in question. Thr 37 and Thr 46 phosphorylation was reported both to have little or no effect on eIF4E binding (Gingras et al. 1999a; Heesom and Denton 1999; Karim et al. 2001), or (at least for Thr 46) to effect a significant reduction in eIF4E-binding affinity (Burnett et al. 1998; Yang et al. 1999; Mothe-Satney et al. 2000b). Significant discrepancies also exist regarding the effect of Ser 65 phosphorylation on eIF4E binding (Lin et al. 1994; Fadden et al. 1997; Yang et al. 1999; Mothe-Satney et al. 2000b; Karim et al. 2001).
Whereas previous studies have relied on the use of transfected, overexpressed mutant proteins (which may not faithfully mimic the behavior of the endogenous protein), we have directly analyzed the order of phosphate addition on the endogenous 4E-BP1 protein. Here, using phosphopeptide mapping and mass spectrometry, we identify Ser 65 and Thr 70 as the principle serum-responsive, rapamycin-sensitive 4E-BP1 phosphorylation sites in HEK 293 cells. Further, using a novel combination of two-dimensional isoelectric-focusing/SDS-PAGE, followed by Western blotting with phosphospecific antibodies, we unambiguously delimit the order of phosphate addition to endogenous 4E-BP1. Phosphate groups are first added to Thr 37 and Thr 46. This priming step is followed by phosphorylation of Thr 70 and, finally, of Ser 65. We also show that phosphorylation of Ser 65 alone, or phosphorylation of both Ser 65 and Thr 70, does not disrupt 4E-BP1 binding to eIF4E. Thus, multiple phosphorylation events (most likely via different kinases) are required to release 4E-BP1 from eIF4E.
Results
Two sets of 4E-BP1 phosphopeptides with different sensitivities to kinase inhibitors
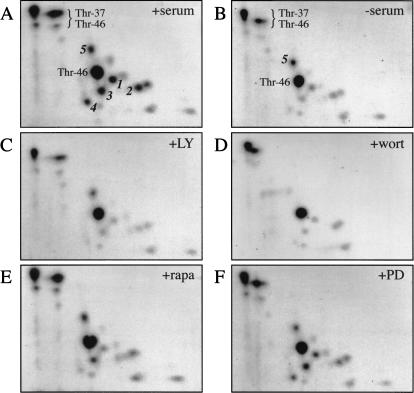
Using two-dimensional phosphopeptide mapping combined with mass spectrometry, we previously identified two 4E-BP1 phosphorylation sites, Thr 37 and Thr 46 (Gingras et al. 1999a). These residues are phosphorylated to a high stoichiometry in the absence of serum, and the phosphorylation state of Thr 37 and Thr 46 is only moderately increased following serum stimulation of HEK 293 cells (Fig. 1; Gingras et al. 1999a). A second group of phosphopeptides (designated 1–4 in Fig. 1A) are detected only at very low levels in the absence of serum (Fig. 1B), but are present at high levels after serum addition (Fig. 1A). To study the signaling pathway(s) mediating these phosphorylation events, the sensitivity of the two groups of phosphopeptides to treatment with various kinase inhibitors (the PI3K inhibitors LY294002 and wortmannin, the FRAP/mTOR inhibitor rapamycin, and the MEK inhibitor PD098059) was investigated. The response of phosphopeptides 1–4 to serum addition is drastically blunted when PI3K or FRAP/mTOR signaling are inhibited by LY294002, wortmannin, or rapamycin (Fig. 1 C–E). [In the experiment presented here, rapamycin did not completely prevent phosphorylation of phosphopeptide 3. There is some variability in the extent of inhibition elicited by the kinase inhibitors from experiment to experiment. Presumably, this is due to instability of the compounds, differences in activity from batch to batch, or to slight variations in cell culture conditions (e.g., cell passage number, level of confluence, batch of serum, etc.). Although we have made every attempt to reduce this variability, minor differences are still observed between different experiments. We have performed phosphopeptide mapping of 4E-BP1 from cells treated with rapamycin many (>10) times, and the conclusion that rapamycin strongly affects phosphorylation of Ser 65 and Thr 70 stems from the sum of these experiments (the extent of inhibition on these phosphopeptides by rapamycin ranges from 4- to 10-fold, whereas the inhibition of phosphorylation at Thr 37 and Thr 46 is ∼1.5-fold; see also von Manteuffel et al. 1997; Gingras et al. 1998, 1999a)]. The ERK inhibitor PD098059 did not significantly affect the phosphorylation of any of the peptides (Fig. 1F). Thus, PI3K and FRAP/mTOR signaling are required for the phosphorylation of peptides 1–4 in response to serum stimulation. The phosphorylation state of phosphopeptide 5 is not consistent from experiment to experiment. In some cases, this phosphopeptide cannot be detected even in the presence of serum (e.g., see Fig. 2D), and it is not reproducibly responsive to serum treatment or sensitive to kinase inhibitor treatment (Gingras et al. 1998, 1999a; see below).
Figure 1.
Sensitivity of 4E-BP1 phosphopeptides to serum deprivation and kinase inhibitor treatment. HEK 293 cells were starved of serum for 30 h. 32P-labeling was performed for 3.5 h. Kinase inhibitors were then added (for 30 min), followed by stimulation for 30 min with 10% dialyzed FBS. Phosphopeptide maps were prepared (see Materials and Methods). Previously identified phosphopeptides containing Thr 37 and/or Thr 46 are indicated. Phosphopeptides exhibiting sensitivity to serum-deprivation or kinase inhibitors are labeled 1–5. The various treatments are indicated in the top, right corner as follows: serum-stimulation (A, +serum), no stimulation (B, −serum), pre-treatment with LY294002 (C, +LY), wortmannin (D, +wort), rapamycin (E, +rapa), or PD098059 (F, +PD). A and B, which served here as controls, were published previously (Gingras et al. 1999a).
Figure 2.
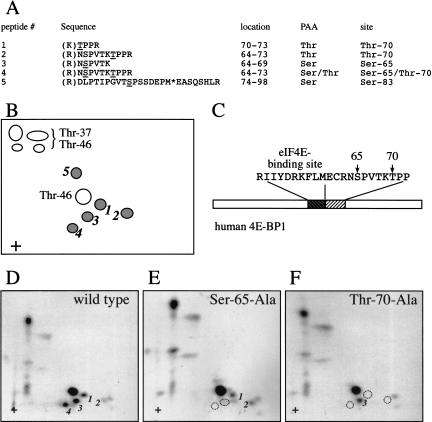
Identification of Ser 65 and Thr 70 as the serum-responsive, kinase inhibitor-sensitive phosphorylation sites. 4E-BP1 was immunoprecipitated from 2 × 108 serum-stimulated cells (1/20 labeled in vivo with [32P]orthophosphate), and subjected to phosphopeptide mapping. Peptides 1–5 (see Fig. 1A) were isolated from the plates and identified by LC-MS/MS. (A) Determination of the sequence of phosphopeptides 1–5, and of the identity of the phosphorylated residues by mass spectrometry. The phosphorylated residue is underlined; (*) oxidized methionine. Trypsin cleavage sites are indicated in parentheses. Phosphoaminoacid analysis (PAA) was performed in parallel. (Identification of phosphopeptides 2 and 4 was reported previously in Gygi et al. 1999a). (B) Schema of the phosphopeptides identified by two-dimensional phosphopeptide mapping. (C) Positions of Ser 65 and Thr 70 in 4E-BP1, relative to the eIF4E-binding site. (D–F) Confirmation of the identity of the phosphopeptides containing Ser 65 and Thr 70. HEK 293T cells expressing HA-tagged 4E-BP1 proteins were subjected to metabolic 32P labeling, and two-dimensional phosphopeptide mapping of the HA–4E-BP1 proteins was performed. Dotted circles indicate the absence of phosphopeptides, as compared with the wild-type protein
Identification of the PI3K- and FRAP/mTORdependent phosphorylation sites
We next used a combination of phosphopeptide mapping and mass spectrometry to identify phosphopeptides 1–5. Several hundred 150-mm plates of HEK 293 cells (at 90% confluence) were deprived of serum for 30 h. Ten dishes of serum-starved cells were metabolically labeled with [32P]orthophosphate. All of the cells were then stimulated with 10% serum for 30 min, and lysed. 4E-BP1 was immunoprecipitated from both the unlabeled and radiolabeled lysates. The radiolabeled and unlabeled immunoprecipitated materials were mixed and subjected to SDS-15% PAGE. After transfer to a nitrocellulose membrane and autoradiography, the membrane fragment harboring 4E-BP1 was excised and incubated with a mixture of trypsin:chymotrypsin (200:1). Digested peptides were washed extensively and subjected to two-dimensional phosphopeptide mapping. Phosphopeptides 1–5 (Figs. 1A, 2B) were isolated from the maps and identified by capillary liquid chromatography-electrospray ionization tandem mass spectrometry (LC-MS/MS). The sequence of the phosphopeptides and identities of the phosphorylated residues are indicated in Figure 2A. Phosphopeptides 1–4 contain phosphorylated Ser 65 and/or Thr 70. These two residues are located immediately downstream of the eIF4E-binding site (Fig. 2C). Phosphopeptide 5 contains phosphorylated Ser 83. Phosphoaminoacid analysis (PAA) of each of the phosphopeptides was conducted in parallel. The phosphoserine (S) and/or phosphothreonine (T) content of each peptide is indicated (Fig. 2A). The identity of the phosphopeptides containing phosphorylated Ser 65 and Thr 70 was further confirmed by two-dimensional phosphopeptide mapping of epitope-tagged wild-type 4E-BP1 (Fig. 2D), and Ser 65-Ala (Fig. 2E) or Thr 70-Ala (Fig. 2F) mutant proteins expressed in HEK 293T cells. Substitution of Ser 65 with an alanine residue resulted in a loss of phosphopeptides 3 and 4, whereas the Thr 70 to alanine mutation resulted in a loss of phosphopeptides 1, 2, and 4.
Phosphospecific antibodies directed against Ser 65 and Thr 70
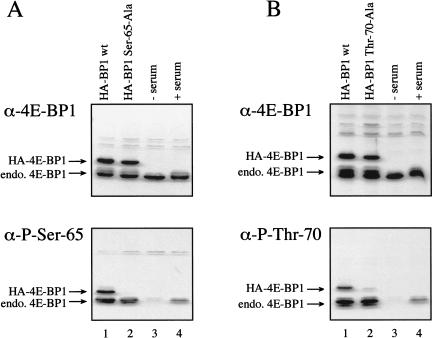
To further investigate the regulation of Ser 65 and Thr 70 phosphorylation, phosphospecific antibodies directed against these sites were produced. To confirm the specificity of these reagents, HA-tagged wild-type 4E-BP1 protein, or 4E-BP1 proteins harboring alanine substitutions at Ser 65 or Thr 70 were expressed in HEK 293T cells, and immunoblotting was performed using an antiserum directed against the entire 4E-BP1 protein (Fig. 3A,B, top), an antibody directed against phospho-Ser 65 (Fig. 3A, bottom), or an antibody directed against phospho-Thr 70 (Fig. 3B, bottom). The HA–4E-BP1 wild-type and mutant proteins were expressed to similar levels (Fig. 3A,B, lanes 1,2). However, the antibody directed against phospho-Ser 65 did not detect the HA–4E-BP1 Ser 65-Ala protein (Fig. 3A, lane 2, bottom). Similarly, the antibody directed against phospho-Thr 70 did not react with the HA–4E-BP1 Thr 70-Ala mutant protein (Fig. 3B, lane 2). Extracts from 293 cells deprived of serum for 30 h (Fig. 3A,B, lanes 3), or deprived of serum for 30 h, then replenished with serum for 30 min (Fig. 3A,B, lanes 4), were also subjected to immunoblotting with these antisera. In both cases, only a very weak signal was detected in the serum-starved cell extracts (lanes 3), whereas strong signals were observed after serum-stimulation (lanes 4). Thus, our phosphospecific antibodies were shown to be specific for the appropriate phosphorylated residues, and confirm that phosphorylation of Ser 65 and Thr 70 is sensitive to serum stimulation. These antibodies were also utilized to confirm the heightened sensitivity of phosphorylation at Ser 65 and Thr 70 to kinase inhibitors (data not shown).
Figure 3.
Phosphospecific antibodies directed against Ser 65 and Thr 70 confirm the serum sensitivity of the phosphorylation at these sites. (A) Western blotting using an antibody to total 4E-BP1 (top), or a phosphospecific antibody directed against Ser 65 (bottom) was performed on extracts from HEK 293T cells transfected with HA–4E-BP1 wt (lane 1) or HA–4E-BP1 Ser 65-Ala (lane 2), or extracts from HEK 293 cells that were serum starved (lane 3), or serum starved, and then serum stimulated for 30 min (lane 4). (B) Western blotting was performed as in A, but using an HA–4E-BP1 Thr 70-Ala protein in lane 2, and a phosphospecific antibody directed against Thr 70 (bottom). The positions of the endogenous 4E-BP1, and the transfected proteins are indicated.
Order of phosphate addition on the endogenous 4E-BP1 protein
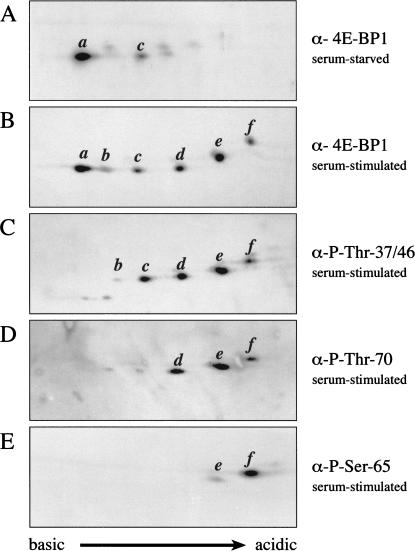
The endogenous 4E-BP1 protein was next analyzed by use of a combination of two-dimensional isoelectric focusing (IEF)/SDS-PAGE and Western blotting. 4E-BP1 from serum-stimulated HEK 293 cells migrates as six isoforms in this system (Fig. 4B). (In some experiments, a seventh, more acidic, isoform is also detected). In cells deprived of serum, only two predominant isoforms are detected (a and c), which migrate toward the basic region of the gel (Fig. 4A). Similarly, only isoforms a and c are detected in cells pretreated with rapamycin or LY294002 prior to serum stimulation (data not shown). Isoform a is unphosphorylated, because it does not incorporate 32P (data not shown). Isoforms b–f incorporate 32P (and are thus predicted to be phosphorylated on one to five residues, respectively).
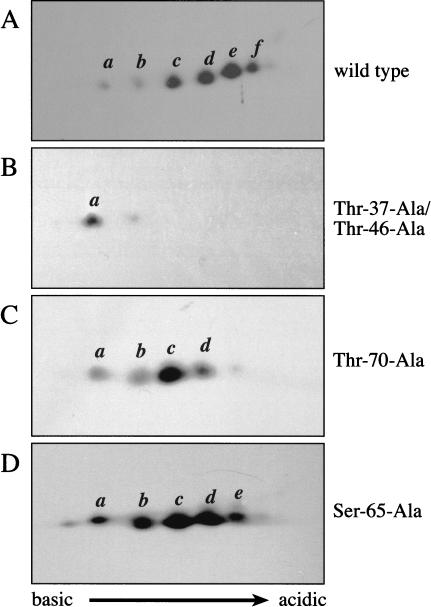
Figure 4.
Phosphorylation of endogenous 4E-BP1 in vivo is an ordered process, culminating in the phosphorylation of Ser 65. Two-dimensional IEF/SDS-PAGE (see Materials and Methods) was performed on lysates from serum-starved (A), or serum-stimulated (B–E) HEK 293 cells. Western blotting was then performed using antisera directed against the entire 4E-BP1 protein (A,B), or a series of phosphospecific antibodies raised against 4E-BP1 Thr 37/Thr 46 (C), Thr 70 (D), or Ser 65 (E). Western blots C–E were subsequently reprobed with the antisera raised against the entire 4E-BP1 protein to confirm the identity of the isoforms.
Our previous phosphopeptide mapping experiments (Gingras et al. 1998, 1999a; Fig. 1) suggested that isoform c (predicted to be the diphosphorylated species), which is present in serum-starved cells, contains both phosphorylated Thr 37 and Thr 46. Interestingly, isoform b (presumably the monophosphorylated protein) is a minor species, both in serum-stimulated cells and in serum-starved or rapamycin-treated cells. This is likely because Thr 37 and Thr 46 are regulated coordinately by FRAP/mTOR, and is consistent with our phosphopeptide mapping data (Gingras et al. 1998, 1999a; Fig. 1), in which equimolar amounts of phospho-Thr 37 and phospho-Thr 46 are detected. Also, when Thr 46 is mutated to alanine, phosphorylation of Thr 37 is dramatically decreased (Gingras et al. 1999a), suggesting that the phosphorylation state of these two sites is intimately linked.
To identify the phosphorylated residues in each of the isoelectric variants, the IEF/SDS-PAGE technique was combined with the use of phosphospecific antibodies. To ensure proper identification of the isoforms, each blot incubated with phosphospecific antibodies was subsequently reprobed with antisera raised against the entire 4E-BP1 protein (data not shown). An antibody directed against phospho-Thr 37 and phospho-Thr 46 recognizes isoforms b–f (all of the phosphorylated isoforms, but not the unphosphorylated a isoform; Fig. 4C). An antibody directed against phospho-Thr 70 reacts avidly with isoforms d–f (but only poorly with isoforms b and c; Fig. 4D. It should be noted that because these blots represent a population of endogenous 4E-BP1 molecules, it is reasonable to assume that even in serum-stimulated cells, some isoforms may be in the process of being dephosphorylated. Thus, the very weak reactivity of isoforms b and c to the phospho-Thr 70 antibody could indicate an ongoing low level of dephosphorylation). The antibody specific to phosphorylated Ser 65 weakly recognizes isoform e, but reacts strongly with isoform f (Fig. 4E). These data thus indicate that (1) Thr 37 and Thr 46 are phosphorylated in the absence of prior phosphorylation, (2) Thr 70 is phosphorylated predominantly only in isoforms already phosphorylated on Thr 37 and Thr 46, and (3) Ser 65 is phosphorylated only in molecules already phosphorylated on Thr 37, Thr 46, and Thr 70. Thus, analysis with phosphospecific antibodies indicates that the order of phosphate addition on endogenous 4E-BP1 in vivo is as follows: Thr 37/Thr 46, Thr 70, and Ser 65. The nature of the fifth phosphorylated isoform detected in our IEF/SDS-PAGE technique remains elusive, but could be Ser 83, or possibly Ser 112 (a site reported to be phosphorylated in adipocytes; Heesom et al. 1998).
Mutational analysis of 4E-BP1 confirms the order of phosphate addition
To further substantiate the conclusions of the IEF/SDS-PAGE experiments, we applied this technique to a series of HA-tagged 4E-BP1 proteins expressed in HEK 293T cells. As expected, the HA-tagged wild-type 4E-BP1 protein resolves into six major isoforms (Fig. 5A). Consistent with previous data (Gingras et al. 1999a), mutation of Thr 37 and Thr 46 to alanine residues results in a striking accumulation of the unphosphorylated isoform a (Fig. 5B). No phosphorylation of Ser 65 or Thr 70 is detected in this mutant, as determined by Western blotting with phosphospecific antibodies (data not shown). Mutation of Thr 70 to alanine results in an almost complete disappearance of isoforms e and f (Fig. 5C). The phosphorylation state of Ser 65, as determined with phosphospecific antibodies, is reduced in the HA–4E-BP1 Thr 70-Ala mutant (although the extent of this reduction varies from experiment to experiment; data not shown). Mutation of Ser 65 to alanine resulted in the disappearance only of isoform f (Fig. 5D), confirming that Ser 65 phosphorylation is not required for phosphorylation of the other sites. Phosphorylated Thr 37/Thr 46 and Thr 70 are detected to similar levels in the wild-type and Ser 65-Ala mutant protein (data not shown). In summary, analysis of a panel of phosphorylation-site mutant proteins confirms our phosphospecific antibody results regarding the order of phosphate addition to the endogenous 4E-BP1 protein. These data also further show that phosphorylation of Thr 37 and Thr 46 is necessary for phosphorylation of the remaining sites. Phosphorylation of Thr 70 is not required for Thr 37 and Thr 46 phosphorylation, but does modulate the phosphorylation of Ser 65. Consistent with it being added last, the presence or absence of a phosphate group on Ser 65 does not influence the phosphorylation of the other sites. Also consistent with these data is the fact that the tryptic peptide encompassing amino acids 64–73 (Figs. 1A, 2A,B) was found by mass spectrometry to contain either phosphorylated Thr 70 alone, or phosphorylated Thr 70 and Ser 65, but not phosphorylated Ser 65 alone.
Figure 5.
Effects of mutating individual phosphorylation sites on 4E-BP1 phosphorylation. HA-tagged 4E-BP1 proteins were expressed in HEK 293T cells. At 48 h after transfection, lysates were prepared and analysed by two-dimensional IEF/SDS-PAGE, as above. Western blots were prepared using an antibody directed against the HA tag.
Phosphorylation of Ser 65 alone is insufficient to mediate release of 4E-BP1 from eIF4E
Our data show that the phosphorylation of 4E-BP1 is an ordered process culminating in the phosphorylation of Ser 65. However, the role of the various phosphorylation events in the regulation of binding to eIF4E remained unclear. Two models may be envisioned, as follows: (1) phosphorylation of Thr 37, Thr 46, and Thr 70 is required only to permit phosphorylation of Ser 65, which would act as a molecular on/off switch, or (2) phosphorylation of these sites, in addition to inducing phosphorylation on Ser 65, also participates in the decrease in affinity for eIF4E, which is observed following phosphorylation of 4E-BP1. 4E-BP1 phosphorylated on Thr 37 and Thr 46 retains the ability to interact with eIF4E (Gingras et al. 1999a). Therefore, it was pertinent to determine whether phosphorylation of Ser 65 alone (or a combination of Ser 65 and Thr 70) is sufficient to elicit eIF4E release. The cocrystal structure of eIF4E complexed with a peptide derived from 4E-BP1 (amino acids 51–67; Marcotrigiano et al. 1999) suggests that Ser 65 phosphorylation could engender an electrostatic repulsion with acidic amino acids (such as Glu 70) on the dorsal surface of eIF4E (Marcotrigiano et al. 1999). Further, 4E-BP1 phosphorylated on Ser 65 is not bound to eIF4E in vivo (data not shown). However, this cannot be taken as proof that isoforms phosphorylated only on this site are unable to bind to eIF4E, as we have shown that 4E-BP1 molecules that are phosphorylated on Ser 65 are also always phosphorylated on several other residues.
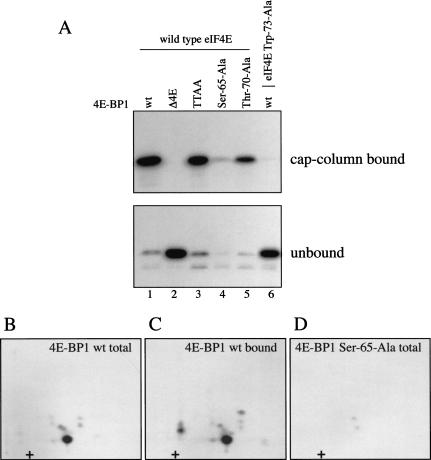
To determine whether Ser 65 phosphorylation alone prevents binding to eIF4E, wild-type and mutant (Δ4E, Thr 37-Ala/Thr 46-Ala, Ser 65-Ala, and Thr 70-Ala) recombinant 4E-BP1 proteins were incubated with recombinant ERK2 in the presence of [γ-32P]ATP. Except for the Ser 65-Ala mutant (which was phosphorylated to <10% of the wild-type protein), the other proteins were phosphorylated to similar extents (data not shown). Thus, the principal phosphorylation site by ERK2 under these conditions is Ser 65 (also see below). Phosphorylated proteins were incubated with an equimolar amount of eIF4E, recovered on an m7GDP-Sepharose (cap-affinity) column (which retains eIF4E and associated proteins), and analyzed by SDS-PAGE followed by autoradioagraphy (Fig. 6A, top, cap-column bound). The unbound fraction was analyzed in parallel (Fig. 6A, bottom, unbound). The phosphorylated 4E-BP1 Δ4E mutant, which is unable to interact with eIF4E, was found exclusively in the unbound fraction (Fig. 6A, lane 2; cf. top and bottom). Similarly, phosphorylated wild-type 4E-BP1 was not coprecipitated with an eIF4E mutant protein (Trp 73-Ala) deficient for 4E-BP1 binding (Fig. 6A, lane 6). 4E-BP1 (whether phosphorylated or not) was also not retained on the cap-affinity column in the absence of eIF4E (data not shown). Therefore, nonspecific interactions with the Sepharose matrix were effectively ruled out. ERK2-phosphorylated wild-type 4E-BP1 interacted efficiently with wild-type eIF4E (Fig. 6A, lane 1). 4E-BP1 proteins harboring mutations at Thr 37 and Thr 46 (TTAA) or at Thr 70 (Thr 70-Ala) were also efficiently phosphorylated by ERK2, and interacted efficiently with wild-type eIF4E (Fig. 6A, lanes 3 and 5). (With the exception of the negative control mutated in the eIF4E-binding site, all of the unphosphorylated proteins, including the Ser 65-Ala mutant, can also interact efficiently with eIF4E. GST fusion proteins were used in this experiment; untagged proteins yielded identical results; data not shown.)
Figure 6.
Phosphorylation of Ser 65 alone is insufficient to effect release from eIF4E. Bacterially expressed wild-type and mutant GST–4E-BP1 proteins were phosphorylated for 15 min with recombinant ERK2 in the presence of [γ-32P]ATP. Labeled 4E-BP1 proteins were incubated for 1 h with equimolar concentrations of recombinant GST–eIF4E proteins. (A) Complexes were recovered on an m7GDP-sepharose column (cap-column bound), and analyzed by SDS-PAGE and autoradiography. Fractions not retained on the cap column (unbound) were analyzed similarly. Δ4E is a 4E-BP1 mutant deleted in the binding site for eIF4E, and TTAA is the double point mutant Thr 37-Ala/Thr 46-Ala. Trp 73-Ala is an eIF4E mutant unable to bind to 4E-BP1. (B–D) Phosphopeptide mapping of ERK2-phosphorylated 4E-BP1. (B) Phosphopeptide map of 4E-BP1 phosphorylated in vitro by ERK2. (C) Phosphopeptide map of 4E-BP1 phosphorylated in vitro by ERK2, and recovered on a cap column. (D) Phosphopeptide map of the Ser 65-Ala mutant phosphorylated in vitro.
To determine whether Ser 65 is phosphorylated in the isoforms bound to eIF4E, wild-type 4E-BP1 labeled in vitro (Fig. 6B), and the same protein isolated after binding to eIF4E, were subjected to phosphopeptide mapping (Fig. 6C; protein from A, lane 1, top). Mapping was also performed on the 4E-BP1 Ser 65-Ala mutant protein (Fig. 6D). The identity of the in vitro-radiolabeled phosphopeptides was determined by mixing these samples with in vivo-labeled endogenous 4E-BP1 (data not shown). These data confirm that Ser 65 is the major site phosphorylated in vitro by ERK2 (Fig. 6B), and, more importantly, show that phosphorylated Ser 65 is clearly found in the fraction of 4E-BP1 bound to eIF4E (Fig. 6C). Thus, phosphorylation of Ser 65 alone is insufficient to abrogate eIF4E binding.
Phosphorylation of both Ser 65 and Thr 70 is not sufficient to effect eIF4E release
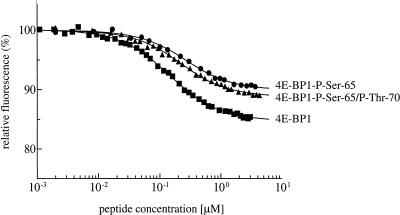
Because serum stimulation increases the phosphorylation state of Ser 65 and Thr 70, it remained possible that phosphorylation of both of these residues could be sufficient to mediate eIF4E release. The study of the effects of Thr 70 phosphorylation has been hampered by the absence of a kinase that can specifically phosphorylate this site to a high stoichiometry in vitro. In addition, with the exception of one report (Karim et al. 2001; see Discussion), no attempt had been made at quantifying the changes in affinity induced by the phosphorylation of 4E-BP1. We determined previously, using isothermic calorimetry, that peptides derived from the eIF4E-binding site (amino acids 51–67) bind to eIF4E with an affinity constant similar to that of the full-length 4E-BP1 protein (with Kds of 50 and 15 nM, respectively; Marcotrigiano et al. 1999). We thus reasoned that synthetic phosphopeptides encompassing the eIF4E-binding site would be a valid model to quantify the effects of phosphorylation at Ser 65 alone, or at Ser 65 and Thr 70. We therefore synthesized peptides encompassing the eIF4E-binding site (Mader et al. 1995; Marcotrigiano et al. 1999), which were unphosphorylated (amino acids 51–67, see Materials and Methods), monophosphorylated at Ser 65 (amino acids 51–67), or diphosphorylated at Ser 65 and Thr 70 (amino acids 51–75). The strength of the interaction between the 4E-BP1 peptides and eIF4E was then determined by fluorescence quenching (Fig. 7). Binding of the 4E-BP1 peptides to the convex dorsal surface of eIF4E leads to quenching of the intrinsic eIF4E fluorescence, mainly due to the interaction between eIF4E Trp 73 (one of the sources of emitted light) and three amino acid side chains in the peptides (Marcotrigiano et al. 1999). The unphosphorylated peptide exhibits a Kd of 105.6 ± 4.4 nM for eIF4E. This value is comparable with that obtained previously for the same peptide (50 nM) by isothermic calorimetry (Marcotrigiano et al. 1999). Consistent with the results presented in Figure 6, phosphorylation of Ser 65 does not disrupt eIF4E binding, but reduces the eIF4E-binding affinity by twofold (Fig. 7; Kd = 210 ± 18 nM). Finally, phosphorylation of both Ser 65 and Thr 70 was also not sufficient to abolish eIF4E binding, and did not alter the affinity of the 4E-BP1 peptide for eIF4E, as compared with the monophosphorylated peptide (Kd = 175 ± 11 nM). Thus, at least in the context of these phosphopeptides, phosphorylation of both Ser 65 and Thr 70 is insufficient to abrogate eIF4E binding. Although it is possible that these peptides may not mimic the behavior of the full length protein precisely, these data suggest that phosphorylation of additional sites, most likely the priming sites Thr 37 and Thr 46, is required to effect eIF4E release.
Figure 7.
4E-BP1 peptides phosphorylated on Ser 65 and Thr 70 interact with eIF4E. Increasing concentrations of an unphosphorylated 4E-BP1 peptide (residues 51–67; ▪), a monophosphorylated peptide (Ser 65; ●), or a diphosphorylated peptide (Ser 65 and Thr 70; ▴) were incubated with eIF4E, and the binding affinity measured using fluorescence quenching of eIF4E. The eIF4E concentration in the experiment shown was 0.1 μM. The peptide concentration range at which the saturation stage is achieved determines the binding constant. It is similar for each of the three peptides, indicating that the binding affinities are nearly equal, with Kd values of ∼100–200 nM. The twofold difference is not significant at the nanomolar level of the binding constant.
Discussion
Using phosphopeptide mapping, mass spectrometry, and a novel combination of two-dimensional isoelectric focusing/SDS-PAGE combined with Western blotting with phosphospecific antibodies directed against various 4E-BP1 phosphorylation sites, we have determined the order of phosphate addition to 4E-BP1 in vivo; Thr 37 and Thr 46 phosphorylation is followed by phosphorylation of Thr 70 and, finally, of Ser 65. We also demonstrated that a combination of phosphorylation events is necessary to abrogate 4E-BP1 binding to e1F4E. The results presented here are in agreement with some previously published data, in which phosphorylation of Ser 65 was shown to be affected by mutation of Thr 37, Thr 46, or Thr 70 (Gingras et al. 1999a; Mothe-Satney et al. 2000a,b). However, our results are at variance with other data presented in the same studies. For example, Mothe-Satney et al. (2000a,b) report that phosphorylation of Thr 70 is independent of prior phosphorylation of Thr 37 and Thr 46. It is important to note that the latter studies relied mainly on the analysis of transfected, overexpressed mutant proteins, which may not faithfully reproduce the behavior of the endogenous protein. Because eIF4E is limiting for translation, overexpression of 4E-BP1 disrupts the delicate balance between eIF4E and its binding partners (including the 4E-BPs and the eIF4G proteins). Overexpression can also lead to mislocalization, or an alteration in upstream signaling events. Furthermore, as 4E-BP1 is an unstructured protein (Fletcher and Wagner 1998; Fletcher et al. 1998; Marcotrigiano et al. 1999), aberrant phosphorylation (i.e., phosphorylation of residues not normally phosphorylated in the endogenous protein) may occur upon expression of large quantities of the protein. In this regard, we have observed phosphorylation of Ser 65 in some experiments involving transfection of a Thr 70-Ala mutant protein (e.g., Fig. 2F), whereas our results with the endogenous protein clearly show that Ser 65 is phosphorylated predominantly only in molecules already phosphorylated on Thr 70. In addition, most previous studies have analyzed the phosphorylation of 4E-BP1 via mobility shift in the one-dimensional SDS-PAGE system; this method does not allow for the analysis of individual phosphorylation sites, because phosphorylation of some of the residues elicits little (if any) shift in electrophoretic mobility. In the current study, we have demonstrated directly that phosphorylation of the endogenous 4E-BP1 occurs in an ordered fashion.
Our data also clearly indicate that phosphorylation of both Thr 70 and Ser 65 is not sufficient to disengage a 4E-BP1 peptide from eIF4E. These results are at variance with reports that have characterized the 4E-BP1/eIF4E interaction utilizing recombinant 4E-BP1 proteins phosphorylated in vitro with ERK, and harboring alanine substitutions at Thr 37, Thr 46, Thr 70, and Ser 83 (Yang et al. 1999; Mothe-Satney et al. 2000b; Karim et al. 2001). By use of the Far-Western technique (a qualitative method used to detect interactions between a nitrocellulose membrane-bound protein and a radiolabeled protein in solution), phosphorylation of either Thr 46 or Ser 65 was reported to result in a decrease in eIF4E binding (Yang et al. 1999; Mothe-Satney et al. 2000b). Also, while this manuscript was in preparation, Karim et al. (2001) reported that phosphorylation of Ser 65 alone leads to a 100-fold decrease in the affinity of 4E-BP1 for eIF4E, as measured using surface plasmon resonance. These authors also report that phosphorylation of Ser 65 reduces eIF4E-binding activity in a pull-down assay (Karim et al. 2001). This is at variance with our data, which were obtained using both full-length proteins and synthetic phosphopeptides. When we performed coprecipitation experiments with the wild-type 4E-BP1 protein phosphorylated by ERK2, we detected only a single major phosphopeptide in the 4E-BP1 fraction bound to eIF4E, which was shown to be phosphorylated on Ser 65. Thus, Ser 65 phosphorylation alone clearly does not lead to a dissociation of 4E-BP1 from eIF4E, under conditions in which the in vivo phosphorylated 4E-BP1 is unable to bind to eIF4E (e.g., Gingras et al. 1998). This conclusion was further substantiated by using peptides monophosphorylated at Ser 65 or diphosphorylated at Ser 65 and Thr 70.
It should be noted that the use of phosphopeptides in this type of study has several advantages over the use of recombinant proteins phosphorylated in vitro by various kinases, as follows: (1) much improved reproducibility from experiment to experiment, (2) phosphorylation only of the desired residue, (3) 100% stoichiometry of phosphorylation, and (4) the possibility of generating large quantities of the phosphopeptide for quantitative measurements. The use of synthetic peptides has also allowed for better control of disulfide bond formation caused by a cysteine residue (Cys 62) located in the vicinity of the eIF4E-binding site. Under nonreducing conditions, oxidation of Cys 62 induces dimer formation (both in the context of the peptides and in the full-length protein). This is especially problematic for the unphosphorylated peptide, which forms dimers more readily than the phosphorylated peptides, presumably because the phosphate groups exert some degree of repulsion to partially prevent dimerization. Remarkably, peptide dimerization was observed to enhance the apparent eIF4E-binding affinity by up to two to three orders of magnitude (for the unphosphorylated peptide; A. Niedzwiecka, unpubl.). Thus, performing affinity measurements without protection against Cys 62 oxidation can give rise to spurious results. It remains possible that peptides do not precisely mimic the behavior of the full-length protein. Ideally, these biochemical measurements should be repeated when synthetic phosphorylated full-length protein becomes available.
The identity of the kinase(s) responsible for phosphorylating 4E-BP1 in vivo is not known. FRAP/mTOR immunoprecipitates can phosphorylate 4E-BP1 in vitro, on Thr 37 and Thr 46 (Burnett et al. 1998; Gingras et al. 1999a). However, a FRAP/mTOR activating antibody was reported to stimulate the phosphorylation of Ser 65 and Thr 70 in vitro, suggesting that FRAP/mTOR could be responsible for phosphorylating all of the 4E-BP1 phosphorylation sites (Brunn et al. 1997a,b; Mothe-Satney et al. 2000a). It was also reported that a kinase activity specific to Ser 65 and Thr 70 could be liberated from a FRAP/mTOR immunoprecipitate, suggesting that a second kinase is present in FRAP/mTOR immunoprecipitates (Heesom and Denton 1999). Thus, whether FRAP/mTOR phosphorylates 4E-BP1 on all or some of these sites remains to be determined.
Whereas FRAP/mTOR immunoprecipitates phosphorylate Thr 37 and Thr 46 in vitro, these two sites exhibit weak rapamycin sensitivity in vivo, at least in the presence of serum (between 1.3- and 1.7-fold; Gingras et al. 1999a). These data correlate well with the mild inhibition of FRAP/mTOR kinase activity following rapamycin treatment (on the order of 1.5-fold; e.g., Scott et al. 1998; Gingras et al. 2001b). Interestingly, in vivo phosphorylation at Thr 37 and Thr 46 is strongly decreased by rapamycin only in cells deprived of serum (Gingras et al. 1999a). How serum protects Thr 37 and Thr 46 from dephosphorylation induced by rapamycin is unknown: Possibly rapamycin activates a phosphatase activity directed against Thr 37 and Thr 46 (see below), but this phosphatase is inactivated by serum. Another possibility is that serum activates a second kinase that is rapamycin insensitive, and which is responsible for the phosphorylation of Thr 37 and Thr 46 in the absence of serum. Further work will be necessary to distinguish between these possibilities.
Despite the fact that FRAP/mTOR does not significantly phosphorylate 4E-BP1 at Ser 65 and Thr 70 in vitro (unless FRAP/mTOR is immunoprecipitated with an activating antibody; Brunn et al. 1997a,b; Mothe-Satney et al. 2000a), FRAP/mTOR signaling clearly plays a critical regulatory role in the phosphorylation of Ser 65 and Thr 70, because these residues display a higher degree of rapamycin sensitivity than Thr 37 and Thr 46 (Gingras et al. 1998). Treatment of cells with the phosphatase inhibitor calyculin prevents 4E-BP1 dephosphorylation in response to rapamycin treatment (Peterson et al. 1999). Thus, it is a distinct possibility that a PP2A-type phosphatase directed against Ser 65 and Thr 70 is activated following rapamycin treatment, and FRAP/mTOR activity is required for repression of this phosphatase. In this respect, genetic screening in Saccacharomyces cerevisiae has suggested that TOR signaling is effected through the repression of PP2A-type phosphatases, which is mediated through regulation of the activity of a conserved phosphatase-binding protein (Tap42p).
Consistent with an important role for the phosphorylation at Thr 37, Thr 46, and Thr 70, mutation of these residues dramatically affects the release of 4E-BP1 from eIF4E following serum stimulation. Mutation of Thr 37/Thr 46 to alanines completely prevents dissociation of the eIF4E/4E-BP1 complex in 293 cells following serum stimulation (Burnett et al. 1998; Gingras et al., unpubl.). Mutation of Thr 70 alone to alanine also largely prevents dissociation (A.-C. Gingras and B. Raught, unpubl.). However, mutation of Ser 65 to alanine does not significantly alter the release (as compared with the wild-type protein) from eIF4E following serum stimulation (A.-C. Gingras and B. Raught, unpubl.). These data agree with the in vivo results presented by Mothe-Satney et al. (2000b), who found that Thr 37-Ala or Thr 46-Ala mutants displayed a higher degree of eIF4E-binding activity than the wild-type protein, whereas the Thr 70-Ala mutant was bound to eIF4E at an intermediate level between the Thr 37-Ala or Thr 46-Ala mutants and the wild-type protein. Mutating Ser 65 to alanine had no detectable effect in the experiments of Mothe-Satney et al. (2000b), in agreement with our studies.
The phosphorylation of several different proteins has been shown to occur in an ordered fashion, but the order of addition on the various phosphorylated residues on the endogenous proteins has only been determined for a few (e.g., the ribosomal S6 protein and the ribosomal protein S6 kinase 1; for review, see Fumagalli and Thomas 2000). The combination of techniques used here (two-dimensional IEF/SDS-PAGE, followed by Western blotting with phosphospecific antibodies) has enabled us to define an order of addition of the phosphate groups onto endogenous 4E-BP1 following serum stimulation of HEK 293 cells. Thus, combined with mass spectrometry, IEF/SDS-PAGE and phosphospecific antibodies can be a useful technique to characterize complex, multi-step phosphorylation events on any protein of interest.
Materials and methods
Construction and expression of 4E-BP1 mutant proteins
The human 4E-BP1 cDNA was used as a template for PCR mutagenesis to mutate Ser 65, Thr 70, and Ser 83 to Ala, Glu, or Asp. Mutated sequences were inserted in-frame into both the cytomegalovirus-based vector pACTAG-2 (which contains an N-terminal 3HA-tag fusion, for expression in mammalian cells), and pGEX-6p1 (which contains an N-terminal GST tag, for bacterial expression; Pharmacia). The murine wild-type eIF4E-coding sequence and a mutant Trp 73-Ala were also inserted in-frame into pGEX-6p1. Inserts were sequenced in their entirety. Other constructs were published previously (Gingras et al. 1999a). Transient transfection of pACTAG-2–4E-BP1 into HEK 293 (ATCC CRL 1573) or HEK 293T cells (ATCC CRL 11268) was performed using Lipofectamine (Life Technologies), according to the manufacturer's instructions. GST fusion proteins were expressed in BL21 bacteria, purified as described (Gingras et al. 1999a), and quantified by Bradford assay and Coomassie staining after SDS-PAGE.
eIF4E purification
For fluorescence measurements, murine eIF4E (residues 28–217 and residues 33–217) was expressed in Escherichia coli (Marcotrigiano et al. 1999), purified from inclusion bodies, folded in a one-step dialysis from 6 M guanidinum hydrochloride, and subjected to anion exchange chromatography on a MonoS column. Protein solutions were buffer exchanged with Ultrafree-15 mL filters with a Biomax 5-kD NMWL membrane (Millipore). Immediately before the spectroscopic measurements, the protein sample was filtered through an Ultrafree-0.5 mL Biomax 100-kD NMWL (Millipore).
Isoelectric focusing/SDS-PAGE
HEK 293 or HEK 293T cells were rinsed twice with cold PBS, scraped, and pelleted. After removal of the PBS, cells were resuspended in 200 μL (per 100-mm plate) of freshly prepared extraction buffer (20 mM Hepes-KOH at pH 7.5, 100 mM KCl, 20 mM β-glycerolphosphate, 1 mM DTT, 0.25 mM Na3VO4, 10 mM NaF, 1 mM EDTA, 1 mM EGTA, 10 nM okadaic acid, and 1 mM PMSF, supplemented with a protease inhibitor cocktail from Boehringer Mannheim). Cells were lysed by three freeze-thaw cycles, debris were pelleted by centrifugation, and the protein concentration in the supernatant was measured by Bradford assay (Bio-Rad). This lysate was diluted directly into Laemmli sample buffer for SDS-PAGE and Western blotting. For two-dimensional isoelectric focusing (IEF)/SDS-PAGE, the lysate (20 μL containing ∼200 μg protein) was mixed with 27 mg of urea and 7 μL of IEF sample buffer (17% v/v pharmalytes at pH 3–10, 4% v/v pharmalytes at pH 2–5.5, 14% v/v β-mercaptoethanol, 35% w/v CHAPS, 1.8% w/v SDS), and incubated at room temperature until the urea was dissolved.
Two-dimensional isoelectric focusing/SDS-PAGE was performed using the Protean II system (Bio-Rad). Briefly, the first-dimension gel (55% w/v urea, 4.257% w/v acrylamide, 0.243% w/v bis-acrylamide, 1.5% w/v CHAPS, 0.5% v/v NP-40, 4% v/v Pharmalytes pH 3–10, 1% v/v Pharmalytes pH 2.5–5) was polymerized in 18 cm × 1.5 mm (inside diameter) glass tubes to 3 cm from the top. After rinsing the surface of the gel, the sample was applied and overlaid with upper chamber buffer (20 mM NaOH). The lower (10 mM H3PO4) and upper chambers were filled with buffer, and the IEF dimension was run at 200 V for 2 h, 500 V for 2 h, and 800 V for 16 h at room temperature. Gels were extruded from the capillary tubes, equilibrated in Laemmli sample buffer for 1 h, and subjected to SDS-15% PAGE (16-cm configuration, 1.5-mm thickness) for the second dimension. Proteins were electrotransferred to nitrocellulose membranes, and analyzed by Western blotting.
Western blotting was performed as described (Gingras et al. 1999a). The phosphospecific antibodies to Ser 65 and Thr 70 were generated at Cell Signaling Technology. The other primary antibodies used for Western blotting in this study were as follows: the anti-HA mouse monoclonal antibody HA.11 (BAbCO), the rabbit polyclonal antisera 11208 directed against 4E-BP1 (Gingras et al. 1996), and the phosphospecific antibody directed against Thr 37 and Thr 46 (Gingras et al. 1999a).
Phosphopeptide mapping of 4E-BP1 and identification of the phosphorylated residues
The procedure utilized for phosphopeptide mapping was essentially as described (Gingras et al. 1998, 1999a; Raught et al. 2000a). Briefly, HEK 293 or HEK 293T cells were labeled in vivo with [32P]orthophosphate, and 4E-BP1 was immunoprecipitated with the anti-4E-BP1 11209 antisera, or with an anti-HA antibody. Immunoprecipitated 4E-BP1 was subjected to SDS-15%PAGE, and transferred to 0.2-μm pore size nitrocellulose membranes (Schleicher & Schuell). Membranes were subjected to autoradiography, and the membrane region harboring 4E-BP1 was excised and subjected to tryptic:chymotryptic (200:1) digestion. Digested products were applied to plastic-backed cellulose-coated thin layer chromatography plates (for Fig. 1; Kodak), or to glass-backed cellulose-coated plates (Figs. 2, 5; Merck). Following electrophoresis and liquid chromatography, plates were dried and subjected to autoradiography. Phosphopeptides were eluted from the plates and analyzed by LC-MS/MS, as described (Watts et al. 1994; Gingras et al. 1999a, Gygi et al. 1999b). Phosphoaminoacid analysis was also performed on phosphopeptides eluted from the plates, as described (van der Geer and Hunter 1994; Gingras et al. 1999a).
Kinase assays and chromatography on m7GDP-agarose
Phosphorylation of 4E-BP1 with recombinant ERK2 (New England Biolabs) was performed for 15 min at 30°C, in the presence of [γ-32P]ATP, according to the manufacturer's instructions. Chromatography on m7GDP-agarose was performed as described (Gingras et al. 1999a). Proteins retained on m7GDP-agarose were washed three times with 20 volumes of buffer (20 mM Hepes-KOH and 75 mM KCl), and analyzed by SDS-PAGE and autoradiography.
Phosphopeptide synthesis and purification
A peptide corresponding to residues 51–67 of mammalian 4E-BP1, RIIYDRKFLMECRNSPV (Pause et al. 1994), was synthesized by Boc protocols for solid-phase peptide synthesis, and cleaved by use of a standard HF procedure (Schnolzer et al. 1992). The phosphopeptide synthesis was performed manually on a Wang resin (Novabiochem) using Fmoc protocols for solid-phase peptide synthesis. Phosphoserine and phosphothreonine were incorporated directly during the synthesis as the corresponding preformed mono-protected N-α-Fmoc-O-benzyl phosphoaminoacid derivatives, purchased from Novabiochem (Wakamiya et al. 1994). The phosphopeptides were cleaved by use of a trifluoroacetic acid:triisopropylsilane:water mixture (TFA:TIS:H2O, 95:2.5:2.5). After cleavage, crude peptides were purified to homogeneity by semipreparative HPLC, and characterized by amino acid analysis (see below) and MALDI-TOF or ESI spectrometry (51–67 4E-BP1 peptide, predicted mass 2141.8, measured mass 2141; P-Ser 65 phosphopeptide, predicted mass 2220.1, measured mass 2221.0; P-Ser 65/P-Thr 70 diphosphopeptide, predicted mass 3180.55, measured mass 3181.0). Peptide concentrations were determined by amino acid analysis after acid gas-phase hydrolysis. Analyses were performed three times for each peptide via conversion of the amino acids to phenylothiocarbamyl derivatives using HPLC (Waters, Millipore), and a PicoTag 3.9 × 150-mm column (Waters, Millipore). All peptides lacked secondary structure in solution, as evidenced by the presence of minima at ∼200 nm recorded on a CD spectropolarimeter (AVIV model 202).
Affinity measurements by fluorescence quenching
Measurements of the affinity of the 4E-BP1 peptides for eIF4E were determined by fluorescence quenching, essentially as described (Blachut-Okrasinska et al. 2000). Before the experiment, eIF4E was saturated with a 50-fold excess of m7GTP. Titration experiments were performed in 50 mM Hepes-KOH (pH 7.20), 100 mM KCl, 1 mM dithiothreitol (DTT), and 0.5 mM disodium ethylenediaminetetraacetate (Na2EDTA). The pH was measured within ±0.01 unit on a Beckman Φ300 pH meter. One-microliter aliquots of increasing concentrations (1 μM to 5 mM) of each peptide were added to 1400 μL of eIF4E solution. Solutions of 4E-BP1 peptides were controlled strictly by use of HPLC and MS for lack of disulfide dimer formation, which can influence the binding affinities (data not shown). The concentration of eIF4E was determined from absorption. Absorption and fluorescence spectra were recorded on Lambda 20 UV/VIS and LS-50B instruments (Perkin-Elmer). The sample was thermostated within ±0.2°C, and the temperature was controlled inside the cuvette with a thermocouple. An excitation wavelength of 290 nm (slit 2.5-nm, auto cut-off filter) and an emission wavelength of 350 nm (slit 2.5 to 4 nm, 290-nm cut-off filter) were used, with a correction for the photomultiplier sensitivity. These conditions ensured the observation of only the tryptophan emission, such that the emission of Tyr 54 in 4E-BP1 was eliminated. Measurements were also run with eIF4E at several concentrations, from 0.05 to 1.0 μM. Fluorescence intensity was monitored by time-synchronized measurements at a single wavelength, with an integration time of 30 sec and a gap of 30 sec for adding the peptide, with magnetic stirring. During the time required for addition of the peptide, the UV xenon flash lamp was switched off, to avoid photobleaching of the sample. A curve for the fluorescence intensity as a function of the total ligand concentration was fitted to the experimental data points by means of a nonlinear, least-squares method, using the program ORIGIN 6.0 (Microcal Software; Blachut-Okrasinska et al. 2000). The dissociation constants were calculated as a weighted average of at least three independent series.
Acknowledgments
We thank Wojciech Augustyniak, Mariusz Tasior, Colin Lister, and Fabienne Vozy for excellent assistance, and Dr. Pawel Mak (Institute of Molecular Biology, Jagiellonian University, Krakow, Poland) for amino acid analysis of the peptides. We thank Dr. Joseph Marcotrigiano for kindly providing the unphosphorylated 4E-BP1 peptide; Drs. Roger Duncan, John Silvius, Joseph Marcotrigiano, and members of the Sonenberg laboratory for useful discussions; and Francis Poulin and Drs. Jie Chen and Thomas Neufeld for critical reading of the manuscript. This work was supported by grants from the National Cancer Institute of Canada and the Howard Hughes Medical Institute to N.S., from the Human Science Frontiers Program to N.S. and S.K.B., from the National Institutes of Health (HG00041) to S.P.G., and from the NSF Science and Technology Center for Molecular Biotechnology to R.A. Work conducted at Warsaw University was supported by the Polish Committee for Scientific Research (K.B.N.) 6 P04A 055 17. A-C.G. is supported by a Doctoral Award from the Canadian Institutes of Health Research (CIHR), B.R. by a postdoctoral fellowship from the CIHR, and M.M. by a Cancer Research Society studentship. N.S. is a CIHR Distinguished Investigator and a Howard Hughes Medical Institute International Scholar.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL nsonen@med.mcgill.ca; FAX (514) 398-1287.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.912401.
References
- Beretta L, Gingras A-C, Svitkin YV, Hall MN, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- Blachut-Okrasinska E, Bojarska E, Niedzwiecka A, Chlebicka L, Darzynkiewicz E, Stolarski R, Stepinski J, Antosiewicz JM. Stopped-flow and Brownian dynamics studies of electrostatic effects in the kinetics of binding of 7-methyl-GpppG to the protein eIF4E. Eur Biophys J. 2000;29:487–498. doi: 10.1007/s002490000096. [DOI] [PubMed] [Google Scholar]
- Brunn GJ, Fadden P, Haystead TAJ, Lawrence JC., Jr The mammalian target of rapamycin phosphorylates sites having a (Ser/Thr)-Pro motif and is activated by antibodies to a region near its COOH terminus. J Biol Chem. 1997a;272:32547–32550. doi: 10.1074/jbc.272.51.32547. [DOI] [PubMed] [Google Scholar]
- Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC, Jr, Abraham RT. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997b;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufner A, Andjelkovic M, Burgering BM, Hemmings BA, Thomas G. Protein kinase B localization and activation differentially affect S6 kinase 1 activity and eukaryotic translation initiation factor 4E-binding protein 1 phosphorylation. Mol Cell Biol. 1999;19:4525–4534. doi: 10.1128/mcb.19.6.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadden P, Haystead TA, Lawrence JC., Jr Identification of phosphorylation sites in the translational regulator, PHAS-I, that are controlled by insulin and rapamycin in rat adipocytes. J Biol Chem. 1997;272:10240–10247. doi: 10.1074/jbc.272.15.10240. [DOI] [PubMed] [Google Scholar]
- Fletcher CM, Wagner G. The interaction of eIF4E with 4E-BP1 is an induced fit to a completely disordered protein. Protein Sci. 1998;7:1639–1642. doi: 10.1002/pro.5560070720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher CM, McGuire AM, Gingras A-C, Li H, Matsuo H, Sonenberg N, Wagner G. 4E binding proteins inhibit the translation factor eIF4E without folded structure. Biochemistry. 1998;37:9–15. doi: 10.1021/bi972494r. [DOI] [PubMed] [Google Scholar]
- Fumagalli S, Thomas G. S6 phosphorylation and signal transduction. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational control of gene expression. Plainview, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 695–718. [Google Scholar]
- Gingras A-C, Svitkin Y, Belsham GJ, Pause A, Sonenberg N. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc Natl Acad Sci. 1996;93:5578–5583. doi: 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A-C, Kennedy SG, O'Leary MA, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes & Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A-C, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: A novel two-step mechanism. Genes & Dev. 1999a;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A-C, Raught B, Sonenberg N. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999b;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- ————— . Control of translation by the target of rapamycin proteins. In: Rhoads RE, editor. Progress in molecular and subcellular biology. Berlin, Germany: Springer-Verlag; 2001a. pp. 143–174. [DOI] [PubMed] [Google Scholar]
- ————— Regulation of translation initiation by FRAP/mTOR. Genes & Dev. 2001b;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- Gygi SP, Han DK, Gingras A-C, Sonenberg N, Aebersold R. Protein analysis by mass spectrometry and sequence database searching: Tools for cancer research in the post-genomic era. Electrophoresis. 1999a;20:310–319. doi: 10.1002/(SICI)1522-2683(19990201)20:2<310::AID-ELPS310>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999b;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: Competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heesom KJ, Denton RM. Dissociation of the eukaryotic initiation factor-4E/4E-BP1 complex involves phosphorylation of 4E-BP1 by an mTOR-associated kinase. FEBS Lett. 1999;457:489–493. doi: 10.1016/s0014-5793(99)01094-7. [DOI] [PubMed] [Google Scholar]
- Heesom KJ, Avison MB, Diggle TA, Denton RM. Insulin-stimulated kinase from rat fat cells that phosphorylates initiation factor-4E binding protein 1 on the rapamycin-insensitive site (serine-111) Biochem J. 1998;336:39–48. doi: 10.1042/bj3360039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey JWB, Merrick WC. Pathway and mechanism of initiation of protein synthesis. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational control of gene expression. Plainview, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 33–88. [Google Scholar]
- Karim MM, Hughes JMX, Warwicker J, Scheper GC, Proud CG, McCarthy JEG. A quantitative molecular model for modulation of mammalian translation by the eIF4E-binding protein 1. J Biol Chem. 2001;276:20750–20757. doi: 10.1074/jbc.M011068200. [DOI] [PubMed] [Google Scholar]
- Kohn AD, Barthel A, Kovacina KS, Boge A, Wallach B, Summers SA, Birnbaum MJ, Scott PH, Lawrence JC, Jr, Roth RA. Construction and characterization of a conditionally active version of the serine/threonine kinase Akt. J Biol Chem. 1998;273:11937–11943. doi: 10.1074/jbc.273.19.11937. [DOI] [PubMed] [Google Scholar]
- Lin TA, Kong X, Haystead TA, Pause A, Belsham G, Sonenberg N, Lawrence JC., Jr PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- Lin TA, Kong X, Saltiel AR, Blackshear PJ, Lawrence JC., Jr Control of PHAS-I by insulin in 3T3-L1 adipocytes. Synthesis, degradation, and phosphorylation by a rapamycin-sensitive and mitogen-activated protein kinase-independent pathway. J Biol Chem. 1995;270:18531–18538. doi: 10.1074/jbc.270.31.18531. [DOI] [PubMed] [Google Scholar]
- Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras A-C, Sonenberg N, Burley SK. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell. 1999;3:707–716. doi: 10.1016/s1097-2765(01)80003-4. [DOI] [PubMed] [Google Scholar]
- Mothe-Satney I, Brunn GJ, McMahon LP, Capaldo CT, Abraham RT, Lawrence JC., Jr Mammalian target of rapamycin-dependent phosphorylation of PHAS-I in four (S/T)P sites detected by phospho-specific antibodies. J Biol Chem. 2000a;275:33836–33843. doi: 10.1074/jbc.M006005200. [DOI] [PubMed] [Google Scholar]
- Mothe-Satney I, Yang D, Fadden P, Haystead TA, Lawrence JC., Jr Multiple mechanisms control phosphorylation of PHAS-I in five (S/T)P sites that govern translational repression. Mol Cell Biol. 2000b;20:3558–3567. doi: 10.1128/mcb.20.10.3558-3567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Belsham GJ, Gingras A-C, Donzé O, Lin TA, Lawrence JC, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- Peterson RT, Desai BN, Hardwick JS, Schreiber SL. Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycin associated protein. Proc Natl Acad Sci. 1999;96:4438–4442. doi: 10.1073/pnas.96.8.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B, Gingras A-C, Gygi SP, Imataka H, Morino S, Gradi A, Aebersold R, Sonenberg N. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J. 2000a;19:434–444. doi: 10.1093/emboj/19.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B, Gingras A-C, Sonenberg N. Regulation of ribosomal recruitment in eukaryotes. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational control of gene expression. Plainview, NY: Cold Spring Harbor Laboratory Press; 2000b. pp. 245–294. [Google Scholar]
- Schnolzer M, Alewood P, Jones A, Alewood D, Kent SB. In situ neutralization in Boc-chemistry solid phase peptide synthesis. Rapid, high yield assembly of difficult sequences. Int J Pept Protein Res. 1992;40:180–193. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- Scott PH, Brunn GJ, Kohn AD, Roth RA, Lawrence JC., Jr Evidence of insulin-stimulated phosphorylation and activation of the mammalian target of rapamycin mediated by a protein kinase B signaling pathway. Proc Natl Acad Sci. 1998;95:7772–7777. doi: 10.1073/pnas.95.13.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Ogawa W, Kitamura T, Hino Y, Kuroda S, Kotani K, Klip A, Gingras A-C, Sonenberg N, Kasuga M. Requirement for Akt (protein kinase B) in insulin-induced activation of glycogen synthase and phosphorylation of 4E-BP1 (PHAS-1) J Biol Chem. 1999;274:20611–20618. doi: 10.1074/jbc.274.29.20611. [DOI] [PubMed] [Google Scholar]
- van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by electrophoresis and chromatography on thin-layer cellulose plates. Electrophoresis. 1994;15:544–554. doi: 10.1002/elps.1150150173. [DOI] [PubMed] [Google Scholar]
- von Manteuffel SR, Gingras A-C, Ming XF, Sonenberg N, Thomas G. 4E-BP1 phosphorylation is mediated by the FRAP–p70s6k pathway and is independent of mitogen-activated protein kinase. Proc Natl Acad Sci. 1996;93:4076–4080. doi: 10.1073/pnas.93.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Manteuffel SR, Dennis PB, Pullen N, Gingras A-C, Sonenberg N, Thomas G. The insulin-induced signalling pathway leading to S6 and initiation factor 4E binding protein 1 phosphorylation bifurcates at a rapamycin-sensitive point immediately upstream of p70s6k. Mol Cell Biol. 1997;17:5426–5436. doi: 10.1128/mcb.17.9.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamiya T, Saruto K, Yasuoka J, Kusumoto S. An efficient procedure for solid-phase synthesis of phosphopeptides by Fmoc strategy. Chem Lett. 1994;4:1099–1102. [Google Scholar]
- Watts JD, Affolter M, Krebs DL, Wange RL, Samelson LE, Aebersold R. Identification by electrospray ionization mass spectrometry of the sites of tyrosine phosphorylation induced in activated Jurkat T cells on the protein tyrosine kinase ZAP-70. J Biol Chem. 1994;269:29520–29529. [PubMed] [Google Scholar]
- Wu X, Senechal K, Neshat MS, Whang YE, Sawyers CL. The PTEN/MMAC1 tumor suppressor phosphatase functions as a negative regulator of the phosphoinositide 3-kinase/Akt pathway. Proc Natl Acad Sci. 1998;95:15587–15591. doi: 10.1073/pnas.95.26.15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Brunn GJ, Lawrence JC., Jr Mutational analysis of sites in the translational regulator, PHAS-I, that are selectively phosphorylated by mTOR. FEBS Lett. 1999;453:387–390. doi: 10.1016/s0014-5793(99)00762-0. [DOI] [PubMed] [Google Scholar]