Abstract
Most neurobehavioral diseases are sexually dimorphic in their incidence, and sex differences in the brain may be key for understanding and treating these diseases. Calbindin (Calb) D28K is used as a biomarker for the well-studied sexually dimorphic nucleus, a hypothalamic structure that is larger in males than in females. In the current study weanling C56BL/6J mice were used to examine sex differences in the Calb protein and message focusing on regions outside of the hypothalamus. A robust sex difference was found in Calb in the frontal cortex (FC) and cerebellum (CB; specifically in Purkinje cells); mRNA and protein were higher in females than in males. Using 2 mouse lines, i.e. one with a complete deletion of estrogen receptor alpha (ERα) and the other with uncoupled gonads and sex chromosomes, we probed the mechanisms that underlie sexual dimorphisms. In the FC, deletion of ERα reduced Calb1 mRNA in females compared to males. In addition, females with XY sex chromosomes had levels of Calb1 equal to those of males. Thus, both ERα and the sex chromosome complement regulate Calb1 in the FC. In the CB, ERα knockout mice of both sexes had reduced Calb1 mRNA, yet sex differences were retained. However, the sex chromosome complement, regardless of gonadal sex, dictated Calb1 mRNA levels. Mice with XX chromosomes had significantly greater Calb1 than did XY mice. This is the first study demonstrating that sex chromosome genes are a driving force producing sex differences in the CB and FC, which are neuoranatomical regions involved in many normal functions and in neurobehavioral diseases.
Key Words: Autism, X inactivation, Sex differences, Calbindin, Fragile X
Introduction
Sex differences in behaviors such as cognition, mood, and motor skills are well documented [1,2,3]. Understanding the underlying neural mechanisms regulating sex differences can offer important insights into neurobehavioral diseases with strong sex biases. The roles of gonadal steroids and their receptors in the development of sexually dimorphic hypothalamic structures are well established [4,5], but few sexual dimorphisms have been characterized outside of the hypothalamus [6]. Moreover, while many sex differences can be attributed to developmental exposure to hormones, a more novel hypothesis, i.e. that sex chromosome genes have a direct action on neural sexual dimorphism, is gaining momentum [7,8]. The current studies specifically address nonhypothalamic sex differences and examine the role of estrogen receptor alpha (ERα) and the sex chromosome complement on Calbindin-D28K (Calb) expression.
Calb, a member of the EF-hand calcium-binding protein family, is highly expressed in the brain and plays an important role in maintaining Ca2+ homeostasis within neurons (for review see Kojetin et al. [9] and Bastianelli [10]). Calb immunoreactivity has been routinely used as a biomarker for the sexually dimorphic nucleus (SDN) in the hypothalamus (males > females) [11,12,13]. The greatest concentrations of Calb are in the cerebellum (CB), where it is expressed exclusively in Purkinje cells [10], and in the cortex; these are regions that play an important role in normal cognition, motor skills, and emotional processing [14,15,16]. This protein has multiple functions. Studies with Calb knockout (KO) mice report distinct deficits in circadian rhythms, motor skills, coordination, and spatial learning in the Morris water maze [17,18,19,20]. In addition, long-term potentiation in the hippocampus is not stable, and in Purkinje cells the time course and amplitude of calcium transients are significantly altered while spine lengths are increased [20,21,22,23]. Finally, in Purkinje cell-specific Calb KO mice motor coordination and visual and vestibular processing are markedly impaired [21].
The current study is the first to show that the Calb gene and protein are expressed in a sexually dimorphic manner outside of the hypothalamus. In particular, sex differences (female > male) in Calb were observed in normal juvenile mouse brains in the CB and the frontal cortex (FC). We capitalized on 2 mouse models to assess the underlying mechanisms regulating sex differences inCalb. Here we examined Calb1 in ERα KO (ERαKO) and wild-type (WT) littermates to determine if this steroid receptor was responsible for the observed sex differences. Next, the roles of the sex chromosomes were evaluated using a mouse model that isolates the effects of gonadal sex and the sex chromosome complement (table 1) [8].
Table 1.
Comparison of normal and FCG mice
| Mouse cross | Nomenclature | Genotype | Gonadal phenotype | X | Y |
|---|---|---|---|---|---|
| WT | F | XX | ovaries | 2 | 0 |
| M | XY | testes | 1 | 1 | |
| FCG | XXF | XX | ovaries | 2 | 0 |
| XYF | XY- | ovaries | 1 | 1 | |
| XXM | XXSry | testes | 2 | 0 | |
| XYM | XYSry | testes | 1 | 1 | |
In the FCG, numbers of X versus Y chromosomes can be separated from gonadal effects.
Methods
Animals
All mice were in a C57BL/6J background and were born, reared, and housed at the University of Virginia School of Medicine Animal Facility in Jordan Hall on a 12:12-hour light:dark cycle (lights on at 6:00 a.m.). Food (Harlan Teklad Mouse/Rat Sterilizable Diet No. 7012) and water were provided ad libitum. Three ‘lines’ of mice were used, all in the C57BL/6J background. These lines were: normal C57BL/6J, ERαKO [24], and the 4 core genotypes (FCG) [8]. All animals were weaned at 20–21 days of age and sacrificed between 21–25 days of age. These young animals were used to compare sexes at a time when gonadal hormone levels were uniformly low in males and females. We have examined testosterone levels and target organ weights in animals from the FCG at this age and there are no genotypic differences [Cox and Rissman, unpubl. data]. The University of Virginia Animal Use and Care Committee approved all animal procedures.
Genotyping
DNA was extracted from mouse ear clippings using a Tissue DNA Extraction and Amplification kit from SIGMA (XNATR). To screen for the presence or absence of the deletion of the ERα gene (Esr1), 2 PCR reactions were run. To screen for the presence or absence of the Y chromosome, the YMT2/B sequence (a member of the Ssty gene family present on the long arm of the Y chromosome) was used. An amplification control myogenin primer set was also used. All of the PCR products were run on a 2% agarose gel.
Brain Tissue Preparation for in situ Hybridization and Protein and RNA Extraction
For in situ hybridization (ISH) studies, normal C57BL/6J (PN21–25) mice were anesthetized using a lethal dose of Euthanasol, and they were transcardially perfused with 4% paraformaldehyde (Polysciences, Inc., Warrington, Pa., USA) in 0.1 M phosphate buffer, pH 7.4. After perfusion, the brains were postfixed overnight prior to vibrotome sectioning (50-μm coronal sections). The sections for ISH were stored in cryoprotectant [30% sucrose, 30% ethylene glycol, and 1% polyvinylpyrrolidone (PVP-40)] at −20°C until they were processed. For protein and RNA collection, mice were sacrificed, their brains were rapidly removed, and the FC, CB, hypothalamus, hippocampus, and amygdala were carefully dissected on ice, frozen on dry ice, and stored at −80°C until further processing for protein and RNA extraction.
Synthesis of Calbindin RNA Probes
T3- or T7-labeled RNA probes were prepared from a PCR-generated Calb1 DNA template with the T7 and T3 promoter in the 5′ ends of the gene. Specifically, a sense primer (Invitrogen, Carlsbad, Calif., USA) containing the T3 promoter (in bold) and Calb1 gene (bp 109–129) (CAGAGATGCAATTAACCCTCACTAAAGGGAGACATCTCTGATCACAGCCTCACAG) and an antisense primer (Invitrogen) containing the T7 promoter (in bold) and Calb1 gene (bp 638–659) (CCAAGCTTCTAATACGACTCACTATAGGGAGATTCCTGCACTGGTAGTAACCTGG) produced a DNA template with both promoters. DNA sequencing prior to use in an in vitro transcription assay verified the template. Digoxigenin-labeled probes were generated and incorporation was measured according to protocols and reagents provided by Roche Applied Science.
In situ Hybridization
Both prehybridization and posthybridization were done following a published protocol [25]. Sections were prehybridized for 60 min at room temperature (RT) followed by 1 h at 37°C in a hybridization mixture containing 0.6 M NaCl, 0.1 M Tris-Cl pH 7.5, 10 mM EDTA, 0.05% NaPP, 0.5 mg/ml yeast total RNA, 0.05 mg/ml yeast tRNA, 1× Denhardt's BSA, 50% formamide, 5% dextran sulphate, 10 μm of the 4 rNTPs, 0.5 mg/ml salmon sperm DNA, and 10 mM DTT. Labeled probe (800–1,000 ng) was added to the hybridization buffer and sections were incubated overnight at 55°C. Posthybridization consisted of washing the sections in 4× SSC/10 mM NaTS at 37°C followed by treatment with RNAse A and RNAse T1 in NTE (0.5 M NaCl, 10 mMTris-Cl pH 7.5, 1 mM EDTA pH 8.0) for 30 min at 37°C. Sections were then rinsed in a series of SSC buffers of decreasing molarity for 20 min each at 37°C followed by a final wash in 0.1× SSC for 60 min at 55°C. For antibody incubation, sections were rinsed in TBST (0.15 M NaCl, 0.1 M Tris-Cl pH 7.5, and 0.1% Tween-20) at RT, incubated in blocking solution (2% Roche Blocking Reagent and 20% heat-inactivated goat serum in TBST), and then incubated with primary antibody digoxigenin-AP (Roche Applied Science) at 1:1,000 in blocking solution for 1 h at RT followed by overnight incubation at 4°C on a shaker. Secondary antibody application consisted of TBST and NTMT [0.1 M NaCl, 0.1 M Tris-Cl (pH 9.5), 50 mM MgCl2, and 0.1% Tween-20] washes followed by incubation in NBT and BCIP (Roche Applied Science) for 30 min to several hours in the dark at RT. The reaction was stopped with 10 mM Tris-Cl pH 7.5 and 1 mM EDTA; sections were mounted on gelatin-coated slides, dehydrated, and coverslipped. The sections were examined with an Olympus BX60 microscope fitted with a Photometrics CoolSNAP charge-coupled device video camera. Four brains from each sex were examined.
Protein Extraction and Immunoblotting
For protein extraction, brain tissues were thawed and homogenized in RIPA buffer. After centrifugation, the total lysate protein concentrations were determined with a bicinchoninic acid protein assay (Pierce Chemical Co.). Proteins were separated on 16% polyacrylamide-SDS gels and transferred to nitrocellulose membranes. After transfer, membranes were blocked and incubated with a polyclonal calbindin antibody (1:20,000; Chemicon) overnight at 4°C. After rinsing, blots were incubated for 1 h with HRP-conjugated anti-rabbit IgG secondary antibody (1:5,000; Vector Laboratories) followed by detection on X-ray film (X-OMAT) with SuperSignal West Pico Chemiluminescent Substrate (Pierce Chemical Co.). The same blots were reprobed with the monoclonal antibody against β-actin at 1:50,000 (Sigma-Aldrich Corp.). The intensities of Calb and β-actin on individual films were measured and analyzed by densitometry with ImageQuant (GE Healthcare Life Sciences). Levels of Calb were normalized to those of β-actin in each sample and the protein amount was expressed as the ratio of calbindin to β-actin. Five brains from each sex were used for these blots.
Quantitative Real-Time PCR
Total RNA was isolated from brain tissues using an RNeasy® Lipid Tissue Mini kit (Qiagen) according to the manufacturer's protocol. The quantity and quality of the RNA was determined using a Bio-Rad SmartSpec Plus spectrophotometer. The cDNA templates were prepared using an AffinityScript QPCR cDNA Synthesis Kit (Agilent Technologies, La Jolla, Calif., USA). The reverse transcription reaction consisted of 0.8 μg total RNA, 0.3 μg random primers, 2× First Strand Master Mix, and AffinityScript RT/RNAse block enzyme mixture in a total volume of 20 μl. Primer annealing occurred at 25°C for 5 min followed by cDNA synthesis for 45 min at 42°C and heat inactivation for 5 min at 95°C. cDNA templates were stored at −20°C prior to use. For calbindin gene expression, real-time PCR was performed using an ABI Prism 275® 7300 Real-Time PCR System with Sequence Detection Software version 1.2.3 (Applied Biosystems, Foster City, Calif., USA). In a 25-μl PCR reaction volume, 5 ng cDNA was mixed with iTaq™ SYBR® Green Supermix with ROX (Bio-Rad) or RT2 Real-Time™ SYBR Green/ROX PCR Master Mix (SABiosciences) and 500 nM primers. Separate β-actin endogenous control reactions were used to normalize the RNA input. Oligonucleotide primers were designed using a consensus sequence from the NCBI genomic alignment database and Primer Express version 2.0 and, they were synthesized by Invitrogen as detailed in table 2. The real-time PCR conditions were 95°C for 3 min (iTaq SYBR Green Supermix) or 10 min (RT2 Real-Time SYBR), followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. After the last PCR cycle, a dissociation melting curve stage was run according to software protocol. Target and endogenous control genes were measured in triplicate for each cDNA sample during each real-time run to avoid intersample variance. For each RNA sample a no-reverse transcriptase reaction was run in parallel to cDNA synthesis and it was measured by quantitative real-time PCR (qPCR) to control for contamination and genomic amplification. Each qPCR reaction was verified for a single PCR product of an expected size with the disassociation melting curve stage. Normalization and quantifications of the Calb1 andβ-actin mRNA were performed using the comparative cycle thresholds (CT) method as described in the ABI PRISM 7700 sequence detection system (user bulletin No. 2). Validation experiments were conducted to test for equally efficient target and endogenous control gene amplifications as described in the user bulletin. Both primers were between 90 and 110% efficient for all amplifications.
Table 2.
Forward and reverse nucleotide sequences of β-actin and Calbindin D28k used to generate primers for qPCR
For estrogen receptor alpha (Esr1), estrogen receptor beta (Esr2), parvalbumin (Pvalb), and neuronal calcium sensor-1 (Ncs-1) gene expression, qPCR was performed using an ABI StepOnePlus real-time PCR system. The following TaqMan® Gene Expression assays from ABI were used to detect the PCR products of interest: Esr1 (Mm00433149_m1*), Esr2 (Mm00599821_m1*), Pvalb (Mm00443100_m1*), and Ncs-1 (Mm00490552_m1*; frequenin homolog). In a 20-μl PCR reaction volume, 5 ng cDNA was mixed with 1 μl of 20× the TaqMan Gene Expression assay of interest and 10 μl of 2× TaqMan Fast Advanced Master Mix. The real-time PCR conditions were 95°C for 20 s followed by 40 cycles of 95°C for 1 s and 60°C for 20 s. Separate β-actin endogenous control reactions (ABI TaqMan Gene Expression assay Mouse ACTB 20×) were run to normalize the RNA input for each cDNA template. Target and endogenous control genes were measured in triplicate for each cDNA sample during each real-time run to avoid intersample variance and a no-reverse transcriptase reaction was run in parallel to cDNA synthesis and was measured by qPCR to control for contamination. Normalization and quantification of the genes of interest and β-actin mRNA were analyzed with StepOne™ software using the comparative cycle thresholds (CT) method. We assayed between 5 and 10 animals of each sex and genotype for these experiments.
Statistical Analyses
Comparisons of Calb protein and mRNA, Pvalb, and Ncs-1 gene expression in brains of C57BL/6J males versus females were conducted with Student's t tests. We used 2-way ANOVA followed by Fisher's LSD tests to compare specific differences between Esr1 and Esr2 in males versus females. We employed 2-way ANOVA and Fisher's LSD tests to compare Calb1 with sex and genotype as the 2 factors in the KO and transgenic mouse experiments. In the ERαKO study the 2 factors were gonadal sex and the presence or absence of ERα. In the FCG experiments gonadal sex and the sex chromosome complement were the 2 factors.
Results
Calbindin mRNA Is More Robust in Females than in Males
Qualitative sex differences in expression were observed in the amount, but not in the location, of Calb1 mRNA throughout the brain. Distinct populations of neurons with robust Calb1 mRNA were noted in regions of the FC, thalamus, hypothalamus, hippocampus, amygdala, and CB. Hippocampal Calb1 mRNA was predominantly expressed in the dentate gyrus and moderate levels were expressed in CA1 neurons. Expression in the CB was particularly robust when compared to other brain regions and, as reported in rats, was specifically confined to the Purkinje neurons [26,27,28]. These findings confirm earlier studies mapping the distribution of Calb1 expression in the rat brain [26,29,30,31,32], but the current study is the first to characterize Calb1 mRNA expression in both sexes of juvenile mouse brain.
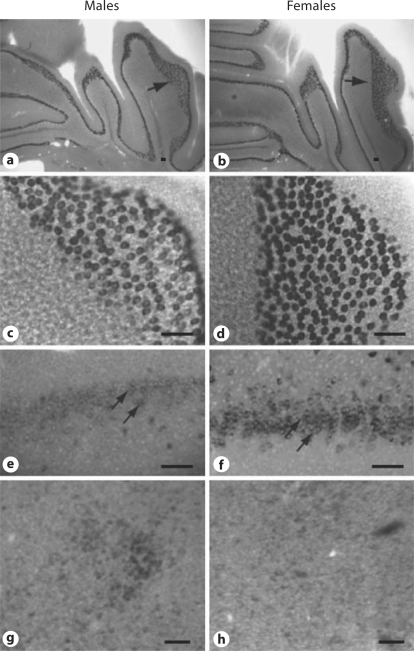
The neuroanatomical localization pattern of Calb1 mRNA expression did not differ between the sexes. However, the amount of mRNA expression, as indicated by the intensity of staining, differed in a regionally specific manner between males and females, particularly in the CB and FC. Examples of Calb1 in these areas in females versus males are shown in figure 1. Although the hybridization patterns are similar in both sexes, the Calb1 signal was stronger in females when compared to males in both the FC and the CB. In contrast, hypothalamic areas such as the SDN of the medial preoptic area (MPOA) exhibit a more robust mRNA signal in male brains when compared to female brains. This corroborates previous findings using immunoreactivity measurements in mice [12]. Other areas of the brain did not reveal marked differences in the intensity of Calb1 mRNA expression in male versus female brains.
Fig. 1.
ISH of calbindin mRNA shows sex differences in the CB (a–d), the FC (e, f), and the SDN of the hypothalamus (g, h). Bright-field photomicrographs of coronal sections show Calb1 in juvenile mouse CB in low (a, b) and high (c, d) magnifications. Photographs of males (a, c, e, g) and females (b, d, f, h). Single arrows in a and b show regions pictured in c and d. Double arrows indicate neurons in the dorsal peduncular area of the FC (bregma 1.42 mm). Scale bars = 100 μm.
Calbindin mRNA Quantified with qPCR Confirms Sex Differences
In the FC, hippocampus, MPOA/hypothalamus, amygdala, and CB (table 3) qPCR was used to quantify differences in Calb1 mRNA levels between males and females. Calb1 mRNA levels in the FC and CB were significantly higher in females when compared to males (p < 0.05). Sex differences were not evident in the MPOA/hypothalamus, the hippocampus, or the amygdala. It is likely that Calb1 mRNA levels in the MPOA/hypothalamus were not dimorphic because the tissue used extended beyond the small confines of the SDN which likely diluted the sex differences.
Table 3.
Sex comparison of Calb1 levels in brain regions
| Area of the brain | Sex | Mean relative mRNA expression ± SEM |
|---|---|---|
| CBa | female | 2.17 ± 0.28 |
| male | 1.23 ± 0.21 | |
| FCa | female | 1.55 ± 0.23 |
| male | 0.84 ± 0.09 | |
| Hippocampus | female | 1.02 ± 0.24 |
| male | 0.61 ± 0.12 | |
| Amygdala | female | 1.40 ± 0.10 |
| male | 1.28 ± 0.19 | |
| Hypothalamus | female | 1.31 ± 0.23 |
| male | 1.29 ± 0.09 | |
Mean (±SEM) relative Calb1 mRNA levels across brain regions and sexes. n = 5–10 mice per region, with equal numbers of males and females.
Significantly greater levels of Calb1 mRNA in females vs. males in these areas (p < 0.05).
To determine whether the sex difference in calbindin expression was a feature shared by other EF-hand calcium-binding proteins in the brain, we examined the mRNA expression of parvalbumin and neural calcium sensor-1 in the CB and FC using qPCR. Both of these proteins are expressed in the FC and in Purkinje cells of the CB [10,33]. The relative mRNA expression of these 2 messages was not significantly different between males and females in either the CB or FC (data not shown). Thus, sex differences in expression are not necessarily a feature of all EF-hand calcium-binding proteins.
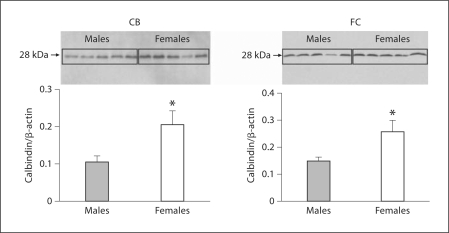
Females Have More Calbindin Protein than Males Do in the CB and Cortex
Calbindin protein concentrations in the FC and CB were quantified using Western blot followed by densitometry (fig. 2). Significantly higher levels of calbindin protein were observed in females when compared to males (p < 0.05).
Fig. 2.
Protein quantification of calbindin reveals sex differences in the cortex and CB. Calbindin protein quantification in CB and FC by Western blots. Each lane represents an individual animal. The ratio of calbindin to β-actin for each sample was measured (n = 5; animals were analyzed for each sex and region). Histograms represent the mean + SEM of the calbindin/β-actin ratio designating the protein level. * Females expressed significantly more calbindin protein (p < 0.05) than did males in both the CB and the FC.
Esr1 and Esr2 Are Expressed at Low Levels in the CB and FC
Because the Calb1 gene has an estrogen-responsive element (ERE) [34] and it is well accepted that estradiol orchestrates many sex neural differences, we examined the expression profile of both Esr1 and Esr2 in the CB and FC. As shown in table 4, both receptors are expressed in the CB and FC of juvenile mice. However, there was no significant difference in expression between males and females in either region. Interestingly, Esr1 showed 7- to 8-fold more expression than Esr2 did in both the CB (F1, 25 = 12.13, p < 0.002) and the FC (F 1, 25 = 31.88, p < 0.001). This is in agreement with previously reported immunoreactivity data for both receptors collected in mouse brain [35,36]. Because the abundance of these messages was so low, we also assayed testes and ovaries, organs known to have higher concentrations of both receptors, as additional controls (table 4).
Table 4.
Relative mRNA expression of estrogen receptors Esr1 and Esr2 as a function of sex in the CB and FC
| CB |
FC |
Controls |
||||
|---|---|---|---|---|---|---|
| females | males | females | males | testes | ovaries | |
| Esrl | 0.74 ± 0.19a | 1.59 ± 0.38a | 3.68 ± 1.00a | 4.33 ± 0.78a | 22.13 | 127.85 |
| Esr2 | 0.15 ± 0.04 | 0.15 ± 0.03 | 0.49 ± 0.13 | 0.47 ± 0.06 | 1.00 | 457.00 |
Mean (±SEM) of the relative gene expression of Esr1 and Esr2 mRNA in the CB and FC. n = 5–10 mice per brain region. Estrogen receptor expression was also examined in the testes and ovaries to serve as positive controls. All expression values were normalized to Esr2 in testes.
Esr1 expression was significantly greater than Esr2 expression in both the CB (p < 0.002) and the FC (p < 0.0001). No significant differences in Esr1 or Esr2 expression were noted between the sexes.
Calbindin mRNA in the FC Is Regulated by ERα and the Sex Chromosome Complement
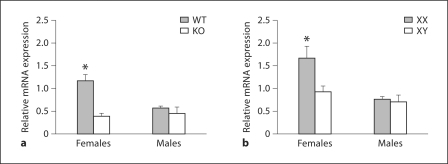
In the FC, ERαKO mice had reduced expression of Calb1 mRNA in females when compared with WT mice (fig. 3); no differences between KO and WT males were observed. In this region significant effects of gonadal sex (F1, 21 = 5.85, p < 0.02), genotype (F1, 21 = 18.1, p < 0.0005), and an interaction between these factors (F1, 21 = 9.2, p < 0.007) were observed. Planned comparisons revealed that WT females had higher levels of Calb1 mRNA expression than did animals in any of the other experimental groups (p < 0.05).
Fig. 3.
In the FC, ERα and the sex chromosome complement influence Calb1 mRNA in females. Mean + SEM Calb1 in the FC of WT and ERαKO (KO) mice (a) and in FCG mice (b). n = 5–6 mice per group. * Significantly different from all other groups (p < 0.05).
A similar pattern was noted in the FC from the FCG mice. A significant main effect of gonadal sex on Calb1 expression (F1, 22 = 10.8, p < 0.004) was found. No main effect of the sex chromosome complement was detected, although a trend towards an interaction between gonadal sex and genotype was observed (F1, 22 = 3.8, p = 0.07). Females had higher levels of Calb1 mRNA expression than did males (p < 0.05). In addition, XX females had higher levels of Calb1 mRNA in the FC than did brain tissues from any of the other experimental groups. These results implicate both ERα and the chromosomal complement as factors associated with Calb1 regulation in the FC of females.
In the CB ERα Affects the Amount of Calb1 mRNA Expressed
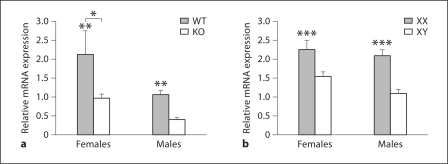
In the CB, expression of Calb1 mRNA in ERαKO and WT mice (fig. 4) was affected by both gonadal sex (F1, 21 = 7.5, p < 0.015) and genotype (F1, 22 = 9.5, p < 0.007), but there was no interaction effect (F = 0.65) between the variables. The effect of gonadal sex was apparently due to overall higher Calb1 mRNA expression in female as compared to male CB. However, both female and male WT mice demonstrated more than 2-fold higher Calb1 mRNA levels in the CB when compared to their ERαKO littermates. Because the sex difference was maintained in ERαKO males and females we conclude that ERα is not critical for the observed sexual dimorphism in the CB.
Fig. 4.
In the CB, ERα modulates Calb1 mRNA in both sexes, but sex chromosome genes modulate the sexual dimorphism. Mean + SEM Calb1 in the CB of WT and ERαKO (KO) mice (a) and FCG mice (b). n = 5–6 mice per group. * Females had significantly greater concentrations of Calb1 mRNA than did males (p < 0.015). ** WT littermates had significantly more Calb1 mRNA than did ERαKO mice (p < 0.007). *** XX mice had significantly more Calb1 than did XY mice (p < 0.0001).
The Sex Chromosome Complement Impacts Sexual Dimorphic Calb1 in the CB
The major determinant of the sexual dichotomous expression of Calb1 in CB was the sex chromosome complement. A significant main effect of the sex chromosome genotype was noted (F1, 22 = 21.7, p < 0.0001) (fig. 4). Specifically, XX females and males showed significantly higher levels of Calb1 than did XY individuals of either sex. This effect was so pronounced that gonadal sex was not a significant factor (F = 2.73), and no interaction between gonadal and chromosomal sex was found (F = 0.65). This result indicates that gonadal sex was not the critical determining factor, but the sex chromosome complement fully explains the observed Calb1 mRNA sex difference in the CB.
Discussion
Robust Sexual Dimorphism Outside the Hypothalamus
Here we show that Calb1 mRNA and protein are sexually dimorphic in 2 nonhypothalamic regions, i.e. the FC and CB. This is the first report that this protein is expressed in a sexually dimorphic manner outside of the hypothalamus, and the direction of this sex difference (female > male) is actually opposite to the higher expression of Calb typically observed in the SDN of male, rather than female, rodents [11,12,13]. There are 2 reported sex differences in genes and/or proteins in rat CB. At PN10 female CB had more mRNA for nerve growth factor than male CB did [37]. In adult rats, Foxp2 immunoreactivity was quantified in Purkinje cells, and a small but significant difference in optical density and cells numbers (males > females) was reported [38]. However, hormone levels were not controlled in this study; thus, it is unclear whether the sex difference was simply a reflection of differences in circulating hormone levels.
Our study specifically identified a novel organizational factor: sex chromosome genes. This finding could be functionally important since several cognitive behaviors are sexually dimorphic in humans and rodents, and both the FC and the CB mediate many complex processes [2,39]. A prime example is the classically conditioned eye blink response which utilizes cerebellar circuitry [2]. In rats, females are faster to learn this response than males, and the Purkinje cells in the CB are essential for the conditioning of this learning [40]. Interestingly, gonadectomized adult mice demonstrate sex differences when performing some motor tasks; females outperformed males on the rotarod and descended a pole more rapidly than males. Given that Calb is expressed only in Purkinje cells [26,27,28] and is associated with sexual dimorphism favoring females, it may play a key role in efficiency in acquiring the eye blink conditioning response and motor skills, and it likely impacts other cognitive and emotional tasks.
Sex Chromosomes Dictate Sex Differences in the CB and FC
The sex chromosome complement significantly impacts Calb1 gene expression, and there are important differences in the way this factor regulates Calb1 in the 2 brain regions under investigation. In the CB, XX females and XX males demonstrated significantly higher levels of Calb1 mRNA compared to mice with an XY genotype; moreover, levels of mRNA did not significantly differ between the XX males and XX females. In contrast, Calb1 mRNA expression in the FC was driven by the sex-chromosomal complement in females only, with XXF having more Calb1 than XYF. In males, both XX and XY mice had low Calb1 levels similar to those of XY females. Thus, the ‘advantage’ that the XX sex-chromosomal complement gave to females was also extended to males in the CB but not in the FC.
Several other neural sex differences have been attributed to the sex chromosome complement. For example, sex differences exist in prodynorphin mRNA in the mouse striatum (XX > XY) [41]. The reverse was noted in embryonic midbrain tyrosine hydroxylase-containing neurons, which are more plentiful in XY than in XX cell cultures [42]. Finally, the same XY > XX difference is apparent in the density of vasopressin in the lateral septum [8,43]. On the other hand, the only cortical sex difference previously examined in FCG is cortical thickness, which is larger in males than in females. Using the FCG model, no-sex chromosome effects were found in this measure [44].
Interactions between ERα and Sex Chromosomes
Our results indicate that both ERα and sex chromosome genes regulate Calb1 in the FC. In this area WT females in the ERαKO mice and XXF in the FCG have more Calb1 than do females with other genotypes. In the CB, because the absence of functional ERα does not eliminate the sex difference, we believe sex chromosomes and ERα play different roles. ERαincreases Calb1 production likely via ERE and this effect is noted in both sexes. In addition, the data from the FCG mice suggest that the normal sex difference in CB is caused by the sex chromosome complement.
Vasopressin is produced in neurons in the bed nucleus of the stria terminalis and these cells project primarily to the lateral septum. Fiber density in the septum is sexually dimorphic with males showing more intense immunoreactivity compared to females [45,46]. Studies of ERαKO mice have demonstrated that when ERα is functional WT males have more vasopressin in the septum than do KO males [47]. In the FCG, XY mice of both sexes have more vasopressin fibers than their XX counterparts with the same gonadal sex [8,43]. Thus, as is the case for Calb1, both the sex chromosome complement and ERα likely underlie this sex difference. Interestingly, the sex differences are reversed in these two systems (Calb females > males and vasopressin females < males). We speculate that ERα interacts with different sex chromosomes in these two cases.
While FC and CB express ERα in juvenile brain, the timing and source of the ligand are unknown at this time. There are regional and sex differences in estradiol in the developing rat brain [48] such that plasma concentrations of steroids may not entirely reveal all actions of steroids on neural architecture. In fact, Purkinje cells are a major site of synthesis for the steroidogenic enzyme P450scc, which generates de novo estradiol (for review see Dean and McCarthy [6] and Tsutsui [49]). The enzyme is developmentally regulated in both sexes, but it shows increased gene expression in the CB of males when compared to females during the early neonatal period [50]. An analysis of neurosteroid concentrations in the CB of C57BL/6 pups showed that 17β-estradiol is higher in females than in males on PN5; the reverse was noted for testosterone and the inactive 17α-estradiol [51]. When local infusions are given to mice at PN5 and brains collected at PN15, ER antagonists reduce Purkinje cell numbers in females. A thorough determination of steroid production in developing mouse cortex and CB, and its association with the Calb1 gene, is essential to unravel these complex effects.
Which Sex Chromosome Genes Regulate Calbindin in the CB?
When a sex chromosome effect is detected in the FCG, several underlying causes are possible. The sex difference may be due to the presence or absence of the Y genes, higher expression of X chromosome genes that escape inactivation in XX mice, or a paternally imprinted gene on the X which is only present in XX mice [7]. Our next challenge is to identify which sex chromosome is responsible for this effect in the CB. If the candidate chromosome is the X, there are several X genes known to escape inactivation in the brain [52]. These candidates, i.e. Kdm6a, Kdm5c, Ddx3x, Usp9x, and Eif2s3x, show robust in situ expression in the CB [53] and studies with FCG mice show that Kdm6a, Kdm5c, Usp9x, and Eif2s3x are expressed at higher levels in brains from XX versus XY individuals of either sex [52]. Both Kdm6a and Kdm5c are histone demethylase enzymes [54]. Interestingly, in PN6 rat cultures from cerebellar cortex, the targeted knockdown of Kdm5c produces marked reductions selectively in dendritic lengths [55]. In progenitor neurons treated in culture, Calb1 is subject to epigenetic regulation by the REST complex on an H3K9Me mark [56]. Therefore, it is possible that either Kdm6a and/or Kdm5c modify Calb1 via similar mechanisms.
Implications for Human Neurobehavioral Disease
Striking sex differences in the incidence, age of onset, and/or severity of a number of neurobehavioral diseases have brought the study of sex differences into the fore-front [1,6]. For example, autism, Parkinson's disease and attention deficit hyperactivity disorder with impulsivity are more prevalent in males, and schizophrenia is typically more severe in males [57,58,59,60,61]. In contrast, Alzheimer's disease, depression and anxiety are more likely to affect women [62,63]. Among the behavioral difficulties patients experience are issues involving executive function, emotionality, reasoning, and motor skills, all of which are mediated by the FC and/or CB functions [64,65]. A reduction in Calb1 gene and/or protein expression has been detected in brains of patients who suffered from an assortment of neurodegenerative diseases [66,67,68], and several neurobehavioral disorders are correlated with decreased size and/or numbers of Purkinje cells [69,70]. Sex differences in these brain regions in normal animals, such as those reported here, could provide clues underlying the etiologies of these diseases.
Acknowledgements
We thank Aileen Wills, Savera Shetty, and Jennifer Lampen for their expert technical assistance. We are grateful to Dr. Pierre Chambon for providing us with the ERαKO heterozygous mice to start our colony. This work was supported by NIH R01NS055218.
References
- 1.Jazin E, Cahill L. Sex differences in molecular neuroscience: From fruit flies to humans. Nat Rev Neurosci. 2010;11:9–17. doi: 10.1038/nrn2754. [DOI] [PubMed] [Google Scholar]
- 2.Dalla C, Shors TJ. Sex differences in learning processes of classical and operant conditioning. Physiol Behav. 2009;97:229–238. doi: 10.1016/j.physbeh.2009.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen-Bendahan CC, van de Beek C, Berenbaum SA. Prenatal sex hormone effects on child and adult sex-typed behavior: methods and findings. Neurosci Biobehav Rev. 2005;29:353–384. doi: 10.1016/j.neubiorev.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Cooke B, Hegstrom CD, Villeneuve LS, Breedlove SM. Sexual differentiation of the vertebrate brain: principles and mechanisms. Front Neuroendocrinol. 1998;19:323–362. doi: 10.1006/frne.1998.0171. [DOI] [PubMed] [Google Scholar]
- 5.McCarthy MM, Wright CL, Schwarz JM. New tricks by an old dogma: mechanisms of the organizational/activational hypothesis of steroid-mediated sexual differentiation of brain and behavior. Horm Behav. 2009;55:655–665. doi: 10.1016/j.yhbeh.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean SL, McCarthy MM. Steroids, sex and the cerebellar cortex: implications for human disease. Cerebellum. 2008;7:38–47. doi: 10.1007/s12311-008-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnold AP. Mouse models for evaluating sex chromosome effects that cause sex differences in non-gonadal tissues. J Neuroendocrinol. 2009;21:377–386. doi: 10.1111/j.1365-2826.2009.01831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Vries GJ, Rissman EF, Simerly RB, Yang LY, Scordalakes EM, Auger CJ, Swain A, Lovell-Badge R, Burgoyne PS, Arnold AP. A model system for study of sex chromosome effects on sexually dimorphic neural and behavioral traits. J Neurosci. 2002;22:9005–9014. doi: 10.1523/JNEUROSCI.22-20-09005.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kojetin DJ, Venters RA, Kordys DR, Thompson RJ, Kumar R, Cavanagh J. Structure, binding interface and hydrophobic transitions of Ca2+-loaded calbindin-D(28K) Nat Struct Mol Biol. 2006;13:641–647. doi: 10.1038/nsmb1112. [DOI] [PubMed] [Google Scholar]
- 10.Bastianelli E. Distribution of calcium-binding proteins in the cerebellum. Cerebellum. 2003;2:242–262. doi: 10.1080/14734220310022289. [DOI] [PubMed] [Google Scholar]
- 11.Budefeld T, Grgurevic N, Tobet SA, Majdic G. Sex differences in brain developing in the presence or absence of gonads. Dev Neurobiol. 2008;68:981–995. doi: 10.1002/dneu.20638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edelmann M, Wolfe C, Scordalakes EM, Rissman EF, Tobet S. Neuronal nitric oxide synthase and calbindin delineate sex differences in the developing hypothalamus and preoptic area. Dev Neurobiol. 2007;67:1371–1381. doi: 10.1002/dneu.20507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sickel MJ, McCarthy MM. Calbindin-D28K immunoreactivity is a marker for a subdivision of the sexually dimorphic nucleus of the preoptic area of the rat: developmental profile and gonadal steroid modulation. J Neuroendocrinol. 2000;12:397–402. doi: 10.1046/j.1365-2826.2000.00474.x. [DOI] [PubMed] [Google Scholar]
- 14.Welborn BL, Papademetris X, Reis DL, Rajeevan N, Bloise SM, Gray JR. Variation in orbitofrontal cortex volume: relation to sex, emotion regulation and affect. Soc Cogn Affect Neurosci. 2009;4:328–339. doi: 10.1093/scan/nsp028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tiemeier H, Lenroot RK, Greenstein DK, Tran L, Pierson R, Giedd JN. Cerebellum development during childhood and adolescence: a longitudinal morphometric MRI study. Neuroimage. 2010;49:63–70. doi: 10.1016/j.neuroimage.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gauthier CT, Duyme M, Zanca M, Capron C. Sex and performance level effects on brain activation during a verbal fluency task: a functional magnetic resonance imaging study. Cortex. 2009;45:164–176. doi: 10.1016/j.cortex.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Servais L, Bearzatto B, Schwaller B, Dumont M, De Saedeleer C, Dan B, Barski JJ, Schiffmann SN, Cheron G. Mono- and dual-frequency fast cerebellar oscillation in mice lacking parvalbumin and/or calbindin D-28k. Eur J Neurosci. 2005;22:861–870. doi: 10.1111/j.1460-9568.2005.04275.x. [DOI] [PubMed] [Google Scholar]
- 18.Farre-Castany MA, Schwaller B, Gregory P, Barski J, Mariethoz C, Eriksson JL, Tetko IV, Wolfer D, Celio MR, Schmutz I, Albrecht U, Villa AE. Differences in locomotor behavior revealed in mice deficient for the calcium-binding proteins parvalbumin, calbindin D-28k or both. Behav Brain Res. 2007;178:250–261. doi: 10.1016/j.bbr.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Kriegsfeld LJ, Mei DF, Yan L, Witkovsky P, Lesauter J, Hamada T, Silver R. Targeted mutation of the calbindin D28K gene disrupts circadian rhythmicity and entrainment. Eur J Neurosci. 2008;27:2907–2921. doi: 10.1111/j.1460-9568.2008.06239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Airaksinen MS, Eilers J, Garaschuk O, Thoenen H, Konnerth A, Meyer M. Ataxia and altered dendritic calcium signaling in mice carrying a targeted null mutation of the calbindin D28k gene. Proc Natl Acad Sci USA. 1997;94:1488–1493. doi: 10.1073/pnas.94.4.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barski JJ, Hartmann J, Rose CR, Hoebeek F, Morl K, Noll-Hussong M, De Zeeuw CI, Konnerth A, Meyer M. Calbindin in cerebellar Purkinje cells is a critical determinant of the precision of motor coordination. J Neurosci. 2003;23:3469–3477. doi: 10.1523/JNEUROSCI.23-08-03469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vecellio M, Schwaller B, Meyer M, Hunziker W, Celio MR. Alterations in Purkinje cell spines of calbindin D-28 k and parvalbumin knock-out mice. Eur J Neurosci. 2000;12:945–954. doi: 10.1046/j.1460-9568.2000.00986.x. [DOI] [PubMed] [Google Scholar]
- 23.Jouvenceau A, Potier B, Poindessous-Jazat F, Dutar P, Slama A, Epelbaum J, Billard JM. Decrease in calbindin content significantly alters LTP but not NMDA receptor and calcium channel properties. Neuropharmacology. 2002;42:444–458. doi: 10.1016/s0028-3908(01)00202-7. [DOI] [PubMed] [Google Scholar]
- 24.Dupont S, Krust A, Gansmuller A, Dierich A, Chambon P, Mark M. Effect of single and compound knockouts of estrogen receptors alpha (ERalpha) and beta (ERbeta) on mouse reproductive phenotypes. Development. 2000;127:4277–4291. doi: 10.1242/dev.127.19.4277. [DOI] [PubMed] [Google Scholar]
- 25.Stornetta RL, Sevigny CP, Schreihofer AM, Rosin DL, Guyenet PG. Vesicular glutamate transporter DPNI/VGLUT2 is expressed by both C1 adrenergic and nonaminergic presympathetic vasomotor neurons of the rat medulla. J Comp Neurol. 2002;444:207–220. doi: 10.1002/cne.10142. [DOI] [PubMed] [Google Scholar]
- 26.Iacopino AM, Rhoten WB, Christakos S. Calcium binding protein (calbindin-D28k) gene expression in the developing and aging mouse cerebellum. Brain Res Mol Brain Res. 1990;8:283–290. doi: 10.1016/0169-328x(90)90041-b. [DOI] [PubMed] [Google Scholar]
- 27.Celio MR. Calbindin D-28k and parvalbumin in the rat nervous system. Neuroscience. 1990;35:375–475. doi: 10.1016/0306-4522(90)90091-h. [DOI] [PubMed] [Google Scholar]
- 28.Amenta F, Cavalotta D, Del Valle ME, Mancini M, Sabbatini M, Torres JM, Vega JA. Calbindin D-28k immunoreactivity in the rat cerebellar cortex: age-related changes. Neurosci Lett. 1994;178:131–134. doi: 10.1016/0304-3940(94)90307-7. [DOI] [PubMed] [Google Scholar]
- 29.Abe H, Watanabe M, Yamakuni T, Kuwano R, Takahashi Y, Kondo H. Localization of gene expression of calbindin in the brain of adult rats. Neurosci Lett. 1992;138:211–215. doi: 10.1016/0304-3940(92)90917-v. [DOI] [PubMed] [Google Scholar]
- 30.Stuart E, Lephart ED. Dimorphic expression of medial basal hypothalamic-preoptic area calbindin-D(28K) mRNA during perinatal development and adult distribution of calbindin-D(28K) mRNA in Sprague-Dawley rats. Brain Res Mol Brain Res. 1999;73:60–67. doi: 10.1016/s0169-328x(99)00236-3. [DOI] [PubMed] [Google Scholar]
- 31.Sequier JM, Hunziker W, Andressen C, Celio MR. Calbindin D-28k protein and mRNA localization in the rat brain. Eur J Neurosci. 1990;2:1118–1126. doi: 10.1111/j.1460-9568.1990.tb00023.x. [DOI] [PubMed] [Google Scholar]
- 32.Wanner I, Baader SL, Brich M, Oberdick J, Schilling K. Subcellular localization of specific mRNAs and their protein products in Purkinje cells by combined fluorescence in situ hybridization and immunocytochemistry. Histochem Cell Biol. 1997;108:345–357. doi: 10.1007/s004180050175. [DOI] [PubMed] [Google Scholar]
- 33.Jinno S, Jeromin A, Kosaka T. Expression and possible role of neuronal calcium sensor-1 in the cerebellum. Cerebellum. 2004;3:83–88. doi: 10.1080/14734220310025187. [DOI] [PubMed] [Google Scholar]
- 34.Gill RK, Christakos S. Regulation by estrogen through the 5′-flanking region of the mouse calbindin-D28k gene. Mol Endocrinol. 1995;9:319–326. doi: 10.1210/mend.9.3.7776978. [DOI] [PubMed] [Google Scholar]
- 35.Mitra SW, Hoskin E, Yudkovitz J, Pear L, Wilkinson HA, Hayashi S, Pfaff DW, Ogawa S, Rohrer SP, Schaeffer JM, McEwen BS, Alves SE. Immunolocalization of estrogen receptor beta in the mouse brain: comparison with estrogen receptor alpha. Endocrinology. 2003;144:2055–2067. doi: 10.1210/en.2002-221069. [DOI] [PubMed] [Google Scholar]
- 36.Merchenthaler I, Lane MV, Numan S, Dellovade TL. Distribution of estrogen receptor alpha and beta in the mouse central nervous system: in vivo autoradiographic and immunocytochemical analyses. J Comp Neurol. 2004;473:270–291. doi: 10.1002/cne.20128. [DOI] [PubMed] [Google Scholar]
- 37.Kornack DR, Lu B, Black IB. Sexually dimorphic expression of the NGF receptor gene in the developing rat brain. Brain Res. 1991;542:171–174. doi: 10.1016/0006-8993(91)91015-s. [DOI] [PubMed] [Google Scholar]
- 38.Hamson DK, Csupity AS, Gaspar JM, Watson NV. Analysis of Foxp2 expression in the cerebellum reveals a possible sex difference. Neuroreport. 2009;20:611–616. doi: 10.1097/WNR.0b013e32832a0a44. [DOI] [PubMed] [Google Scholar]
- 39.Andreano JM, Cahill L. Sex influences on the neurobiology of learning and memory. Learn Mem. 2009;16:248–266. doi: 10.1101/lm.918309. [DOI] [PubMed] [Google Scholar]
- 40.Nakanishi S. Genetic manipulation study of information processing in the cerebellum. Neuroscience. 2009;162:723–731. doi: 10.1016/j.neuroscience.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 41.Chen X, Grisham W, Arnold AP. X chromosome number causes sex differences in gene expression in adult mouse striatum. Eur J Neurosci. 2009;29:768–776. doi: 10.1111/j.1460-9568.2009.06610.x. [DOI] [PubMed] [Google Scholar]
- 42.Carruth LL, Reisert I, Arnold AP. Sex chromosome genes directly affect brain sexual differentiation. Nat Neurosci. 2002;5:933–934. doi: 10.1038/nn922. [DOI] [PubMed] [Google Scholar]
- 43.Gatewood JD, Wills A, Shetty S, Xu J, Arnold AP, Burgoyne PS, Rissman EF. Sex chromosome complement and gonadal sex influence aggressive and parental behaviors in mice. J Neurosci. 2006;26:2335–2342. doi: 10.1523/JNEUROSCI.3743-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Markham JA, Jurgens HA, Auger CJ, De Vries GJ, Arnold AP, Juraska JM. Sex differences in mouse cortical thickness are independent of the complement of sex chromosomes. Neuroscience. 2003;116:71–75. doi: 10.1016/s0306-4522(02)00554-7. [DOI] [PubMed] [Google Scholar]
- 45.De Vries GJ, Wang Z, Bullock NA, Numan S. Sex differences in the effects of testosterone and its metabolites on vasopressin messenger RNA levels in the bed nucleus of the stria terminalis of rats. J Neurosci. 1994;14:1789–1794. doi: 10.1523/JNEUROSCI.14-03-01789.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han TM, De Vries GJ. Organizational effects of testosterone, estradiol, and dihydrotestosterone on vasopressin mRNA expression in the bed nucleus of the stria terminalis. J Neurobiol. 2003;54:502–510. doi: 10.1002/neu.10157. [DOI] [PubMed] [Google Scholar]
- 47.Scordalakes EM, Rissman EF. Aggression and arginine vasopressin immunoreactivity regulation by androgen receptor and estrogen receptor alpha. Genes Brain Behav. 2004;3:20–26. doi: 10.1111/j.1601-183x.2004.00036.x. [DOI] [PubMed] [Google Scholar]
- 48.Amateau SK, Alt JJ, Stamps CL, McCarthy MM. Brain estradiol content in newborn rats: sex differences, regional heterogeneity, and possible de novo synthesis by the female telencephalon. Endocrinology. 2004;145:2906–2917. doi: 10.1210/en.2003-1363. [DOI] [PubMed] [Google Scholar]
- 49.Tsutsui K. Neurosteroids in the purkinje cell: biosynthesis, mode of action and functional significance. Mol Neurobiol. 2008;37:116–125. doi: 10.1007/s12035-008-8024-1. [DOI] [PubMed] [Google Scholar]
- 50.Lavaque E, Mayen A, Azcoitia I, Tena-Sempere M, Garcia-Segura LM. Sex differences, developmental changes, response to injury and cAMP regulation of the mRNA levels of steroidogenic acute regulatory protein, cytochrome p450scc, and aromatase in the olivocerebellar system. J Neurobiol. 2006;66:308–318. doi: 10.1002/neu.20221. [DOI] [PubMed] [Google Scholar]
- 51.Biamonte F, Assenza G, Marino R, D'Amelio M, Panteri R, Caruso D, Scurati S, Yague JG, Garcia-Segura LM, Cesa R, Strata P, Melcangi RC, Keller F. Interactions between neuroactive steroids and reelin haploinsufficiency in Purkinje cell survival. Neurobiol Dis. 2009;36:103–115. doi: 10.1016/j.nbd.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 52.Xu J, Disteche CM. Sex differences in brain expression of X- and Y-linked genes. Brain Res. 2006;1126:50–55. doi: 10.1016/j.brainres.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 53.Xu J, Watkins R, Arnold AP. Sexually dimorphic expression of the X-linked gene Eif2s3x mRNA but not protein in mouse brain. Gene Expr Patterns. 2006;6:146–155. doi: 10.1016/j.modgep.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 54.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 55.Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 56.Mehedint MG, Niculescu MD, Craciunescu CN, Zeisel SH. Choline deficiency alters global histone methylation and epigenetic marking at the Re1 site of the calbindin 1 gene. FASEB J. 2010;24:184–195. doi: 10.1096/fj.09-140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wooten GF, Currie LJ, Bovbjerg VE, Lee JK, Patrie J. Are men at greater risk for Parkinson's disease than women? J Neurol Neurosurg Psychiatry. 2004;75:637–639. doi: 10.1136/jnnp.2003.020982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McGrath J, Saha S, Chant D, Welham J. Schizophrenia: a concise overview of incidence, prevalence, and mortality. Epidemiol Rev. 2008;30:67–76. doi: 10.1093/epirev/mxn001. [DOI] [PubMed] [Google Scholar]
- 59.Honda H, Shimizu Y, Imai M, Nitto Y. Cumulative incidence of childhood autism: a total population study of better accuracy and precision. Dev Med Child Neurol. 2005;47:10–18. doi: 10.1017/s0012162205000034. [DOI] [PubMed] [Google Scholar]
- 60.Merikangas KR, He JP, Brody D, Fisher PW, Bourdon K, Koretz DS. Prevalence and treatment of mental disorders among US children in the 2001–2004 NHANES. Pediatrics. 2010;125:75–81. doi: 10.1542/peds.2008-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mazzocco MM. Advances in research on the fragile X syndrome. Ment Retard Dev Disabil Res Rev. 2000;6:96–106. doi: 10.1002/1098-2779(2000)6:2<96::AID-MRDD3>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 62.Candore G, Balistreri CR, Grimaldi MP, Vasto S, Listi F, Chiappelli M, Licastro F, Lio D, Caruso C. Age-related inflammatory diseases: role of genetics and gender in the pathophysiology of Alzheimer's disease. Ann NY Acad Sci. 2006;1089:472–486. doi: 10.1196/annals.1386.008. [DOI] [PubMed] [Google Scholar]
- 63.Zahn-Waxler C, Shirtcliff EA, Marceau K. Disorders of childhood and adolescence: gender and psychopathology. Annu Rev Clin Psychol. 2008;4:275–303. doi: 10.1146/annurev.clinpsy.3.022806.091358. [DOI] [PubMed] [Google Scholar]
- 64.Abrahams BS, Geschwind DH. Connecting genes to brain in the autism spectrum disorders. Arch Neurol. 2010;67:395–399. doi: 10.1001/archneurol.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ackermann H. Cerebellar contributions to speech production and speech perception: psycholinguistic and neurobiological perspectives. Trends Neurosci. 2008;31:265–272. doi: 10.1016/j.tins.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 66.Iacopino AM, Christakos S. Specific reduction of calcium-binding protein (28-kilodalton calbindin-D) gene expression in aging and neurodegenerative diseases. Proc Natl Acad Sci USA. 1990;87:4078–4082. doi: 10.1073/pnas.87.11.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sutherland MK, Wong L, Somerville MJ, Yoong LK, Bergeron C, Parmentier M, McLachlan DR. Reduction of calbindin-28k mRNA levels in Alzheimer as compared to Huntington hippocampus. Brain Res Mol Brain Res. 1993;18:32–42. doi: 10.1016/0169-328x(93)90171-k. [DOI] [PubMed] [Google Scholar]
- 68.Kiyama H, Seto-Ohshima A, Emson PC. Calbindin D28K as a marker for the degeneration of the striatonigral pathway in Huntington's disease. Brain Res. 1990;525:209–214. doi: 10.1016/0006-8993(90)90866-a. [DOI] [PubMed] [Google Scholar]
- 69.Bauman ML, Kemper TL. The neuropathology of the autism spectrum disorders: what have we learned? Novartis Found Symp. 2003;251:112–122. discussion 122–128, 281–297. [PubMed] [Google Scholar]
- 70.Fatemi SH, Halt AR, Realmuto G, Earle J, Kist DA, Thuras P, Merz A. Purkinje cell size is reduced in cerebellum of patients with autism. Cell Mol Neurobiol. 2002;22:171–175. doi: 10.1023/a:1019861721160. [DOI] [PubMed] [Google Scholar]