Abstract
Killer immunoglobulin-like receptors (KIRs) recognize class I major histocompatibility complex molecules and participate in the calibration of activation thresholds during human natural killer (NK) cell development. The stochastic expression pattern of the KIR repertoire follows the product rule, meaning that the probability of the coexpression of two or more different KIRs equals the product of the individual expression frequencies for those KIRs. The expression frequencies of individual KIRs are independent of major histocompatibility complex class I and are instead established and maintained by a dynamic, yet ill-defined, transcriptional program. Here, we review recent advances in our understanding of the architecture of the regulatory regions within KIR genes and discuss a potential role for non-coding RNA in KIR transcriptional regulation during NK cell development. Understanding the molecular mechanisms that underlie KIR expression may help guide us in the design of novel, rational strategies for the use of NK cells in transplantation and immunotherapy.
Key Words: Natural killer cell, Killer immunoglobulin-like receptor, Transcription, Antisense RNA, Promoter
Genetic and Epigenetic Regulation of Killer Immunoglobulin-Like Receptor Genes
Killer immunoglobulin-like receptor (KIR) messenger RNAs, originally designated as ‘natural killer (NK)-associated transcripts’, were discovered through subtractive hybridization in 1995 [1,2], and the sequences were subsequently mapped to human chromosome 19q13.4 [3]. Fourteen KIR genes and two pseudogenes have been identified, and it is now apparent that significant allelic and haplotypic variability has evolved within the KIR locus through a variety of genetic events including extensive intergenic sequence exchange, deletion/duplication and unequal crossing over [4,5]. KIR genes with a variegated expression pattern are >91% identical with respect to their core promoter regions, suggesting that a single mechanism controls the expression of the majority of KIR genes [6]. The remarkably high level of promoter sequence homology observed amongst individual genes within the KIR locus begs the question of how a complex, stochastic expression pattern is established. Although we are still far from answering this question, a few clues have been uncovered.
One of the first significant clues related to the mechanisms controlling KIR transcription was the discovery that non-expressed KIR alleles are methylated within a small cytosine-phospho-guanine island surrounding the transcriptional start site, while transcriptionally active KIR alleles lack these epigenetic marks [7,8,9]. An analysis of peripheral blood NK cells also revealed that variegated KIRs, such as KIR3DL1, have a predominantly monoallelic expression pattern [8]. DNA methylation appears to be the central epigenetic regulator of KIR transcription, as N-terminal histone modifications are similar between active and repressed KIR genes [9,10]. A major question yet to be answered is whether specific KIR genes are targeted for demethylation during NK cell development or whether demethylation occurs in a nonspecific manner across the KIR locus, and clonal expression patterns are subsequently established.
Another major insight into how KIR transcription is regulated came from the identification of two variants of the KIR2DL5 gene, now known as KIR2DL5A and KIR2DL5B, which were both identified in the genomic DNA of a single donor. Messenger RNA could only be detected for KIR2DL5A, and expression correlated with an intact acute myeloid leukemia gene 1 (AML1) binding site [11]. Intriguingly, the KIR2DL5B promoter was active in reporter assays carried out in the NK3.3 cell line despite its lack of a functional AML1 binding site, and DNA demethylation induced by pharmacological agents led to expression of the endogenous KIR2DL5B gene [12]. Together, these results suggest that AML1 plays a central role in establishing an open chromatin confirmation permissive to active KIR transcription during human NK cell development.
Mechanistic insights into KIR transcriptional regulation have also been inspired by the discovery of probabilistic transcriptional switches within the 5′ regulatory regions of murine Ly49 genes, which are functionally homologous to KIR but evolutionarily distinct. The 5′ regulatory regions of Ly49 genes include a distal promoter that is active in immature NK cells [13] and exhibits probabilistic, bidirectional transcriptional activity [14]. Forward transcription from the distal promoter correlates with stable Ly49 expression, while reverse transcription leads to the production of non-coding antisense transcripts and the absence of Ly49 expression [14]. Analyses of the intergenic regions of KIR genes have also revealed the presence of multiple promoters that coordinate probabilistic gene transcription, though the mechanism appears to be quite distinct.
The KIR Distal Promoter
A novel distal promoter element was identified upstream of all KIR genes analyzed, and it was characterized in detail for KIR3DL1 and KIR2DL4[15]. KIR3DL1 distal transcripts initiate at position −406 [[16]; unpubl. data], and promoter activity was detected in intergenic fragments extending up to position −1347 relative to the KIR3DL1 start codon. The distal promoter is not tissue specific, as it is active in both NK cells and non-NK cell lines. Adjacent L1 and Alu elements within the distal promoter harbor multiple putative transcription factor binding sites that are necessary for transcriptional activity [15].
Transposable elements have a long evolutionary history and have played a substantial role in shaping the landscape of the human genome. A previous analysis of the apolipoprotein(a), apo(a), gene provides evidence for the ability of transposable elements to function as regulatory elements to control the expression of nearby genes. Like KIR, the apo(a) gene is part of a cluster of highly homologous genes and pseudogenes. The apo(a) gene contains a 228-bp core promoter region and a 1.8-kb distal enhancer region approximately 20 kb upstream of the apo(a) transcription start site that is able to confer a >10-fold activation of the apo(a) gene in in vitro assays. The distal enhancer region resides within a LINE element and contains binding sites for Sp1, CRE-BP, Ets-1 and several other transcription factors that are required for optimal activity of the enhancer [17]. As is the case with KIR genes, the apo(a) conventional core promoter is tissue specific, while the distal enhancer region is not [15,17].
One of the predicted transcription factor binding sites within the distal KIR promoter L1M5 element is c-Myc. The KIR3DL1/S1, KIR2DL1/S1, KIR2DL1 and KIR2DS3 c-Myc sites are located approximately 1,150 bases upstream of their respective proximal translational start sites. Myc sites for KIR2DL2/S2, KIR2DL3 and KIR3DL2 also exist at position −1150. However, for this set of genes, a C→T polymorphism at the first position of the site abrogates c-Myc binding, rendering the site nonfunctional. Nevertheless, because of a 406-base deletion and a 317-base insertion within the 5′ intergenic region of the KIR2DL2/S2, KIR2DL3 and KIR3DL2 genes, a functional, consensus c-Myc site exists within an Alu element at position −1190. Chromatin immunoprecipitation assays carried out with primary human NK cells revealed that interleukin (IL)-15 stimulation induces c-Myc binding to the KIR distal promoter and directly drives distal transcription [18].
The acquisition of ‘IL-15 responsiveness’ is a key step during NK cell development, as IL-15 is necessary for both NK cell maturation and survival [19]. CD122, which is the β-chain for the IL-2/IL-15 receptor complex, is detectable on the surface of CD56bright NK cells and mature CD56dim cells, but not on cells transitioning through earlier stages of NK cell development [20]. Therefore, it is likely that IL-15 induces c-Myc expression and drives transcription from KIR distal promoters once cells reach the CD56bright stage. KIR distal promoters in developmental intermediates likely have some activity independent of IL-15, as KIR distal transcripts were detected in CD34–CD7+CD56– NK cell precursors [18]. Transcript levels from the KIR3DL1 distal promoter are significantly higher in CD56dim KIR3DL1+ cells when compared with CD56dim KIR3DL1– cells, suggesting that the ‘opening’ of the proximal promoter has a positive feedback effect on the activity of the distal promoter [18,21]. The mechanism by which distal promoters facilitate KIR transcription is still open to speculation.
One possibility is that distal KIR promoters participate in the establishment of active histone signatures across the KIR locus during NK cell development. A comparative analysis of chromatin modifications within the KIR locus demonstrated that while there is no discernable association between KIR expression and ‘active’ chromatin modifications, there are lineage-specific differences between KIR-expressing and KIR-non-expressing cell types. Active histone signatures, defined as high levels of H4K8 acetylation and low levels of H3K9 dimethylation, were restricted to NK and CD8+ T cells, which both express KIR. A pattern of histone modifications associated with inactive loci was observed within the KIR locus of CD34+ hematopoietic progenitor cells and B and CD4+ T cells, suggesting that a lineage-specific transition takes place and ‘primes’ the KIR locus for expression [10].
One model for how distal KIR promoters may participate in the lineage-specific transition is through the recruitment of transcriptional cofactors that possess histone acetyltransferase activity. Intriguingly, a putative p300/CBP site exists within the distal promoters of all KIR genes. p300/CBP is a large protein known to interact with a variety of DNA-binding transcription factors and has the remarkable ability to acetylate all four core histones in nucleosomes [22]. Therefore, distal promoters could act as nucleation points where histone acetylation is initiated and subsequently spread across the KIR locus. Certain epigenetic marks, such as histone acetylation, can spread over great distances. The presence of the β-globin locus control region on stably maintained episomes in cell lines results in histone hyperacetylation over the entire minichromosome [23]. Once a permissive chromatin landscape is established, IL-15-induced c-Myc binding and transcription from the distal promoter may act to allow transcription from proximal KIR promoters that then become demethylated. It is interesting to note that although consensus c-Myc binding sites contain an internal cytosine-phospho-guanine site (CACGTG), the c-Myc binding sites in the KIR distal promoters do not (CACATG), suggesting that distal transcription is not influenced by DNA methylation.
The KIR Proximal Promoter
Core KIR proximal promoters span approximately 270 bases and contain a multitude of overlapping transcription factor binding sites [24]. A comprehensive in vitro linker-scanning mutagenesis approach showed that a high degree of functional redundancy exists between binding sites, with multiple mutations needed to completely abrogate transcription [25]. Peripheral blood NK cells exhibit a predominantly monoallelic KIR expression pattern, and DNA methylation within proximal promoters strongly correlates with stable transcriptional silencing of individual alleles [7,8,9].
The phenomenon of monoallelic gene expression is common in the immune system and likely evolved to ensure that most individual cells express only one member of a family of receptors encoded by highly homologous genes. This results in each cell possessing only one out of many possible specificities. This ability is clearly important for NK cells, which utilize KIR to distinguish subtle differences in major histocompatibility complex expression on target cells. An important clue with respect to the mechanism by which stochastic and monoallelic expression patterns are established within the KIR locus was uncovered with the discovery of probabilistic, bidirectional activity within the proximal promoter.
Antisense transcripts were cloned from several KIR genes by a modified 3′ rapid amplification of cDNA ends. A canonical polyadenylation signal site 409 bases upstream of the start of translation is used for the majority of KIR3DL1, KIR2DL1, KIR2DS1, KIR2DL2, KIR2DS2, KIR3DS1 and KIR3DL2 antisense transcripts. This polyadenylation signal is absent from KIR2DL2, KIR2DS2and KIR3DL2. Instead, antisense transcripts from these genes use a polyadenylation signal site 295 bases upstream of the translational start site. Antisense transcription initiates between positions −149 to −175 relative to the start of translation, and the proximal promoter is active in both the forward and reverse orientations. Intriguingly, KIR antisense transcripts were only detected by RT-PCR in cells that lack surface KIR expression [21]. Probabilistic expression from bidirectional proximal promoters is influenced by polymorphisms, particularly those within putative binding sites for the Sp1 and YY1 transcription factors. The ratio of forward to reverse promoter activities for individual KIR alleles positively correlates with the frequency of gene expression, and antisense transcript levels negatively correlate with KIR transcription [26].
The observation that distal and antisense transcripts overlap across the proximal KIR promoters led to the hypothesis that a small RNA might be processed in this region and might directly participate in transcriptional silencing of individual KIR alleles [27]. Examples of transcriptional gene silencing by siRNAs and microRNAs have been reported previously [28,29]. Unexpectedly, when both antisense and sense transcripts spanning the KIR3DL1 or KIR2DL1 promoter were transfected together into HEK293 cells, a single 28-base RNA processed from the antisense transcript was identified from three independent small RNA libraries. The sequence of the KIR3DL1 28-base RNA corresponds to positions −173 to −201 within the proximal promoter relative to the start of translation. Notably, this 28-base product could not be detected if antisense transcripts were transfected alone, suggesting that the formation of double-stranded RNA (dsRNA) is necessary for processing the small RNA. The existence of dsRNA across KIR promoters in primary NK cells was confirmed by an S1 nuclease protection assay using RNA isolated from peripheral blood CD56+ cells. The 28-base RNA could also be readily cloned from primary peripheral blood CD56+ NK cells [16].
The only known species of single-stranded RNA with a 24- to 30-base length is PIWI-interacting RNA (piRNAs), and a defining characteristic of mammalian piRNAs is the presence of a 2′-O-methyl group at the 3′ terminal ribose in place of a hydroxyl group [30,31]. The 28-base RNA from the KIR antisense transcript is resistant to periodate oxidation and β-elimination, strongly suggesting that it contains a protective group at its 3′ terminus and belongs to the PIWI or a PIWI-like family of small RNAs. The overexpression of either full-length KIR3DL1 antisense transcripts or the 28-base RNA alone in developing human NK cells leads to a significant reduction in receptor expression, which correlates with promoter methylation [16].
piRNAs were originally identified as critical mediators of transposon control in Drosophila germ cells [32], and piRNAs were once assumed to be restricted to the germ line. However, an Argonaute-3-independent pathway that involves PIWI proteins appears to function in somatic cells, suggesting that piRNAs may have distinct roles outside of the germ line [33]. Few studies of mammalian piRNAs or proteins belonging to the PIWI family have been reported to date.
Four members of the PIWI-like family, PIWIL1, PIWIL2, PIWIL3 and PIWIL4, have been identified in humans. PIWIL4 is the only family member that exhibits a ubiquitous expression pattern, and the overexpression of PIWIL4 in HEK293T cells causes a substantial increase in the dimethylation states of multiple H3K9 sites within the p16Ink4a gene [34]. PIWIL4 may modify histones by recruiting heterochromatin protein 1A (HP1a) to specific chromosomal sites. The N-terminal chromo domain of HP1 binds to the N-terminal tail of histone H3 when it is dimethylated on lysine 9. Once bound to chromatin, the shadow domain of HP1 can recruit histone methyl transferases, which propagates the H3K9me2 mark to allow heterochromatin spreading and transcriptional repression [35]. HP1a has been identified as a binding partner of PIWI proteins through the use of yeast two-hybrid screens, and the introduction of a mutation that disrupts the PIWI-HP1a interaction negatively affects the epigenetic silencing abilities of PIWI proteins in Drosophila somatic cells [36]. PIWIL4 protein is expressed in human CD56+ peripheral blood NK cells [16], and an intricate two-way relationship exists between histone modifications and DNA methylation. For example, natural antisense transcripts across the promoter of the p15 tumor suppressor gene trigger transcriptional gene silencing through heterochromatin formation and DNA methylation [37]. However, further investigation is required to determine whether the 28-base RNA associated with KIR represses transcription through this mechanism.
The KIR Intron 2 Promoter
An additional layer of complexity with regard to transcription within the KIR locus was recently uncovered with the discovery of a promoter within intron 2 of many KIR genes that produces a spliced antisense transcript complementary to exons 1 and 2 as well as the proximal promoter. The expression of this antisense transcript is restricted to cells at early progenitor stages of NK cell development and is not found in mature NK cell lines or in CD56+ peripheral blood NK cells. The KIR3DL1 and KIR2DL1 intron 2 promoters contain functional binding sites for the myeloid zinc finger 1 transcription factor, which may be involved in initiating transcription from the intron 2 promoters in NK cell progenitors. Remarkably, when intron 2 antisense transcripts and corresponding sense transcripts were cotransfected into HEK293 cells, a 28-base small RNA derived from the sense strand was identified that corresponds to exon 2 of the KIR3DL1 gene. The functional significance of this small RNA remains to be tested [Sharma et al., in prep.].
An Integrated Model for KIR Transcriptional Regulation
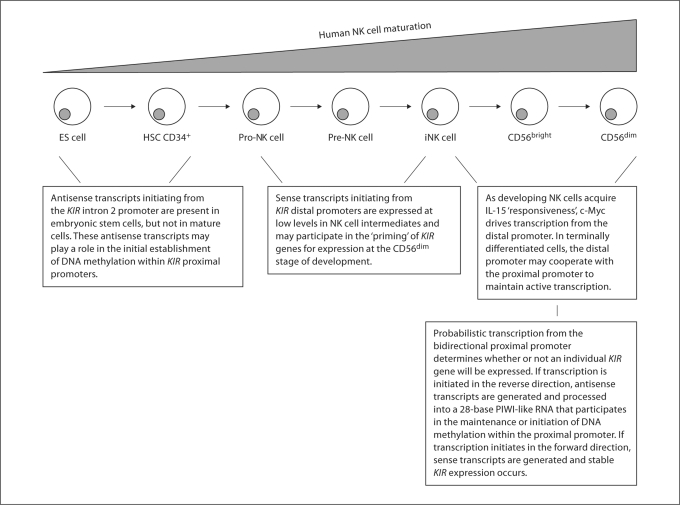
The transcriptional regulation of KIR genes appears to be much more complex than previously appreciated, and there is still much to learn. We propose the following model based on our studies of the dynamics of intergenic transcription during human NK cell development and the work of other investigators in the field: the initial regulatory events within the KIR locus likely occur early in development, perhaps at the embryonic stem cell stage when antisense transcripts generated from the intron 2 promoter can first be detected. These transcripts may be involved in establishing DNA methylation within proximal KIR promoters, which has been observed in CD34+ hematopoietic progenitor cells [10]. As these cells become further differentiated towards the NK cell lineage, the downregulation of myeloid zinc finger 1 could stop transcription from the intron 2 promoter.
As cells transition into the CD56bright stage and become responsive to IL-15, KIR distal transcripts are generated as a direct result of c-Myc binding to the distal promoter. These distal transcripts, or the act of transcription itself, may then participate in ‘priming’ KIR genes for transcription through chromatin modifications. Demethylation of proximal KIR promoters appears to take place as CD56bright NK cells differentiate into CD56dim cells and acquire surface KIR as the result of a mechanism that has not been completely defined but may require the AML1 transcription factor.
We propose that during the terminal differentiation of human NK cells, probabilistic transcription from the bidirectional proximal promoter determines whether or not a particular KIR gene will be expressed. If transcription initiates in the sense direction, no dsRNA is generated, and the proximal KIR promoter remains open, allowing for stable gene transcription. If transcription initiates in the antisense direction, dsRNA is formed over the proximal promoter between antisense transcripts and distal transcripts. The 28-base RNA is then processed from the antisense strand and participates in the maintenance of DNA methylation or de novo methylation within the proximal promoter, leading to stable transcriptional silencing (fig. 1). Because antisense transcripts are generated at very low levels [16,21], we suggest that they do not diffuse away from the promoter where they were transcribed, and the silencing mechanism occurs locally, in cis. This would allow for the specificity of action necessary in a region with high sequence homology.
Fig. 1.
We propose that transcriptional regulation of the KIR locus is a dynamic process that occurs throughout human NK cell development. Shown here are the stages of human NK cell development as previously reported [19] and our proposed model for sequential transcriptional events within the KIR locus. ES cell = Embryonic stem cell; HSC = hematopoietic stem cell; iNK cells = immature NK cells.
Conclusion
Several studies published within the past 5 years have convincingly shown that KIR genes are an important factor in the resolution of multiple infectious diseases and autoimmune disorders and in the survival of patients receiving transplants for the treatment of acute myelogenous leukemia [38,39]. As researchers continue to narrow down the critical KIR-human leukocyte antigen interactions that affect clinical outcomes, there will be a need to more deeply understand KIR regulation with the goal of manipulating the KIR repertoire for therapeutic purposes. We hope that future work will build on the existing model for KIR regulation and open doors for targeted manipulation of the expression of this clinically relevant gene family.
References
- 1.Colonna M, Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 2.Wagtmann N, Biassoni R, Cantoni C, Verdiani S, Malnati MS, Vitale M, Bottino C, Moretta L, Moretta A, Long EO. Molecular clones of the p58 NK cell receptor reveal immunoglobulin-related molecules with diversity in both the extra- and intracellular domains. Immunity. 1995;2:439–459. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 3.Suto Y, Maenaka K, Yabe T, Hirai M, Tokunago K, Takok K, Juji T. Chromosomal localization of the human natural killer cell class I receptor family genes to 19q13.4 by fluorescence in situ hybridization. Genomics. 1996;35:270–272. doi: 10.1006/geno.1996.0355. [DOI] [PubMed] [Google Scholar]
- 4.Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci USA. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin MP, Bashirova A, Traherne J, Trowsdale J, Carrington M. Cutting edge: expansion of the KIR locus by unequal crossing over. J Immunol. 2003;171:2192–2195. doi: 10.4049/jimmunol.171.5.2192. [DOI] [PubMed] [Google Scholar]
- 6.Trowsdale J, Barten R, Haude A, Stewart CA, Beck S, Wilson MJ. The genomic context of natural killer cell receptor extended gene families. Immunol Rev. 2001;181:20–38. doi: 10.1034/j.1600-065x.2001.1810102.x. [DOI] [PubMed] [Google Scholar]
- 7.Santourlidis S, Trompeter HI, Weinhold S, Eisermann B, Meyer KL, Wernet P, Uhrberg M. Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J Immunol. 2002;169:4253–4261. doi: 10.4049/jimmunol.169.8.4253. [DOI] [PubMed] [Google Scholar]
- 8.Chan HW, Kurago ZB, Stewart CA, Wilson MJ, Martin MP, Mace BE, Carrington M, Trowsdale J, Lutz CT. DNA methylation maintains allele-specific KIR gene expression in human natural killer cells. J Exp Med. 2003;197:245–255. doi: 10.1084/jem.20021127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan HW, Miller JS, Moore MB, Lutz CT. Epigenetic control of highly homologous killer Ig-like receptor gene alleles. J Immunol. 2005;175:5966–5974. doi: 10.4049/jimmunol.175.9.5966. [DOI] [PubMed] [Google Scholar]
- 10.Santourlidis S, Graffmann N, Christ J, Uhrberg M. Lineage-specific transition of histone signatures in the killer cell Ig-like receptor locus from hematopoietic progenitor to NK cells. J Immunol. 2008;180:418–425. doi: 10.4049/jimmunol.180.1.418. [DOI] [PubMed] [Google Scholar]
- 11.Vilches C, Gardiner CM, Parham P. Gene structure and promoter variation of expressed and non-expressed variants of the KIR2DL5 gene. J Immunol. 2000;165:6416–6421. doi: 10.4049/jimmunol.165.11.6416. [DOI] [PubMed] [Google Scholar]
- 12.Gómez-Lozano N, Trompeter HI, de Pablo R, Estefania E, Uhrberg M, Vilches C. Epigenetic silencing of potentially functional KIR2DL5 alleles: implications for the acquisition of KIR repertoires by NK cells. Eur J Immunol. 2007;37:1954–1965. doi: 10.1002/eji.200737277. [DOI] [PubMed] [Google Scholar]
- 13.Saleh A, Makrigiannis AP, Hodge DL, Anderson SK. Identification of a novel Ly49 promoter that is active in bone marrow and fetal thymus. J Immunol. 2002;168:5163–5169. doi: 10.4049/jimmunol.168.10.5163. [DOI] [PubMed] [Google Scholar]
- 14.Saleh A, Davies GE, Pascal V, Wright PW, Hodge DL, Cho EH, Lockett SJ, Abshari M, Anderson SK. Identification of probabilistic transcriptional switches in the Ly49 gene cluster: a eukaryotic mechanism for selective gene activation. Immunity. 2004;21:55–66. doi: 10.1016/j.immuni.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Stulberg MJ, Wright PW, Dang H, Hanson RJ, Miller JS, Anderson SK. Identification of distal KIR promoters and transcripts. Genes Immun. 2006;8:124–130. doi: 10.1038/sj.gene.6364363. [DOI] [PubMed] [Google Scholar]
- 16.Cichocki F, Lenvik T, Sharma N, Yun G, Anderson SK, Miller JS. Cutting edge: KIR antisense transcripts are processed into a 28-base PIWI-like RNA. J Immunol. 2010;185:2009–2012. doi: 10.4049/jimmunol.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z, Boffelli D, Boonmark N, Schwartz K, Lawn R. Apolipoprotein(a) gene enhancer resides within a LINE element. J Biol Chem. 1998;273:891–897. doi: 10.1074/jbc.273.2.891. [DOI] [PubMed] [Google Scholar]
- 18.Cichocki F, Hanson RJ, Lenvik T, Pitt M, McCullar V, Li H, Anderson SK, Miller JS. The transcription factor c-Myc enhances KIR gene transcription through direct binding to an upstream distal promoter element. Blood. 2009;113:3245–3253. doi: 10.1182/blood-2008-07-166389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy MK, et al. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med. 2000;191:771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freud AG, Yokohama A, Becknell B, Lee MT, Mao HC, Ferketich AK, Caligiuri MA. Evidence for discrete stages of human natural killer cell differentiation in vivo. J Exp Med. 2006;203:1033–1043. doi: 10.1084/jem.20052507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies GE, Locke SM, Wright PW, Li H, Hanson RJ, Miller JS, Anderson SK. Identification of bidirectional promoters in the human KIR genes. Genes Immun. 2007;8:245–253. doi: 10.1038/sj.gene.6364381. [DOI] [PubMed] [Google Scholar]
- 22.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 23.Madisen L, Krumm A, Hebbes TR, Groudine M. The immunoglobulin heavy chain locus control region increases histone acetylation along linked c-myc genes. Mol Cell Biol. 1998;18:6281–6292. doi: 10.1128/mcb.18.11.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart CA, Bergen JV, Trowsdale J. Different and divergent regulation of the KIR2DL4 and KIR3DL1 promoters. J Immunol. 2003;170:6073–6081. doi: 10.4049/jimmunol.170.12.6073. [DOI] [PubMed] [Google Scholar]
- 25.Presnell SR, Zhang L, Ramilo CA, Chan HW, Lutz CT. Functional redundancy of transcription factor-binding sites in the killer cell Ig-like receptor (KIR) gene promoter. Int Immunol. 2006;18:1221–1232. doi: 10.1093/intimm/dxl043. [DOI] [PubMed] [Google Scholar]
- 26.Li H, Pascal V, Martin MP, Carrington M, Anderson SK. Genetic control of variegated KIR gene expression: polymorphisms of the bi-directional KIR3DL1 promoter are associated with distinct frequencies of gene expression. PLoS Genet. 2008;4:e1000254. doi: 10.1371/journal.pgen.1000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pascal V, Stulberg MJ, Anderson SK. Regulation of class I major histocompatibility receptor expression in natural killer cells: one promoter is not enough! Immunol Rev. 2006;214:9–21. doi: 10.1111/j.1600-065X.2006.00452.x. [DOI] [PubMed] [Google Scholar]
- 28.Morris KV, Chan SWL, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 29.Kim DH, Sætrom P, Snøve O, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirino Y, Mourelatos Z. Mouse PIWI-interacting RNAs are 2'-O-methylated at their 3′ termini. Nat Struct Mol Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- 31.Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T. The 3′ termini of mouse PIWI-interacting RNAs are 2'-O-methylated. Nat Struct Mol Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- 32.Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li C, Vagin W, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugimoto K, Kage H, Aki N, Sano A, Kitagawa H, Nagase T, Yatomi Y, Ohishi N, Takai D. The induction of H3K9 methylation by PIWIL4 at the p16Ink4a locus. Biochem Biophys Res Commun. 2007;359:497–502. doi: 10.1016/j.bbrc.2007.05.136. [DOI] [PubMed] [Google Scholar]
- 35.Vermaak D, Malik HS. Multiple roles for heterochromatin protein 1 genes in Drosophila. Annu Rev Genet. 2009;43:467–492. doi: 10.1146/annurev-genet-102108-134802. [DOI] [PubMed] [Google Scholar]
- 36.Lin H, Yin H. A novel epigenetic mechanism in Drosophila somatic mediated by Piwi and piRNAs. Cold Spring Harb Symp Quant Biol. 2008;73:273–281. doi: 10.1101/sqb.2008.73.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulkarni S, Martin MP, Carrington M. The Yin and Yang of HLA and KIR in human disease. Semin Immunol. 2008;20:343–352. doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cooley S, Tractenberg E, Bergemann TL, Saeteurn K, Klein J, Le CT, Marsh SG, Guethlein LA, Parham P, Miller JS, Weisdorf DJ. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute mylogenous leukemia. Blood. 2009;113:726–732. doi: 10.1182/blood-2008-07-171926. [DOI] [PMC free article] [PubMed] [Google Scholar]