Abstract
The Ddc1/Rad17/Mec3 complex and Rad24 are DNA damage checkpoint components with limited homology to replication factors PCNA and RF-C, respectively, suggesting that these factors promote checkpoint activation by “sensing” DNA damage directly. Mec1 kinase, however, phosphorylates the checkpoint protein Ddc2 in response to damage in the absence of all other known checkpoint proteins, suggesting instead that Mec1 and/or Ddc2 may act as the initial sensors of DNA damage. In this paper, we show that Ddc1 or Ddc2 fused to GFP localizes to a single subnuclear focus following an endonucleolytic break. Other forms of damage result in a greater number of Ddc1–GFP or Ddc2–GFP foci, in correlation with the number of damage sites generated, indicating that Ddc1 and Ddc2 are both recruited to sites of DNA damage. Interestingly, Ddc2 localization is severely abrogated in mec1 cells but requires no other known checkpoint genes, whereas Ddc1 localization requires Rad17, Mec3, and Rad24, but not Mec1. Therefore, Ddc1 and Ddc2 recognize DNA damage by independent mechanisms. These data support a model in which assembly of multiple checkpoint complexes at DNA damage sites stimulates checkpoint activation. Further, we show that although Ddc1 remains strongly localized following checkpoint adaptation, many nuclei contain only dim foci of Ddc2–GFP, suggesting that Ddc2 localization may be down-regulated during resumption of cell division. Lastly, visualization of checkpoint proteins localized to damage sites serves as a useful tool for analysis of DNA damage in living cells.
Keywords: DDC1, DDC2, DNA damage checkpoint, localization, chromosome
The eukaryotic DNA damage checkpoint machinery both delays cell cycle progress and promotes repair processes in response to genotoxic stress. This response ensures that only an intact, fully replicated genome will be inherited by cells after mitotic division. The damage checkpoint is capable of responding to many types of genetic lesions, including double-strand DNA breaks (DSBs), UV and gamma radiation-induced damage, chemically-modified DNA, and errors in DNA replication. Early DNA processing events that give rise to single-stranded DNA (ssDNA) (Garvik et al. 1995; Lee et al. 1998) and/or other repair intermediates may provide common substrates for recognition by the DNA damage checkpoint. How this signal (or signals) is detected by the checkpoint machinery remains unknown.
In the budding yeast Saccharomyces cerevisiae, it has been suggested that Rad24, Ddc1, Rad17, Mec3, and Rad9 act as “sensors” of DNA damage (for reviews, see Weinert 1998; Lowndes and Murguia 2000). Rad17 and Ddc1, which exist in a complex with Mec3 (Paciotti et al. 1998), contain limited homology to PCNA (Caspari et al. 2000; Venclovas and Thelen 2000), the processivity factor for DNA polymerase. PCNA forms a homotrimeric ring-like clamp around dsDNA, and is loaded onto DNA in a reaction catalyzed by the RF-C complex (for review, see Mossi and Hubscher 1998). The RAD24 gene contains homology to all five subunits of RF-C (Griffiths et al. 1995). Moreover, Rad24 is found in a complex containing Rfc2–Rfc5 in which Rad24 has replaced Rfc1 (Green et al. 2000). These data suggest that the Ddc1/Mec3/Rad17 complex acts as a damage-specific DNA clamp, and perhaps loading of these proteins is catalyzed by a modified version of RF-C co-opted by Rad24. Once bound to DNA, this damage-specific clamp could become competent to recruit additional checkpoint factors, subsequently activating the checkpoint response. Additional evidence that Rad9 and Rad24 are recruited to DNA damage sites stems from experiments that measure ssDNA generation at damaged telomeres. In cdc13-1 cells, telomeres are recognized as DNA damage and undergo 5′ → 3′ endonucleolytic degradation (Garvik et al. 1995). cdc13-1 cells deleted for RAD24 are partially defective for this degradation, whereas deletion of RAD9 results in increased 5′ → 3′ degradation of telomeric sequences (Lydall and Weinert 1995). rad24Δ, rad9Δ, and rad24Δ cells exhibit similar levels of degradation, suggesting that Rad9 antagonizes a pro-endonucleolytic function of Rad24, and implicating both proteins in activities at sites of damage.
Mec1 is a PI3K-like kinase that has also been proposed to sense DNA damage. Mec1 is required for phosphorylation and activation of the Rad53 (Sanchez et al. 1996; Sun et al. 1996) and Chk1 (Sanchez et al. 1999) protein kinases, which in turn target the cell cycle machinery to enact checkpoint arrest. Ddc1 (Paciotti et al. 1998) and Rad9 (Emili 1998; Sun et al. 1998) are also phosphorylated following damage in a Mec1-dependent, Rad24-dependent manner, indicating that putative sensor proteins are a target of Mec1. Mec1, and its Schizosaccharomyces pombe homolog Rad3, is required for damage-inducible phosphorylation of the checkpoint protein Ddc2 (S.p. Rad26) (Edwards et al. 1999; Paciotti et al. 2000; Rouse and Jackson 2000; Wakayama et al. 2001). This phosphorylation requires no other known checkpoint genes. In addition, Ddc2 (also called Lcd1 and Pie1) contains sequence similarity to the RF-C subunit Rfc5. Therefore, Mec1/Ddc2 function either in a parallel pathway, or upstream of Rad24, Rad17, Mec3, and Ddc1.
Rad24, Rad17, Mec3, Ddc1, Rad9, Mec1, and Ddc2 are all reasonable candidates for damage-sensing molecules. Homologs of some of these genes have been shown to relocalize in response to DNA damage. The mammalian homolog of Ddc1 (hRad9) becomes perinuclear (Komatsu et al. 2000) and more tightly chromatin-associated (Burtelow et al. 2000) after treatment with the DNA methylating agent MMS or ionizing radiation (IR), respectively. Moreover, the Mec1 homolog, ATR, forms nuclear foci that colocalize with BRCA1 after IR treatment (Tibbetts et al. 2000). In this study, we provide direct evidence of the recruitment of checkpoint proteins to DNA lesions in live cells. Ddc1 fused to GFP forms a single subnuclear focus on induction of a single telomeric double-strand break generated by the HO endonuclease, whereas induction of damage at multiple sites in cdc13-1 and cdc9-1 cells results in the formation of two to five and >10 foci, respectively. These foci can be observed within 0.5 h of damage induction, and increase in intensity with time, persisting in all cells even after checkpoint adaptation. Therefore, recruitment of Ddc1 protein is rapid during checkpoint activation and perdures into subsequent cell cycles. We show that damage-inducible Ddc1 localization requires the checkpoint genes RAD17, MEC3, and RAD24, but not MEC1, DDC2, RAD9, or RAD53. Similarly, we found that Ddc2–GFP is relocalized following cdc13-1 or HO break-induced damage in a manner identical to Ddc1 protein.
In contrast to Ddc1, formation of Ddc2–GFP foci depends on MEC1 and no other checkpoint gene tested. Ddc2 foci increased in intensity at early time points following an HO break, but many cells viewed at late time points, during and after checkpoint adaptation, exhibited faint Ddc2 foci. We have also shown that Ddc2 associates with DNA breaks in chromatin IP experiments. Finally, Rad24, Rad9, or Rad53 fusions to GFP show either no or weak recruitment (relative to Ddc1 and Ddc2) to damage sites. Together, these data constitute a model in which independently recruited checkpoint complexes are concentrated at damage sites, allowing them to physically interact to promote activation of the DNA damage checkpoint.
Localization of checkpoint proteins provides a method for visualization and analysis of damage in living cells. For instance, an intrachromosomal DSB results in a single nuclear Ddc1 focus, suggesting that broken DNA ends remain associated. Ddc1–GFP also allows observation of spontaneous DNA damage in repair-deficient strains; most rad52 cells were shown to contain one or more Ddc1 foci in the absence of induced damage. In this manner, checkpoint protein localization may serve as an indicator of genomic instability in living cells.
Results
Ddc1 is recruited to DNA damage sites
To determine whether localization of Ddc1 is regulated by DNA damage, we examined haploid cells expressing Ddc1 fused to GFP in the presence and absence of an irreparable DSB. The Ddc1–GFP fusion protein is expressed under its own promoter and is the only copy of Ddc1 in this strain. A microcolony assay (see Materials and Methods) was used to determine that the fusion protein is fully proficient for the DNA damage checkpoint (data not shown). A DSB is generated by the site-specific HO endonuclease under control of the galactose-inducible promoter in a strain containing an HO cleavage site at the telomere of chromosome VII. The endogenous HO site (used for mating type switching in yeast) is deleted in this strain, so that expression of HO will introduce a single DSB into the genome. On induction of HO in asynchronous cultures, ∼90% of cells had undergone cleavage by 4 h, as assayed by Southern blotting (Fig. 1a). When cells were grown in glucose (noninducing) media, Ddc1–GFP localization was diffusely nucleoplasmic (Fig. 1b), as confirmed by DAPI staining (data not shown). Similar Ddc1–GFP nucleoplasmic localization was seen when HO was induced (by galactose) in cells lacking an HO site (data not shown). On shifting to galactose-containing media, we observed the appearance of a single focus in each nucleus, such that 76% (74/97) of nuclei contained a Ddc1–GFP focus after 3 h, and >95% (81/85) of cells contained a focus by 6 h (Fig. 1c). Ddc1–GFP foci were less intense at earlier time points and grew continually brighter during a 16-h time course (Fig. 1b). We believe this is most likely caused by continuous resection of the 5′ strand of the DSB (Lee et al. 1998). Though the extent of resection observed at irreparable breaks is probably exaggerated, it is likely to occur at sites of reparable damage as well. Resection would allow multiple Ddc1 molecules to bind to one break site if Ddc1 were recognizing either ssDNA or other repair intermediates (see Discussion). We observed that 8% of uninduced cells (14/167) contained faint Ddc1 foci (Fig. 1b, inset, t = 0; Fig. 1c). These foci may result from spontaneous DNA damage.
Figure 1.
Ddc1 forms a single subnuclear focus after a dsDNA break. (a) Southern blot of DNA proximal to the HO site at the telomere of chromosome VII. To monitor the rate of HO-induced cleavage, log-phase cells were transferred from raffinose to galactose media to induce the HO endonuclease and samples were collected at time points indicated. The uncleaved band is heterogeneous in size because it contains telomeric sequences. The cleaved fragment disappears at late time points because of gradual degradation. (b) Visualization of Ddc1–GFP. Cells at the zero time point were grown in glucose media to repress HO expression; cells at 3-, 6-, 12-, and 16-h time points were grown in raffinose media, then transferred to galactose media for the indicated time. Inset at t = 0 representative of occasional Ddc1–GFP foci in HO-uninduced cells. At the 12-h and 16-h time points, multiple focal planes were merged to demonstrate that Ddc1–GFP foci are seen in all nuclei. The arrowheads in the 12-h time point indicate the presence of an adapted anaphase cell containing a single focus in each nucleus. (c) Fraction of cells containing Ddc1–GFP foci as a time course of HO induction. Data combined from all focal planes within a field. (d) Time course of adaptation following an HO break. Cells were grown in raffinose media, transferred to galactose media for 2 h, sonicated, and plated to raffinose plus galactose plates. Microcolonies containing more than one cell (>2 cell bodies) were considered adapted.
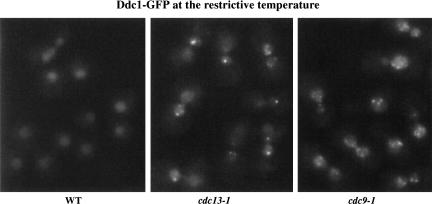
The observation that a single telomeric DSB results in a single Ddc1–GFP focus is consistent with the model that Ddc1 is recruited to sites of DNA damage. We reasoned that the generation of multiple sites of DNA damage should result in the formation of multiple Ddc1–GFP foci, and would rule out the possibility that Ddc1 localizes to another subnuclear structure following DNA damage. To address this issue, we used two conditional mutations, cdc13-1 and cdc9-1, which generate different types of DNA damage and should result in distinct patterns of Ddc1–GFP localization. Cdc13 associates with telomeres (Nugent et al. 1996; Bourns et al. 1998) and is thought to protect telomere ends. cdc13-1 cells undergo extensive telomere degradation at the nonpermissive temperature, resulting in a DNA damage checkpoint-dependent arrest (Garvik et al. 1995). In S. cerevisiae, telomeres are reported to cluster into four to seven discrete foci per focal plane as visualized by Rap1–GFP fluorescence and by in situ hybridization against telomeric sequences (Gotta et al. 1996). We first confirmed by Rap1–GFP fluorescence in living cells that telomeres maintain this clustering pattern of four to seven foci in cdc13-1 arrested cells at 34°C (data not shown). When we analyzed Ddc1–GFP fluorescence in cdc13-1 cells at 34°C, we observed an average of 2.3 foci per nucleus per focal plane (Fig. 2), with most cells exhibiting two to five foci. Ddc1 foci are observed by 0.5 h (data not shown), and all cells scored contain foci by 2 h (144/144). A Rad17–GFP fusion exhibited a similar localization pattern in most cdc13-1 cells (although the foci were less pronounced in some of the cells), whereas a Rad24–GFP fusion did not show significant relocalization (see below). The number of Ddc1–GFP foci observed is fewer than that observed for Rap1–GFP localization, perhaps because not all telomeres are damaged in cdc13-1 cells. It has been reported that only ∼5% of telomeric DNA becomes single stranded in cdc13-1-arrested cells (Garvik et al. 1995).
Figure 2.
Ddc1 forms multiple nuclear foci following cdc13-1-induced and cdc9-1-induced DNA damage. Log phase cultures of cdc13-1 DDC1–GFP and cdc9-1 DDC1–GFP cells were shifted to 34°C and 36°C, respectively, and visualized after 2 h. Also shown are asynchronously growing wild-type control cells containing DDC1–GFP alone after shift to 34°C for 2 h.
When cells harboring a conditional allele of DNA ligase (cdc9-1) are grown at the nonpermissive temperature, Okazaki fragments generated during DNA replication are not efficiently ligated (Johnston and Nasmyth 1978), resulting in a DNA damage checkpoint arrest (Weinert and Hartwell 1993). After shifting cdc9-1 cells to 36°C for 2 h, Ddc1–GFP localization became punctate in nuclei of all cells (93/93) (Fig. 2). The large number of foci and their heterogeneous intensity made them difficult to score, but most cells exhibited >10 prominent foci of Ddc1–GFP per nucleus. The pattern of Ddc1 localization seen in cdc9-1 cells is distinct from that observed following a single DSB or cdc13-1-induced telomeric damage, consistent with the greater extent of damage thought to be generated in cdc9-1 cells.
Ddc1 localization persists through adaptation to the damage checkpoint
In response to a telomeric HO break, yeast cells arrest in mitosis for ∼8–10 h before undergoing a process called “checkpoint adaptation” and re-entering the cell cycle (Sandell and Zakian 1993; Toczyski et al. 1997; Lee et al. 1998). Recent evidence suggests adaptation results from a down-regulation of checkpoint signaling, including a reduction in Rad53 activity (Pellicioli et al. 2001). We determined the rate of adaptation in our strain to examine Ddc1 localization after adaptation to the HO break. Cells were assayed for adaptation by a microcolony assay on galactose-containing plates (see Materials and Methods). Sixty percent of cells (177/300) adapt by 8 h after HO induction, and >98% of cells (295/300) are adapted by 11 h (Fig. 1d). At the 11-h time point, over a third of cells (111/300) have divided a second time to give rise to four-cell microcolonies. When we examine Ddc1 localization during an adaptation time course, we observe Ddc1 foci in >98% of cells at 9, 12, and 16 h after HO induction (Fig. 1c). Therefore, loss of Ddc1 localization does not precede adaptation.
Rad17, Mec3, and Rad24 are required for damage-inducible Ddc1 localization
To determine whether Ddc1 requires other checkpoint components for its localization after DNA damage, we examined Ddc1–GFP fluorescence in checkpoint mutant strains. In cells deleted for RAD24, RAD17, or MEC3, Ddc1–GFP foci are not observed in either cdc13-1 cells at the nonpermissive temperature (Fig. 3) or HO break-induced cells (data not shown). Western blot analysis of Ddc1–GFP in rad24, rad17, and mec3 mutants showed that the Ddc1–GFP fusion protein levels are similar in these mutants to the levels seen in wild-type cells (data not shown). Therefore, Rad24, Rad17, and Mec3 are not only required for the DNA damage checkpoint but are essential for detectable subnuclear localization of Ddc1–GFP after DNA damage.
Figure 3.
Ddc1 requires Rad24, Rad17, and Mec3 for damage-inducible localization. cdc13-1 DDC1–GFP cells deleted for RAD17, MEC3, RAD24, MEC1, DDC2, RAD53, or RAD9 were raised to 34°C for 2 h before visualization of GFP. Single focal planes for each sample are shown.
We tested whether damage-inducible localization of Ddc1 relies on the proposed sensor molecules Mec1, Ddc2, and Rad9, as well as the Rad53 kinase. cdc13-1 (Fig. 3) or HO break-induced cells (data not shown) deleted for MEC1, DDC2, RAD9, or RAD53 were all competent for damage-inducible localization of Ddc1–GFP. The same number of Ddc1–GFP foci were observed in these mutants on damage induction (two to five foci after cdc13-1-induced damage and one focus after an HO break) as in checkpoint-proficient cells. The intensity of Ddc1 foci was not noticeably reduced in any of these mutants, although there were slight variations between experiments. Tel1 is a Mec1-related kinase that is redundant with Mec1 for some damage-inducible phosphorylation events (Emili 1998). Ddc1–GFP was able to form foci in cdc13-1 mec1 tel1 mutants, ruling out a redundant requirement for mec1 and tel1 in Ddc1 localization (data not shown).
Ddc2–GFP localizes to DNA damage in cdc13-1 and HO break-induced cells, and its localization requires Mec1 but not other known checkpoint genes
We examined the localization of other checkpoint proteins by fusing GFP protein to the carboxyl terminus of Rad24, Rad17, Mec3, Rad9, Ddc2, and Rad53 proteins. Whereas Rad24–GFP and Rad17–GFP suffered slight checkpoint defects in cdc13-1-damaged cells by microcolony assay, a Mec3–GFP fusion protein was completely defective for checkpoint arrest (data not shown). As discussed above, we observed that this hypomorphic Rad17–GFP fusion in cdc13-1 cells at the nonpermissive temperature yielded a similar localization pattern to Ddc1–GFP (data not shown). Rad24–GFP showed a diffuse nuclear localization in cdc13-1 cells growing asynchronously at 23°C or arrested at 35°C for 2 h, possibly exhibiting subnuclear foci at the nonpermissive temperature in a small fraction of cells (data not shown).
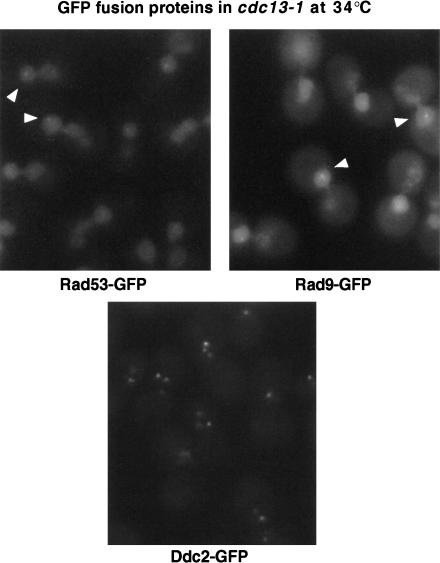
Ddc2–GFP, Rad9–GFP, and Rad53–GFP fusion proteins were each fully proficient for checkpoint function by microcolony assay (data not shown) and were tested for damage-inducible localization in cdc13-1 cells. Rad9–GFP and Rad53–GFP were both nuclearly localized at the permissive temperature for cdc13-1. On shifting to the nonpermissive temperature for 2 h, a fraction of cdc13-1 RAD9–GFP or cdc13-1 RAD53–GFP cells showed very faint foci (Fig. 4, arrowheads), but most cells showed no relocalization at all. In contrast, Ddc2–GFP was diffusely nuclear at 23°C but formed bright subnuclear foci in cdc13-1 cells shifted to 35°C. These foci could be detected by 0.5 h after temperature shift, and were observed in >98% of cells by 2 h (165/167) (Fig. 4). Similar to Ddc1–GFP localization in cdc13-1 cells, nuclei contained an average of 2.1 Ddc2–GFP foci, with a maximum of five foci per nucleus. Therefore, Ddc2–GFP localizes to telomeres as rapidly as Ddc1–GFP in response to cdc13-1-induced telomeric damage. Ddc2–GFP also formed two to three foci in most cells following treatment with the DNA-damaging agent Zeocin, a bleomycin derivative (data not shown).
Figure 4.
Localization of other checkpoint proteins fused to GFP in cdc13-1-damaged cells. GFP fusions generated for Rad53, Rad9, and Ddc2 were expressed under the endogenous promotor for each protein in cdc13-1 cells. Log phase cultures were shifted to 34°C for 2 h before visualization of GFP fusions. Arrowheads indicate the presence of occasional faint foci in cells expressing Rad53–GFP and Rad9–GFP.
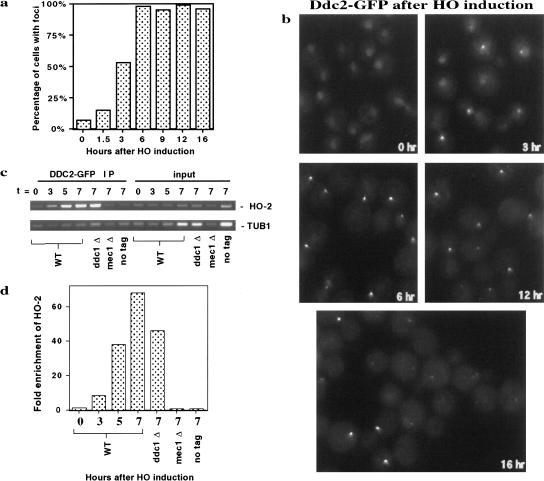
Ddc2–GFP localizes to a single subnuclear focus in cells containing an HO-induced break (Fig. 5), as was observed for Ddc1–GFP. Whereas 7% of uninduced cells (25/370) contain a distinguishable Ddc2–GFP focus, 53% of cells (100/190) contained a Ddc2–GFP focus by 3 h after induction of HO endonuclease and >98% of cells (311/317) contained a Ddc2–GFP focus by 6 h. Interestingly, when we examined the intensity of Ddc2–GFP foci with time during the experiment, we saw that Ddc2–GFP foci were of increasing intensity until 6–9 h of HO induction, after which an increasing fraction of cells exhibited only faint Ddc2 foci (Fig. 5; 12-h and 16-h time points). Notably, the loss of intensity of Ddc2 foci correlates with the time at which cells undergo adaptation to an HO break in this strain. An adaptation time course of the Ddc2–GFP-tagged strain after an HO break (data not shown) was identical to that shown for Ddc1–GFP (Fig. 1d). Ninety-six percent of cells coexpressing Ddc1–GFP and Ddc2–GFP also showed a single focus of fluorescence after induction of an HO break, suggesting that these two proteins are recruited to a single DNA locus. The 4% of cells showing two foci is likely attributable to spontaneous DNA damage and is indistinguishable from what was seen with Ddc1–GFP or Ddc2–GFP alone. In summary, the damage-inducible localization patterns of Ddc2 in cdc13-1 and HO break-induced cells indicates that it is recruited to DNA damage in addition to Ddc1.
Figure 5.
Ddc2–GFP forms a single focus in HO break-induced cells. (a) Fraction of cells containing Ddc2–GFP foci as a time course of HO induction. Data combined from all focal planes within a field. (b) Visualization of Ddc2–GFP foci following an HO break. HO endonuclease was induced in cells as described in Figure 1. Cells grown in glucose to repress HO expression are presented as the t = 0 time point. A representative focal plane for 0, 3, 6, 12, and 16 h after HO induction is shown. (c) Chromatin immunoprecipation of Ddc2–GFP at an HO break site. The t = 0 represents cells grown in glucose to repress HO induction. Cells collected at each indicated time point were fixed and analyzed by chromatin IP. Ddc2–GFP and cross-linked DNA were immunoprecipitated using affinity-purified anti-GFP antibodies. After reversal of cross-linking and DNA purification, PCR was performed using HO-2 primers directed to unique DNA sequence 0.4 kb from the HO cleavage site. Primers to amplify unlinked TUB1 sequence were used as an internal control in each PCR reaction. Input DNA was isolated from fixed cells at each time point before immunoprecipitation, and treated in parallel with IP samples in preparation for PCR. Control strains deleted for DDC1 or MEC1 or lacking a Ddc2–GFP tag were grown and harvested at the 7-h time point and treated in parallel to the wild-type Ddc2–GFP strain. (d) The HO-2 and TUB1 bands in c were quantitated and the ratio of HO-2:TUB1 band intensities calculated for each IP and input DNA sample. Fold enrichment was calculated as the ratio of HO-2:TUB1 in the IP samples to its respective input DNA control and graphed as shown.
Although the correlation between the number of Ddc2 (and Ddc1) foci and the number of damage sites strongly suggests that Ddc2 associates with damaged DNA, we wished to demonstrate this directly using chromatin IPs. Cells expressing Ddc2–GFP were collected throughout a time course of HO induction, fixed with formaldehyde, sonicated, and immunoprecipitated with anti-GFP antibody. DNA was extracted from the immunoprecipitates and PCR amplified (Fig. 5c). PCR reactions allowed simultaneous amplification of either an internal negative control (TUB1) and a PCR product 0.4-kb from the HO site (HO-2) (Fig. 5c,d) or a different internal negative control for nonspecific DNA binding (ACT1) and a PCR product 1.0 kb from the HO site (HO-1; data not shown). These data show enrichment for the two HO proximal PCR products but not the unlinked controls. By 3 h, HO sequences are eightfold enriched, increasing to nearly 70-fold enrichment by 7 h. Two hours after HO induction is the earliest time point at which we could observe HO proximal association in other experiments (data not shown). PCR amplification of the input DNA showed no preference for the HO site-proximal PCR products, indicating that cutting does not, in and of itself, induce a preference for amplification of the HO site-proximal products. In fact, the relative ability to amplify the break-proximal sequences from input DNA decreased with duration of HO induction, possibly because of degradation of both DNA strands at the HO break site (see Lee et al. 1998). Consistent with the epistasis data for Ddc2 localization, Mec1 but not Ddc1 was required for Ddc2 association with the break site. A side-by-side experiment performed using Ddc1–GFP showed no preferential precipitation of HO-proximal sequences (data not shown). Moreover, we were unsuccessful in our attempts to observe Ddc1 localization by other procedures requiring cross-linking (immunofluorescence or chromatin spreads), a technical challenge that might be related to the topological nature of the binding of a PCNA-like complex to DNA.
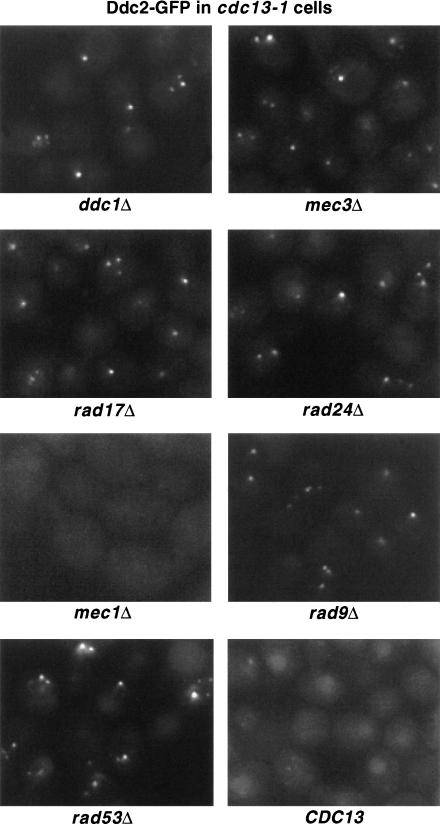
In contrast to Ddc1–GFP localization, MEC1 but not DDC1, MEC3, RAD17, or RAD24 is largely required for Ddc2–GFP recruitment to cdc13-1-induced telomeric damage (Fig. 6). However, mec1 cells did show very weak Ddc2–GFP foci in 5% of cells. In addition, Ddc2–GFP relocalized normally in cdc13-1 cells deleted for RAD9 or RAD53 (Fig. 6). Therefore, RAD9 and RAD53 are unimportant for localization of either Ddc1 or Ddc2 in response to DNA damage, whereas Ddc1 and Ddc2 have mutually exclusive requirements for Mec3/Rad17/Rad24 or Mec1, respectively. It should be noted that our epistasis analysis for Ddc1–GFP and Ddc2–GFP does not distinguish between effects on the establishment and the maintenance of protein localization at damage sites.
Figure 6.
MEC1 is required for Ddc2 localization following cdc13-1-induced damage. Log phase cdc13-1 DDC2–GFP cells deleted for DDC1, RAD17, MEC3, RAD24, MEC1, RAD53, or RAD9 were raised to 35°C for 2 h before visualization of GFP. Ddc2–GFP localization in CDC13 checkpoint-proficient cells at 35°C is shown at bottom right. Single focal planes for each sample are shown.
Visualization of broken chromosome fragments and spontaneous damage in living cells
Damage-induced recruitment of checkpoint proteins provides a valuable tool for visualizing sites of DNA damage in living cells. We wished to use Ddc1–GFP localization to determine the fate of the two chromosomal fragments generated after introduction of an intrachromosomal DSB. If the broken ends of the cleaved chromosome remain associated following endonucleolytic cleavage, we expected to observe a single focus of Ddc1–GFP, whereas if the two halves separate we predicted observation of two foci of Ddc1–GFP localization. Our prior experiments demonstrate that a single Ddc1 focus is formed following cleavage of a chromosome at its telomere. In that experiment, an acentric fragment ∼0.4 kb in size containing telomeric sequences was generated by the HO break. Because of 5′ → 3′ resection at DSBs, a fragment this size is likely to be rapidly degraded, and was not expected to be detectable by Ddc1 recruitment. In support of this, DNA sequences located 0.7 kb from an HO site are degraded by 2–3 h after HO induction (Lee et al. 1998). To determine whether an intrachromosomal HO break resulted in one or two Ddc1 foci, we constructed a strain containing a single HO site on chromosome VII that would generate chromosomal fragments 320 kb and 780 kb in size on cleavage. Following HO induction, we observed the formation of a single Ddc1–GFP focus in 89% of cells (215/243) by 6 h (Fig. 7a). Six percent of cells (18/243) contained two foci, comparable to that observed when a telomeric break was generated (7%: 6/85). A likely explanation is that these cells contain both an HO break and spontaneous DNA damage (Fig. 1c, t = 0 time point), as 8% (14/167) of undamaged cells exhibited a Ddc1 focus. An HO break introduced at the TRP5 locus on chromosome VII yielded similar results to those above (data not shown). Therefore, intrachromosomal HO cleavage gives rise to a single Ddc1 focus in the majority of cells.
Figure 7.
Ddc1 localization shows that the two ends of an intrachromosomal DSB remain associated, and that cells lacking Rad52 contain high levels of unrepaired damage. (a) Cells containing an HO site near the chromosome VII SRM1 locus were grown in raffinose, transferred to galactose media, and Ddc1–GFP was visualized after 6 h. (b) Log-phase rad52 cells in the absence of induced DNA damage were visualized for Ddc1–GFP.
The observation that 8% of undamaged cells exhibit dim Ddc1–GFP foci led us to examine whether we could also use Ddc1–GFP localization to detect spontaneously arising DNA damage in cells. It has been reported previously that rad52 cells, which are defective for recombinational repair, exhibit reduced plating efficiency (Toczyski et al. 1997) and increased spontaneous chromosome loss (Galgoczy and Toczyski 2001). The decreased viability of this strain may result from spontaneous generation of DSBs that cannot undergo recombinational repair. On microscopic examination, rad52 cells are heterogeneous in size and many are large-budded, a morphological indicator of checkpoint arrest. Nearly 60% (88/153) of cells showed one or more Ddc1–GFP foci of varying intensity (Fig. 7b). Only 13% of rad52 cells, however, are unable to form colonies (Toczyski et al. 1997), indicating that spontaneous DNA damage detected by Ddc1 localization is either not lethal in both daughters or is repaired by a less efficient, Rad52-independent pathway.
Discussion
To be effective as a mechanism that preserves genomic integrity, the DNA damage checkpoint must be extremely sensitive in its ability to detect DNA damage. Introduction of a single DSB results in a prolonged cell cycle arrest before mitosis. Our results demonstrate that multiple molecules of both Ddc1 and Ddc2 recognize a single DSB site, as many molecules of each protein must be recruited to this site to be visualized microscopically in living cells. The rapid assembly of multiple checkpoint complexes at a single damage site may be the mechanism by which the DNA damage checkpoint achieves its high sensitivity. Recruitment of Ddc1 and Ddc2 to damage sites occur independently of each other. We propose that DNA damage activates the checkpoint by providing a scaffold on which the Mec1/Ddc2 kinase complex and the Ddc1/Rad17/Mec3 complex can interact.
The DNA damage checkpoint proteins Ddc1 and Ddc2 are recruited to sites of DNA damage
Although there have been clues that DNA lesions are detected by the checkpoint machinery, direct evidence that checkpoint factors are recruited to the physical sites of DNA damage had not been directly demonstrated previously. In mammalian cells, localization of the Ddc1 homolog hRad9 has been examined following DNA damage, with somewhat conflicting results. hRad9 is reported to become predominantly perinuclear on treatment with the DNA methylating agent MMS, and to interact with nuclear membrane-associated anti-apoptotic Bcl-2 proteins (Komatsu et al. 2000). Another report describes the conversion of hRad9 to a salt inextractable nuclear form following irradiation of human cells that was detected by indirect immunofluorescence as punctate nuclear staining in permeabilized cells (Burtelow et al. 2000). These results suggest that hRad9 becomes associated with one or more subnuclear structures following DNA damage. Our data indicate the following: (1) Many Ddc1 molecules are recruited to sites of damage in a manner dependent on Rad24, Rad17, and Mec3; (2) Ddc1 molecules are continually recruited with time, as indicated by increasing intensity of Ddc1 foci following damage; and (3) Ddc1 recruitment persists after down-regulation of the checkpoint during adaptation. Because these phenomena can be visualized in living cells, the caveats of fixation-dependent immunofluorescent techniques are avoided.
We demonstrate that damage-inducible Ddc1 recruitment requires RAD24, RAD17, and MEC3. Considering the homology of these proteins to RFC and PCNA, our data support a model in which Ddc1, Rad17, and Mec3 form a complex that loads onto DNA in a reaction catalyzed by Rad24 and other RF-C components. Preliminary data indicate that Rad17 is also localized to DNA damage. We detected no major changes in Rad24–GFP localization in response to damage, although Rad24 is required for proper localization of Ddc1–GFP in a manner consistent with its proposed catalytic role. Rfc1 and Rad24 may act as specificity factors for the RF-C complex. Whereas RF-C catalyzes loading of the PCNA homotrimer during DNA replication, a Rad24-containing variant of RF-C could catalyze loading of a trimeric complex containing Ddc1, Rad17, and Mec3 to genetic lesions. One function of this complex may be to facilitate 5′ → 3′ endonucleolytic processing (for review, see Weinert 1998). Interestingly, we find that Rad24 is not required for general nucleoplasmic localization of Ddc1, whereas the S. pombe homolog of Rad24 (S.p. Rad17) is required for nuclear localization of S.p. Hus1 (S.c. Mec3), a member of the S. pombe PCNA-like checkpoint complex (Caspari et al. 2000).
The S. pombe checkpoint gene Rad26 was identified recently to possess a homolog in budding yeast, Ddc2. Homologs in higher eukaryotes have not yet been identified. Interestingly, Ddc2 was shown to contain sequence similarity to Rfc5 and Rad50, a subunit of the RF-C complex and a recombinational repair protein, respectively (Rouse and Jackson 2000). Ddc2 associates with Mec1 independently of DNA damage, and shares identical mutant phenotypes with MEC1, including its requirement for DNA damage and replication checkpoints and its sml1-suppressed lethality. These data suggest that Ddc2 may function as a regulatory subunit of the Mec1 kinase. Taken together with the result that Ddc2 is phosphorylated after damage in a manner dependent on Mec1 but no other known checkpoint proteins, Mec1 and Ddc2 have been proposed to act as sensors of DNA damage (Paciotti et al. 2000).
In this study, we have shown that Ddc2 is recruited to cdc13-1-induced telomeric damage and to a single HO-induced break. In the absence of DNA damage, Ddc2 localization is diffusely nuclear. Introduction of an HO-induced break results in formation of a single intense Ddc2–GFP focus in the nucleus such that the diffuse nucleoplasmic pool of Ddc2 becomes practically undetectable. It appears as if the majority of the Ddc2 protein in the nucleus is recruited to the HO break site. This recruitment is facilitated by MEC1, in agreement with the hypothesis that these proteins function as a single complex. Although both Ddc1 and Ddc2 were found to localize to damage sites, it was difficult to detect localization of Rad24, Rad9, or Rad53 under the same conditions of DNA damage. This result is informative in that if any of these proteins function at sites of DNA damage, they must be present either transiently or substoichiometrically to Ddc1 and Ddc2.
We have demonstrated in this study that both Ddc1 and Ddc2 are recruited to DNA damage sites. A caveat of our experimental approach is that we do not observe significant localization of either protein until 30 min after damage induction in cdc13-1 cells and 2 h in HO break-induced cells. Checkpoint-dependent phosphorylation of Ddc1, Rad9, and Rad53 has been reported as early as 15 min after treatment with UV and IR (Sanchez et al. 1996; Sun et al. 1996, 1998; Emili 1998; Paciotti et al. 1998). We were not able to demonstrate that checkpoint factors associate with damage sites with equally rapid kinetics. We posit, however, that localization of checkpoint factors to damage sites is required for rapid checkpoint activation and that we are limited by the sensitivity of live fluorescence techniques for three reasons: (1) We observed slow cleavage kinetics by HO endonuclease in our system. Notably, even detection of checkpoint activation by Rad53 phosphorylation in response to a single HO break in asynchronously growing cells does not become prominent until 2–4 h after HO induction (Pellicioli et. al. 2001). (2) Our data suggest that Ddc1 and Ddc2 molecules are continually recruited with time, which is likely the result of continuous 5′ → 3′ strand resection at an irreparable HO break. (Lee et al. 1998). To observe Ddc1 and Ddc2 localization by fluorescence microscopy in live cells, the number of fluorescent molecules at the break site must exceed the level of background fluorescence, which is mainly attributable to unbound Ddc1 or Ddc2 molecules. (3) We observe Ddc1 and Ddc2 foci in 7%–8% of undamaged cells, presumably because of spontaneous DNA damage. Therefore, we are unable to attribute focus formation to the induction of DNA damage until the fraction of cells with foci is significantly higher.
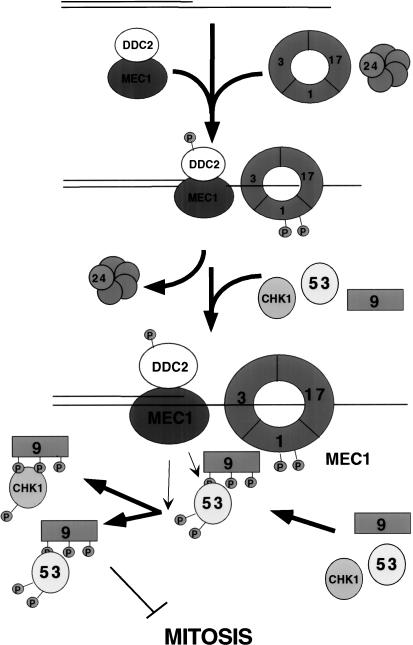
The substrate recruitment model for the activation of the DNA damage checkpoint pathway
Our data show that damage-induced localization of Ddc1 and Ddc2 occur with similar kinetics following either cdc13-1 or HO break-induced damage. Recruitment of Ddc1 to damage requires RAD17, MEC3, and RAD24, but not MEC1 or DDC2. In contrast, efficient relocalization of Ddc2 after DNA damage requires MEC1, but not DDC1, RAD17, MEC3, or RAD24. Taken together, this suggests a model in which there are two independent damage-recognizing branches of the checkpoint pathway (Fig. 8). In this model (the Substrate Recruitment model), association with DNA damage allows the Mec1/Ddc2 complex to be brought into close proximity with the Ddc1/Rad17/Mec3 complex. The Mec1/Ddc2 kinase complex is believed to act as the central regulator of checkpoint signaling, as it is required for both the DNA damage checkpoint and the incomplete replication checkpoint. Mec1 is also required for all known phosphorylation events that occur following DNA damage, including the phosphorylation of key cell cycle targets (for review, see Weinert 1998). Mec1 is capable of phosphorylating Ddc2 in the absence of other checkpoint factors in response to damage (Paciotti et al. 2000). Our data are consistent with the model that Mec1 kinase activity is stimulated by recruitment of Ddc2/Mec1 to damage sites. In vitro experiments have shown that the ATM kinase, a mammalian homolog of Mec1, is stimulated by pre-incubation with DNA (Smith et al. 1999). We propose, however, that stimulation of Mec1 kinase activity is not sufficient to activate the checkpoint, as Ddc1/Rad17/Mec3 and Rad24 are required for Mec1-dependent phosphorylation of known checkpoint targets other than Ddc2. We suggest a model in which the Ddc1/Rad17/Mec3 complex recruits Mec1 substrates, allowing Mec1 to phosphorylate targets of the DNA damage checkpoint signal transduction cascade (Fig. 8). Targets recruited by a Ddc1-containing complex may include Rad9, Rad53, and the checkpoint kinase Chk1 (Sanchez et al. 1999). The finding that Rad9 and Rad53 are not required for Ddc1 or Ddc2 localization and are not themselves strongly localized is consistent with a model in which these proteins act transiently at sites of damage, downstream of Ddc1 and Ddc2. Our observation that Ddc1 is able to re-localize on induction of damage in a mec1 strain indicates that Mec1-dependent Ddc1 phosphorylation is not required for damage recognition. Instead, Ddc1 phosphorylation may aid in its ability to recruit other factors to the damage site. Our model for checkpoint activation is reminiscent of many other signaling systems that act by concentrating signaling components to a common site.
Figure 8.
The substrate recruitment model: Checkpoint activation by the concentration of checkpoint factors on a DNA strand. Mec1/Ddc2 and the Ddc1/Mec3/Rad17 complex are recruited to DNA damage independently, promoting their interaction. Multiple Ddc1/Rad17/Mec3 complexes are loaded by the Rad24/RF-C complex and may function to recruit the substrates of Mec1.
Localization of Ddc1 and Ddc2 during adaptation to the damage checkpoint
When we examined Ddc1 localization over an extended time course of HO induction, we determined that Ddc1 foci are maintained in >98% of cells as they undergo adaptation to the DNA damage checkpoint. Adaptation correlates with down-regulation of the damage checkpoint, as evidenced by decreased Rad53 activity during adaptation to an irreparable HO break (Pellicioli et al. 2001). Loss of Rad53 activity does not result from a decrease in Ddc1 localization, as Ddc1 foci increased in intensity in all cells through a 16-h time course of HO induction. During this time, HO-break-induced cells have adapted and divided several times. Ddc1 recruitment is unlikely to disappear transiently during adaptation as no drop in the fraction of cells exhibiting Ddc1 foci is observed. Therefore, Rad53 activity is down-regulated downstream of damage recognition by Ddc1.
In contrast, Ddc2–GFP foci do not increase in intensity throughout a 16-h time course. Following HO induction, Ddc2–GFP foci appear maximal in intensity by 6 h, after which a subpopulation of cells begin to exhibit only faint foci of Ddc2–GFP. Despite this observation, >95% of cells maintain Ddc2–GFP foci between 6 and 16 h of HO induction. Though a drop in Ddc2–GFP focus intensity correlates with the timing of adaptation in these cells, we cannot be certain whether Ddc2 recruitment is a cause or an effect of adaptation. In any case, we can reasonably conclude that unlike Ddc1, the Ddc2 protein is not continually accumulated at sites of DNA damage.
Ddc1–GFP localization as a tool for visualizing DNA damage in living cells
Because Ddc1–GFP foci are maintained at sites of DNA damage, Ddc1–GFP localization provides a novel approach to the analysis of DNA damage events in real time. Though we predicted that we would only see a single focus after a telomeric break, we were uncertain of the number of foci that would be observed after an intrachromosomal break. Both sides of the break should generate a checkpoint signal in the form of ssDNA resulting from 5′ → 3′ strand resection. We observed only a single focus of Ddc1 following an intrachromosomal break, suggesting that the broken ends remain associated (although the resolution of this assay precludes us from determining whether some separation has occurred). It will be interesting to determine which proteins are required for this association.
Ddc1–GFP localization also serves as an indicator of spontaneous DNA damage in cells. Faint Ddc1–GFP foci are observed in undamaged wild-type cells at a frequency of 8%. Similarly, Ddc2–GFP foci are observed at a frequency of 7% in asynchronously growing cells. Presumably this frequency is fixed because of the equilibrium between rates of spontaneous damage and subsequent repair. Rad52 is required for recombination-based repair, the primary pathway used for DSB repair. rad52 cells exhibit elevated rates of spontaneous chromosome loss (Galgoczy and Toczyski 2001). When examined for Ddc1 localization, 60% of rad52 cells contain one or more Ddc1–GFP foci. Only 13% of asynchronously growing rad52 cells, however, are unable to form colonies (Toczyski et al. 1997), indicating that spontaneous DNA damage detected by Ddc1 localization is either not lethal in both daughters or is repaired by a less efficient, Rad52-independent pathway. Use of Ddc1–GFP provides a real-time read out for genomic instability with resolution at the single-cell level in a genetically tractable system.
Materials and methods
Yeast strains and constructs
cdc13-1 and cdc9-1 experiments were conducted in A364a: MatA cdc13-1 his3 ura3 leu2 trp1 and MatA cdc9-1 his7 leu2 ura3 can1, respectively. Control strains were wild type for CDC13 or CDC9. The following checkpoint mutants were generated by gene replacement: rad24::TRP1, mec3::URA3, rad17::LEU2, rad9::LEU2, mec1::TRP1, and rad53::URA3. The lethality of MEC1 and RAD53 deletions was suppressed by the sml1 mutation (Zhao et al. 1998).
The strain containing the telomeric HO site at adh4::HO site::HIS3, yJM01, used in telomeric break experiments was derived from the disomic LS20 strain constructed in Sandell and Zakian (1993), however, all strains in this paper are monosomic. The genotype of yJM01 was: MatΔ can1 ade2 trp1 his3 ura3 leu2 lys5 cyh2 ade3::galHO adh4:: HIS3::HO site. The following checkpoint mutants were generated by gene replacement: rad17::TRP1, rad24::TRP1, mec3::TRP1, sml1::URA3, mec1::TRP1, and sml1::URA3 rad53::TRP1. Intrachromosomal DSB strains contained HO sites either interrupting TRP5 on chromosome VII (see Galgoczy and Toczyski 2001) or near the SRM1 locus. The intrachromosomal break near SRM1 was generated by PCR amplification of a cassette containing the TRP1 gene and an HO site from Mata using oligonucleotides with 5′ homology to a noncoding region near SRM1. The TRP1/HO site cassette template was contained in pDG3 (Galgoczy and Toczyski 2001). The oligonucleotides used were MIDHO5, 5′-ATTTGCATAGTCAGTGTGGCGTGCATTCTCCACAGAA TCTAACTAATCAAcaatcttgatccggagcttt-3′ and MIDHO3, 5′-TGTTCTATTTTTTATTCGCGAAAATGATGCAACACCCA GTCACGATACATtcaaccactctacaaaacca-3′, which are homologous with either nucleotide 323929 to 323978 in chromosome VII plus 20 nucleotides in pDG3 or 324028 to 323979 in chromosome VII plus 20 nucleotides in pDG3, respectively. RAD52 was disrupted in yJM20 for the spontaneous damage experiment, as in Sandell and Zakian (1993).
The above strains were transformed with integrating plasmid pJAM108, containing a carboxy-terminal DDC1 fragment fused to GFP(S65T). The DDC1 fragment used for cloning was generated by Vent (Roche) High-fidelity PCR of yeast genomic DNA, using primers 5′-ggagctccaccgcggtagaattggttgaagttactgacagtaac-3′ and 5′-cgggatccgtcaaatataccccttggcttttctacttgtg-3′. This fragment was digested with SacII and BamHI and ligated into the GFP vector (donated by Jodi Nunnari, University of California, Davis). For integration at DDC1, pJAM108 was linearized with BsmI. Plasmid-based GFP tagging constructs for Rad24, Rad17, Mec3, and Rad53 were constructed similarly to pJAM108 to generate pJAM100, pJAM104, pJAM102, and pJAM106, respectively. A pRS306-based GFP-tagging construct for Rad9 was donated by W. Lim (University of California, San Francisco), and carboxy-terminal RAD9 sequences were introduced by PCR to generate pJK1. Plasmid information and specific primer sequences used for each protein tag are available on request. Ddc2 was carboxy-terminally tagged with GFP(S65T) using a PCR-based integration approach as described in Longtine et al. (1998). For viewing telomere localization in cdc13-1 cells, the Rap1–GFP construct (generously provided by Yasushi Hiraoka, K.A.R.C., Kobe, Japan) was integrated into the cdc13-1 strain above.
Chromatin immunoprecipitation
The chromatin IP technique was carried out essentially as described (Strahl-Bolsinger et al. 1997). Sonication of formaldehyde-fixed extracts resulted in DNA fragments ∼1 kb in size. Immunoprecipitations were performed for 2 h at 4°C using affinity-purified polyclonal anti-GFP antibodies (generously donated by A. Rudner, Harvard University, Boston, MA) bound to protein A agarose beads (Bio-Rad). An aliquot of each sample (input DNA) was not subjected to immunoprecipitation but was prepared and analyzed by PCR in parallel to immunoprecipitated samples. Cross-links were reversed by incubation overnight at 65°C, followed by DNA purification and semi-quantitative PCR. Primers pairs HO-1 and HO-2 are specific for the HO break site introduced at the telomere of chromosome VII in strains described above. Internal control primers act1 and tub1 were used to amplify unlinked DNA sequences at the ACT1 and TUB1 loci. All primer sequences are available on request. The predicted fragment sizes are approximately as follows: HO-1, 300 bp; HO-2, 285 bp; ACT1, 500 bp; TUB1, 150 bp. PCR products were run on 2% agarose gels containing 0.05 μg/mL EtBr, visualized and quantitated using the Bio-Rad Gel Doc 2000 and Bio-Rad Quantity One 4.0.3 software. Fold enrichment of HO-proximal sequences was calculated as the ratio of HO site:control ratio of each IP sample with its corresponding input sample.
Microcolony checkpoint assays
Positive transformants for each GFP-tagging construct were assayed by PCR and tested for checkpoint proficiency by a plate-based microcolony assay in cdc13-1 and/or HO break-induced cells. Microcolony assays in cdc13-1 cells were performed by incubating log phase cultures at 36°C for 1 h, followed by sonication and plating to prewarmed rich plates. Plates were incubated at 36°C for 2–4 h, and the number of cell bodies per microcolony were counted (n = 100) and compared with cdc13-1 and the appropriate cdc13-1 checkpoint mutant control treated in parallel. Microcolony checkpoint assays in HO-break-induced cells were performed by pregrowing cells in YM1—2% raffinose at 30°C, shifting cells to YM1—2% galactose liquid media for 2 h before sonication and plating to YM1-raffinose/galactose plates. Plates were incubated at 30°C and the number of cell bodies/microcolony counted (n = 100) at multiple time points between 2 and 8 h after plating, and compared with wild-type and appropriate checkpoint mutant controls treated in parallel.
HO cleavage assay
A time course of HO cleavage at the adh4::HO site::HIS3 in yJM20 was assessed as follows (rates of cleavage were originally reported in Sandell and Zakian 1993). Cells were grown at 30°C in YM1—2% raffinose, transferred to YM1—2% galactose and harvested at indicated time points. Genomic DNA was quantitated at OD260, 5 μg of each sample was digested with PstI, and analyzed by electrophoresis followed by Southern blotting and probing for sequences integrated on the CEN proximal side of the HO site.
Checkpoint adaptation assay
yJM20 cells were grown in preinduction media (YM1–raffinose) at 30°C and shifted to YM1 containing 2% glucose or 2% galactose for 2 h. Samples were sonicated, plated to rich plates containing glucose or raffinose and galactose, respectively, and microcolonies scored (n = 300) for cell number at the time points indicated in Figure 1. Adapted cells were scored as >2 cell bodies.
Microscopy
Microscopy was performed using an Olympus IX70 flourescence microscope with a 100×, 1.4 NA PlanApo Olympus Oil Immersion objective. GFP flourescence was detected using a Chroma FITC filter set (excit. 485/20 nm, emiss. 515/30 nm), and captured with a Hamamatsu C4742–95 CCD camera. Data were visualized using Metamoph 4.5 imaging software by Universal Imaging Corporation. Fields of cells were photographed in four to seven focal planes in succession to gather data through the depth of each nucleus. Exposure time, gain, and binning functions were held constant. Figures were prepared in Adobe Photoshop, keeping processing parameters constant within an experiment.
Acknowledgments
We thank J. Li and F. Schaufele for generous technical assistance and use of microscope equipment; D. Galgoczy and J. Kaye for generation of several strains used in this study; and members of the J. Li and D. Morgan labs for many scientific discussions. We gratefully acknowledge S. Vidwans, A. Rudner, A. Carroll, and B. Thornton for helpful comments on the manuscript. We also acknowledge institutional support from the UCSF Cancer Center and support from National Institutes of Health grant GM59691-01.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL toczyski@cc.ucsf.edu; FAX (415) 502-3179.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.903501.
References
- Bourns BD, Alexander MK, Smith AM, Zakian VA. Sir proteins, Rif proteins and Cdc13p bind Saccharomyces telomeres in vivo. Mol Cell Biol. 1998;18:5600–5608. doi: 10.1128/mcb.18.9.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtelow MA, Kaufmann SH, Karnitz LM. Retention of the human Rad9 checkpoint complex in extraction-resistant nuclear complexes after DNA damage. J Biol Chem. 2000;275:26343–26348. doi: 10.1074/jbc.M001244200. [DOI] [PubMed] [Google Scholar]
- Caspari T, Dahlen M, Kanter-Smoler G, Lindsay HD, Hofmann K, Papadimitriou K, Sunnerhagen P, Carr AM. Characterization of Schizosaccharomyces pombe Hus1: A PCNA-related protein that associates with Rad1 and Rad9. Mol Cell Biol. 2000;20:1254–1262. doi: 10.1128/mcb.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RJ, Bentley RJ, Carr AM. A Rad3–Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat Cell Biol. 1999;1:393–398. doi: 10.1038/15623. [DOI] [PubMed] [Google Scholar]
- Emili A. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol Cell. 1998;2:183–189. doi: 10.1016/s1097-2765(00)80128-8. [DOI] [PubMed] [Google Scholar]
- Galgoczy DJ, Toczyski DP. Checkpoint adaptation precedes spontaneous and damage-induced genomic instability in yeast. Mol Cell Biol. 2001;21:1710–1718. doi: 10.1128/MCB.21.5.1710-1718.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser S. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C, Erdjument-Bromage H, Tempst P, Lowndes N. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr Biol. 2000;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- Griffiths D, Barbet N, McCready S, Lehmann A, Carr A. Fission yeast rad17: A homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 1995;14:5812–5823. doi: 10.1002/j.1460-2075.1995.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L, Nasmyth K. Saccharomyces cerevisiae cell cycle mutant cdc9 is defective in DNA ligase. Nature. 1978;274:891–893. doi: 10.1038/274891a0. [DOI] [PubMed] [Google Scholar]
- Komatsu K, Miyashita T, Hang H, Hopkins KM, Zheng W, Cuddeback S, Yamada M, Lieberman HB, Wang HG. Human homologue of S. pombe Rad9 interacts with BCL-2/BCL-xL and promotes apoptosis. Nat Cell Biol. 2000;2:1–6. doi: 10.1038/71316. [DOI] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lowndes N, Murguia J. Sensing and responding to DNA damage. Curr Opin Genet Dev. 2000;10:17–25. doi: 10.1016/s0959-437x(99)00050-7. [DOI] [PubMed] [Google Scholar]
- Lydall D, Weinert T. Yeast checkpoint genes in DNA damage processing: Implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- Mossi R, Hubscher U. Clamping down on clamps and clamp loaders. Eur J Biochem. 1998;254:209–216. [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- Paciotti V, Lucchini G, Plevani P, Longhese MP. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein ddc1p, which physically interacts with mec3p. EMBO J. 1998;17:4199–4209. doi: 10.1093/emboj/17.14.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciotti V, Clerici M, Lucchini G, Longhese MP. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes & Dev. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE. Regulation of Sacchoromyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol Cell. 2001;7:293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- Rouse J, Jackson SP. LCD1: An essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J. 2000;19:5801–5812. doi: 10.1093/emboj/19.21.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Bachant J, Wang H, Hu F, Liu D, Tetzlaff M, Elledge SJ. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science. 1999;286:1166–1171. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- Sandell LL, Zakian VA. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- Smith GC, Cary RB, Lakin ND, Hann BC, Teo SH, Chen DJ, Jackson SP. Purification and DNA binding properties of the ataxia-telangiectasia gene product ATM. Proc Natl Acad Sci. 1999;96:11134–11139. doi: 10.1073/pnas.96.20.11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Sun Z, Fay DS, Marini F, Foiani M, Stern DF. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes & Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- Sun Z, Hsiao J, Fay DS, Stern DF. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- Tibbetts R, Cortez D, Brumbaugh K, Scully R, Livingston D, Elledge S, Abraham R. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes & Dev. 2000;14:2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toczyski DP, Galgoczy DJ, Hartwell LH. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell. 1997;90:1097–1106. doi: 10.1016/s0092-8674(00)80375-x. [DOI] [PubMed] [Google Scholar]
- Venclovas C, Thelen MP. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama T, Kondo T, Ando S, Matsumoto K, Sugimoto K. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:755–764. doi: 10.1128/MCB.21.3.755-764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. DNA damage checkpoints update: Getting molecular. Curr Opin Genet Dev. 1998;8:185–193. doi: 10.1016/s0959-437x(98)80140-8. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993;134:63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]