Abstract
Due to its genetic, molecular, and behavioral tractability, Drosophila has emerged as a powerful model system for studying molecular and cellular mechanisms underlying the development and function of nervous systems. The Drosophila nervous system has fewer neurons and exhibits a lower glia:neuron ratio than is seen in vertebrate nervous systems. Despite the simplicity of the Drosophila nervous system, glial organization in flies is as sophisticated as it is in vertebrates. Furthermore, fly glial cells play vital roles in neural development and behavior. In addition, powerful genetic tools are continuously being created to explore cell function in vivo. In taking advantage of these features, the fly nervous system serves as an excellent model system to study general aspects of glial cell development and function in vivo. In this article, we review and discuss advanced genetic tools that are potentially useful for understanding glial cell biology in Drosophila.
Keywords: Glial cell biology, Drosophila, genetic tools, GAL4/UAS, MARCM
INTRODUCTION
Nervous systems are composed of two major types of cells, neurons and glia. Neurons are wired into circuits to process information and control an organism’s behavior, whereas glia provide the necessary support for neural development, wiring and function. Studying both neurons and glial cells, as well as their interactions, is essential for the elucidation of nervous system development and function. However, glial studies have lagged behind neuronal studies, largely because the roles of glial cells were underestimated. Furthermore, in vertebrates, glial cells outnumber neuronal cells and existing tools are insufficient for in vivo analysis of such large population of cells.
Drosophila, with its relatively simple nervous system, its significantly smaller population of glial cells, and the growing number of genetic tools for flies, provides a good alternative to uncover essential roles of glial cells in nervous system development and function. Tools that have proven to be particularly useful include genetic cell-labeling markers, which enable specific subsets of neurons and glia to be labeled, and the binary gene expression system, which is widely used to label and manipulate both neurons and glia.
Glial cells were first systematically characterized in Drosophila in the embryonic central nervous system (CNS) using ultrastructural analysis (Jacobs and Goodman 1989). Every glial cell in the Drosophila embryonic CNS has been characterized (Ito et al. 1995; Jacobs et al. 1989; Klambt and Goodman 1991; Schmid et al. 1999; Schmidt et al. 1997). Genetic ablation of embryonic glial cells causes neuronal loss and neural wiring defects, revealing indispensable roles for glia in neuronal viability and morphogenesis (Booth et al. 2000; Hidalgo and Booth 2000). In spite of the significantly lower glia:neuron ratio in flies, the sophistication of glial cell organization in adult fly brains is on par with vertebrate brains (Awasaki et al. 2008). These glial cells play vital roles in brain functions. For example, disruption of glial activity alters various adult behaviors, including circadian rhythm, courtship behavior and longevity (Buchanan and Benzer 1993; Ewer et al. 1992; Grosjean et al. 2008; Kretzschmar et al. 1997; Suh and Jackson 2007). Loss of glial function in mature fly brains also induces neural degeneration (Kretzschmar et al. 1997; Xiong et al. 1994).
A set of research tools has enabled sophisticated studies of Drosophila glial cell development and function. In particular, the GAL4/UAS binary gene expression system has made modern genetic molecular studies of glia and neuron-glia interactions possible. This system greatly facilitates ectopic induction of any cloned gene in specific patterns in vivo. Once GAL4 drivers expressing GAL4 in a specific subset of cells are identified, expression of any gene that is placed downstream of the UAS (upstream activation sequence) can be specifically induced in these cells (Brand and Perrimon 1993). UAS-driven expression of a reporter protein (e.g. GFP) allows one to analyze cell morphology and development. Furthermore, specific cellular or genetic functions can be disrupted or activated by expressing various effector molecules. Therefore, glial cells can be not only labeled but also manipulated by glia-specific GAL4 drivers. For example, glia-specific GAL4 drivers have allowed scientists to uncover essential roles of glial cells in axon guidance during Drosophila embryonic CNS development. Ablation of longitudinal glial cells by expression of ricin toxin with subtype glial GAL4 drivers causes a guidance defect of pioneer axons (Hidalgo and Booth 2000). The axon guidance defect in mutants of the slit gene, which encodes the repulsive axon guidance ligand, is rescued by the selective expression of Slit with the midline glia GAL4 driver but not with the pan-neural GAL4 driver (Kidd et al. 1999).
For more elaborate analyses of cell types, new genetic tools that allow single-cell labeling and temporal control of GAL4 activity have been developed. In addition, other binary gene expression systems have been established in Drosophila, enabling independent transgene induction in distinct cell types (e.g. glia and neurons). By combining these new tools with the identification of diverse glial subtype-specific GAL4 drivers, more sophisticated experiments can be performed to visualize and manipulate glia in vivo, even at the single-cell level. These technical advances, as discussed here, will rapidly increase our understanding of glial cell biology.
LABELLING GLIAL CELLS WITH THE GAL4/UAS SYSTEM
GAL4 Driver Lines
In order to label, monitor and manipulate a specific subtype of glial cells in a particular developmental or functional state with the GAL4/UAS system, it is necessary to obtain various GAL4 lines that selectively express GAL4 in each glial cell of interest. Several useful GAL4 lines that label specific subsets of glia have been widely used to study glial development and functions in the embryonic and larval central nervous system (CNS), (e.g. ftz-n-GAL4, glioblasts and longitudinal glia; (Lin et al. 1995), sim-GAL4 and slit-GAL4, midline glia; (Scholz et al. 1997), htl-GAL4, a part of longitudinal glia; (Shishido et al. 1997), repo-GAL4, all glia except for midline glia (Sepp et al. 2001)).
By searching genes that are expressed in specific subsets of glial cells, it is possible to isolate regulatory elements from these genes to further generate glial subtype-specific drivers. Large-scale systematic screens to identify glial genes regulated by the glial cell missing (gcm) gene have been performed in the Drosophila embryo. In Drosophila, gcm governs the derivation of all embryonic glial cells except for the midline glia (Hosoya et al. 1995; Jones et al. 1995; Vincent et al. 1996). Loss of gcm causes deficits in glial production while ectopic gcm transforms neurons to glia, indicating that gcm controls the expression of genes required for glial cell specification. Based on this notion, expression profiling of glial genes was performed using a genome wide micro array approach by comparing animals having loss of and gain of gcm activity (Altenhein et al. 2006; Egger et al. 2002; Freeman et al. 2003). Several genes specifically expressed in a subset of glia were identified and their functions were analyzed (Bainton et al. 2005; Beckervordersandforth et al. 2008; Daneman and Barres 2005; Doherty et al. 2009; Freeman et al. 2003; Mayer et al. 2009; Schwabe et al. 2005). For example, the astrocyte-like glia-specific GAL4 line, arlm (astrocytic leucine-rich repeat molecule)-GAL4 has recently been shown to selectively label astrocyte-like glia in the adult brain (Doherty et al. 2009). Certainly, future gene expression profiling in specific subtypes or functional states of glial cells will help to establish additional useful glial GAL4 drivers. Once the relevant regulatory elements for glial subtype-specific genes are isolated, they could be also used for other binary gene expression systems (see below). Therefore, isolation of the cis-regulatory elements for the specific subtypes or states of glial cells will provide further experimental flexibility and the power to dissect the important function of each discrete glial subset.
A classical approach to isolate tissue-specific GAL4 lines is screening of GAL4 enhancer trap lines. GAL4 lines that label specific subtypes of glial cells in the embryonic CNS and the adult brain were isolated from large sets of GAL4 collections (Awasaki et al. 2008; Ito et al. 1995) (Fig. 1). Using these GAL4 lines to label subsets of glia has contributed substantially to our fundamental knowledge about the variety and organization of fly glia. In addition, by using a transposon replacement technique, it is possible to convert lacZ enhancer trap lines into the GAL4 driver lines (Sepp and Auld 1999). In fact, gcm-GAL4, which labels glial precursors, was established with this technique and has been used to study glial development (Chotard et al. 2005; Paladi and Tepass 2004).
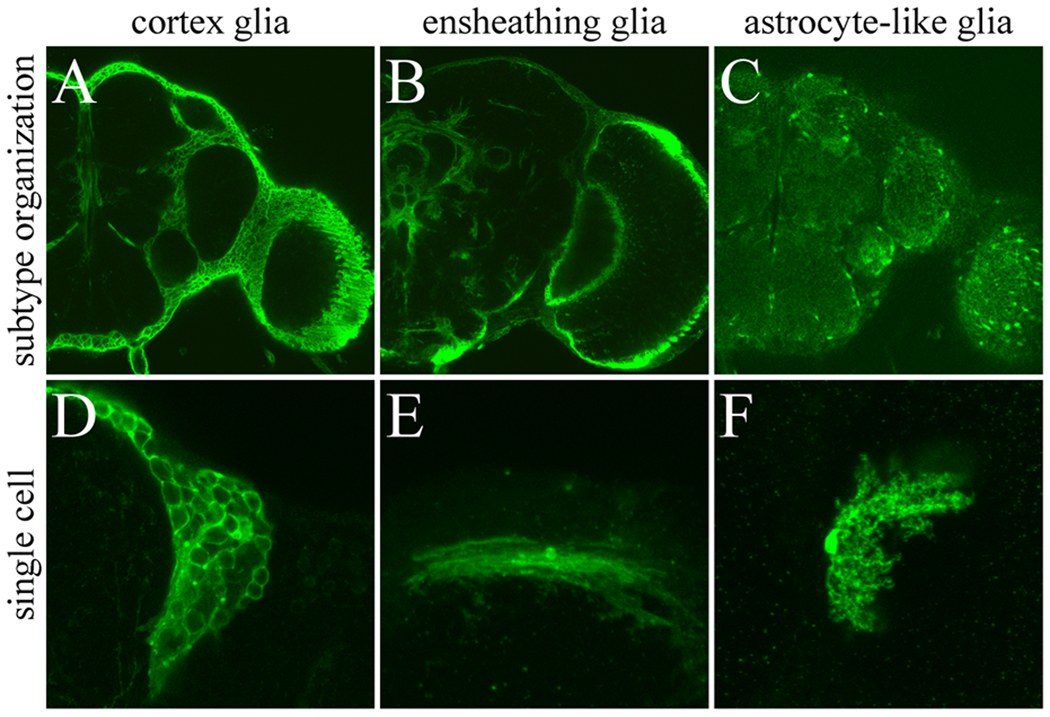
Fig. 1.
GAL4 lines label specific subtypes of glial cells in adult fly brain. Adult cortex glia (A), ensheathing glia (B), and astrocyte-like glia (C) labeled with glial subtype-specific GAL4 lines driving expression of GFP (A) and mCD8::GFP (B and C). Entire subtype glial organization (A–C) and single cell of each subtype glia labeled by UAS-FLP-out (D–F) are shown.
NP2222 (A and D), NP6520 (B and E), and NP1243 (C and F) were used for GAL4 drivers.
Visualizing and Monitoring Glial Cells with UAS-reporters
Virtually, any kind of cell marker can be expressed with the GAL4/UAS system. When glial GAL4 lines drive membrane tagged reporters, such as UAS-GAP::GFP (UAS-myristylated-GFP), UAS-mouse-CD8::GFP and UAS-rat-CD2::RFP, glial membranes are labeled with GFP or RFP (Lee and Luo 1999; Ritzenthaler et al. 2000; Yu et al. 2009). Furthermore, expression of non-membrane tethered fluorescent proteins, such as UAS-GFP and UAS-RFP can label the cytosol of glial cells. Glial nuclei are labeled with reporter proteins that contain a nuclear localization signal (nls), like UAS-nls-GFP, UAS-nls-LacZ and UAS-H2B-YFP (Bellaiche et al. 2001; Ito et al. 1997; Shiga et al. 1996).
In addition to general cell markers, various cellular organelles can be labeled or monitored through expression of organelle markers. Various reporters fused with specific subcellular localization motifs will label discrete organelles. Previously, mitochondria, the golgi apparatus, endosomes and autophagosomes have been successfully visualized and analyzed with the GAL4/UAS system in Drosophila (i.e. UAS-mito-GFP for mitochondria (Cox and Spradling 2003), UAS-Mannosidase II-eGFP for the golgi apparatus (Zheng et al. 2008), UAS-Rab4-RFP for endosomes (Sweeney et al. 2006), and UAS-GFP-LC2 and UAS-GFP-Atg5 for autophagosomes (Rusten et al. 2004)). These organelle-specific markers will serve as powerful tools to understand the intracellular dynamics of glial cell biology.
Labeling Single Glial Cells
Each subtype of glia possesses a characteristic cellular morphology, which could be reflecting its function. To reveal the morphology of individual glial cells, it is possible to use sophisticated genetic strategies in the fly to label single or a small subsets of GAL4-positive glial cells (Figs. 1D–F). Two kinds of tools are useful for this type of single cell analysis, the FLP-out system (Struhl and Basler 1993; Wong et al. 2002) and the MARCM (mosaic analysis with a repressible cellular marker) system (Lee and Luo 1999). The site-specific recombination system of the S. cervesia 2µm plasmid, which consists of a gene encoding a recombinase (FLP) and its target recombination sequences, FRT (FLP recombination target) sites, are used in both of these strategies (Golic 1991).
In the FLP-out system, a gene or a stop-codon sequence is placed between the two FRTs. This “FRT cassette” is then put between UAS and a reporter gene (UAS-FLP-out) or a promoter and GAL4 (GAL4-FLP-out) (Ito et al. 1997; Struhl and Basler 1993). Recombination between the two FRTs occurs in some cells by induction of transient FLP (e.g. under the control of the heat-shock promoter (Golic 1991)). This removes the FRT cassette (FLP-out) and allows cells to express the reporter gene or GAL4. Therefore, only a small subset of the GAL4 positive (or potentially GAL4 positive) cells is stochastically labeled with the reporter when FLP is mildly induced. While the UAS-FLP-out system can be driven by any GAL4 line, only the specific reporter gene that is located downstream FRT cassette can be expressed (Wong et al. 2002). Conversely, the GAL4-FLP-out system can drive any reporter as well as any effector genes, making this a more versatile technique to simultaneously visualize and manipulate single cells.
While classic mitotic recombination is induced by X-rays or γ rays to generate mosaic animals, FLP-dependent mitotic recombination between FRT sites has been developed as a more controllable and easy-to-use approach (Xu and Rubin 1993). To label one sister cell induced by mitotic recombination, the MARCM system utilizes GAL80, which antagonizes GAL4 (Lee and Luo 1999). GAL80 suppresses expression of the UAS-linked genes even in the presence of GAL4. The FLP-induced mitotic recombination between the FRT sequences removes the GAL80 gene in one of the sister cells, allowing UAS-linked genes to be expressed in these GAL80 minus cells. When the mitotic recombination is induced in the dividing cells, only one fraction of progeny would be labeled stochastically.
Compared to the FLP-out system, however, MARCM requires greater effort to assemble multiple components (i.e. transgenes). In addition, the proper conditions to induce mitotic recombination must be determined because this labeling is depending on the cell proliferation pattern during development. Therefore, the FLP-out system is a more convenient and simpler method to visualize single cell morphology. Although, the MARCM system provides certain notable benefits for cell lineage and mutant analysis (see below).
Using a similar FLP-out concept, the multicolor fluorescent labeling system, called Brainbow, has been developed in mice (Livet 2007). This system induces stochastic expression of multiple fluorescent-protein markers from a single transgene using a Cre-mediated excision between pairs of incompatible lox sites. Under the control of a single promoter, cells are stochastically labeled with multicolor fluorescent molecules. If this technology is applied in the GAL4/UAS system, it will be feasible to label GAL4 positive glial cells with different fluorescent reporters, a useful tool to explore untouched questions such as glial tiling.
MANIPULATION OF GLIAL CELLS
Manipulation of Gene Functions
By using the GAL4/UAS system, it is possible to manipulate particular gene and cellular functions in a specific subset of glial cells with effector molecules. In Drosophila, many mutant proteins that disrupt or activate certain gene or cellular functions have been identified, including dominant-negative and constitutively-active forms of receptors and signaling molecules. By expressing such mutant proteins’ effectors, it is possible to induce suppression or over-activation of specific molecular cascades in glial cells. Multiple effector and reporter molecules can be co-expressed under the same GAL4 driver. Using this strategy, essential functions of EGF, FGF and PDGF signaling in glia on cell-proliferation, apoptosis, wrapping and migration have been identified (Bergmann et al. 2002; Franzdottir et al. 2009; Griffiths and Hidalgo 2004; Hidalgo et al. 2001; Learte et al. 2008; Sepp and Auld 2003). In addition, a human glioma model has recently been successfully established by constitutive co-activation of two signaling pathways, EGFR-Ras and PI3K in Drosophila glia with the GAL4/UAS system (Read et al. 2009; Witte et al. 2009).
The gene silencing technique using RNA interference (RNAi) has been exploited in many fields of experimental biology. In Drosophila, the transgenic RNAi approach, which induces expression of double strand RNA (dsRNA) targeting 300 to 600bp of unique gene fragment under the control of UAS, has been established (Fortier and Belote 2000; Kennerdell and Carthew 2000; Lam and Thummel 2000; Martinek and Young 2000). Using this technique, it is possible to disrupt the activity of individual genes with spatial and temporal control. This is extremely beneficial to analyze the function of a particular gene in specific types of cells. The first use of this approach to study glial cell biology in flies was conducted with UAS-vein-RNAi to show that a subset of longitudinal glia in the embryo requires the binding of Vein to EGFR to maintain glial survival (Hidalgo et al. 2001). Also, using this method, glial requirement of engulfing genes in the clearance of degenerating axons was demonstrated (Awasaki et al. 2006; MacDonald et al. 2006; Ziegenfuss et al. 2008). Furthermore, an impressive collection of transgenic RNAi lines is publically available via three public stock centers, Harvard (3,800 lines), Mishima (11,000 lines), and Viena (22,000 lines) (Dietzl et al. 2007; Ni et al. 2009). Collectively, about 90% of fly genes in the genome can be knocked-out with these libraries. It is, therefore, feasible to identify genes that affect glial development or function by screening transgenic RNAi lines using specific glial GAL4 lines.
Although these RNAi lines might have non-specific targeting (off-targeting) effects, the specificity of the targeting can be verified with the advanced RNAi technique, the microRNA (miRNA)-based-RNAi. In this method, 21 or 22bp miRNA targeting to the particular gene is expressed by the GAL4/UAS system (Chen et al. 2007; Haley et al. 2008). Compared to the conventional RNAi approach, in which hundreds of bp of dsRNAs are expressed, higher specificity is expected. Finally, the effects of miRNA can be rescued by co-expression of a miRNA-resistant gene, which possesses altered codons in the miRNA targeting sites. These tools should facilitate identification of genes that regulate glial development and function.
Manipulation of Cellular Functions
One of the simplest methods to understand the role of a specific type of cell is to perform cell type-specific ablation. In the fly, it is possible to ablate cells by the induction of apoptosis with ectopic expression of the cell death genes, reaper, hid (head involution defectives) and grim (Chen et al. 1996; Grether et al. 1995; White et al. 1996). Whereas the expression of these cell death genes does not always induces apoptosis because of the susceptibility and state of cells, several subtypes of glial cells in the developing CNS and brain are removed by the expression of the cell death genes, rpr and hid (Booth et al. 2000; Spindler et al. 2009). Induction of the ricin toxin can also ablate cells (Moffat et al. 1992). In fact, the function of embryonic glial cells has been analyzed by specifically ablating glia with ricin (Booth et al. 2000; Hidalgo et al. 1995). Therefore, it is possible to explore the neuron-glia interaction even in the adult nervous system by ablating a specific glial subtype with the temporal control of the transgene expression (see below). Conversely, it is also possible to suppress apoptosis specifically in glial cells by the targeted expression of Drosophila apoptosis inhibitor protein, DIAP, or by the baculovirus apoptosis preventing molecule, P35. In the embryonic CNS, some glial subtypes undergo apoptosis during development to adjust their numbers to neurons (Fichelson and Gho 2003; Hidalgo et al. 2001; Kinrade et al. 2001; Sonnenfeld and Jacobs 1995; Stemerdink and Jacobs 1997). In addition, mutation of cell death genes suppressed glial apoptosis, resulting in an increased number of glial cells (Rogulja-Ortmann et al. 2007). To further the understanding of apoptosis of particular glial subtypes later in development, targeted expression of an apoptosis inhibitor would be useful. By using these reagents, it is also possible to examine whether certain glial cell death is caused by apoptosis or other mechanisms.
Glial cell-specific ablation would give insight into the requirement of glial cells in certain aspects of nervous system development and function. However, it is difficult to know what kind of glial cellular function is required for these roles. The effector molecules that affect specific cellular function can also be useful to analyze glial cells. To manipulate neural activity, the temperature-sensitive allele of Shibire (Shibirets) has been used to study synaptogenesis and behavior (Kitamoto 2001; Kitamoto 2002; Waddell et al. 2000). Shibirets inhibits endocytosis under the restrictive-temperature, resulting in the suppression of synaptic transmission. Although it is not clear that fly glia possess glial transmitters similar to those of mammals, expression of the Shibirets can suppress all Shibire-dependent cellular functions, including endocytosis. It has been shown that phagocytic function of glial cells is efficiently suppressed by Shibirets in the developing and adult nervous systems at the restrictive-temperature (Awasaki and Ito 2004; Doherty et al. 2009; Hebbar and Fernandes 2010). In Drosophila, Tetanus toxin Light chain (TeTxLC) is also used to block synaptic transmission. In the synapse, TeTxLC cleaves Synaptobrevin, which is essential for exocytosis of synaptic vesicles, resulting in inhibition of the neurotransmitter release (Sweeney et al. 1995). To address the issue of glial transmission in the fly, these effectors will be useful and valuable tools.
Temporal control of the transgene expression
The role of specific genes in unique glial cell types can be assessed using glia-specific GAL4 drivers. However, the expression pattern of some GAL4 lines is dynamic throughout development. Thus, constitutive expression of effectors with GAL4 driver lines can produce misleading results. To analyze genetic and cellular functions in developing or mature glial cells, it is necessary to manipulate expression of effectors only at the appropriate developmental stage. Temporal regulation of transgenes allows us to disrupt or alter the function of a gene of interest during later stages of glial development but retain normal function of the gene during the critical stages of early gliogenesis. Several new tools enable temporal control of transgene expression.
The temporal and regional gene expression targeting (TARGET) system enables activation of GAL4 only at the restrictive temperature (McGuire et al. 2003). By constitutively expressing a temperature-sensitive repressor for GAL4, GAL80ts, the TARGET system can regulate expression of UAS-gene under temperature control. One advantage of this strategy is that any GAL4 driver and UAS-gene can be easily combined to control both the spatial and temporal expression of transgenic effectors.
The GeneSwitch system also enables the control of gene expression in a temporal manner (Osterwalder et al. 2001). In this system, the ligand-binding domain of the mutant progesterone receptor is fused to GAL4 to generate a ligand-inducible chimeric GAL4 activator. This chimeric molecule undergoes a conformational change only in the presence of hormone, resulting in binding to the UAS sequence and activation of transcription of reporter or effector genes. Unlike the TARGET system, which utilizes any available GAL4 driver line, the GeneSwitch system requires establishment of new transgenic lines that possess a GeneSwitch cassette under the cell-type specific promoter. Enhancer trap screening with the GeneSwitch cassette has been conducted and there are several readily available GeneSwitch lines that label glial cells following hormonal induction (Nicholson et al. 2008).
Multiple binary gene expression systems
By using the GAL4/UAS system and its variants, gene and cellular functions can be manipulated in glial cells with spatial and temporal control. The cell-autonomous effects of such manipulations can be analyzed by the co-expression of reporter molecules with the same GAL4 driver. On the other hand, to analyze extrinsic effects on glial growth and function, other labeling systems are required. For the analysis of neuron-glia interactions, it is necessary to assess the effects of glia on neurons by using an independent labeling system other than GAL4/UAS (e.g. antibody staining, LacZ enhancer trap lines, promoter GFP lines, etc). It also has been shown that proper development and function of glial cells is controlled by neurons. How can these neuron-glia interactions be manipulated or visualized? Assessment of the interaction is largely dependent on methods that will allow us to monitor extrinsic effects. Multiple binary gene expression systems, which can manipulate and label different types of cells independently, are ideal for analyzing neuron-glia interactions. Below, we describe two types of GAL4-independent binary gene expression systems that have been recently established in Drosophila and allow the simultaneous labeling and manipulation of different cell types in vivo.
First, in the LexA/lexAop binary gene expression system the DNA binding domain of E. coli LexA repressor (LexA) is fused to the VP16 acidic activation domain, LexA::VP16, or GAL4 activation domain, LexA::GAD, to activate gene expression (Lai and Lee 2006) (Fig. 2). Reporter and effector genes are placed downstream of the LexA binding sequences (lexAop). LexA::GAD is suppressed by GAL80, while LexA::VP16 is unaffected, which provides the option of regulating this system in a temporal manner. Another binary expression system is the Q system, which utilizes regulatory genes from the Neurospora qa gene cluster (Potter et al. 2010). The transcriptional activator QF induces expression of genes placed downstream of QUAS and is repressed by the repressor QS.
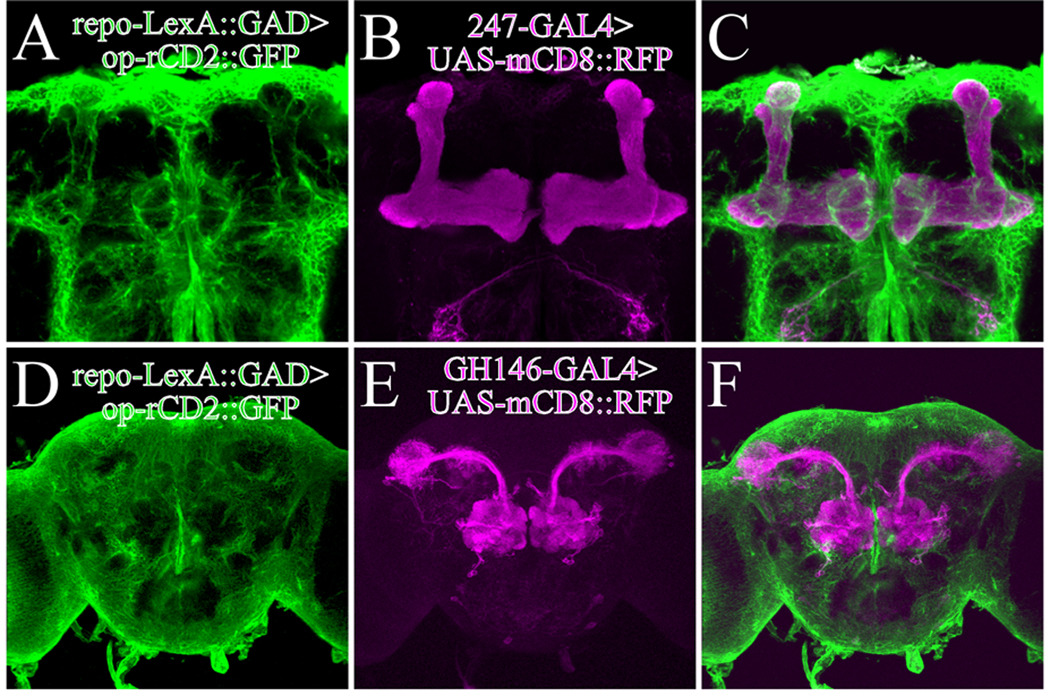
Fig. 2.
Glial cells and neuronal subset can be independently labeled with two binary systems. Glial cells labeled with repo-LexA::GAD driven rCD2::GFP (Green) (A and D). Mushroom body neurons labeled with 247-GAL4 driven mCD8::RFP (B) and olfactory projection neurons labeled with GH146-GAL4 driven mCD8::RFP (C). Merged images are shown in [C] and [F].
Theoretically, any GAL4 driver can be adapted to generate a LexA or QF driver by using the transposon replacement technique or by borrowing the key cell-type specific gene regulatory fragments; notably, making the reporter and effector lines (i.e. lexAop or QUAS lines) is very straightforward. Therefore, analysis using these multiple binary expression systems has great potential for studying not only neuron-glia but also glia-glia interactions. In addition, despite the complexity of the genetics, it is feasible to combine three systems together to label axons, dendrites and glial cells in the same organism. This approach might provide tremendous opportunity to study the tripartite synapse (Halassa et al. 2007), which is unexplored in Drosophila.
It is also possible to tightly refine transgene expression with the multiple binary expression system. By using combinations of drivers and suppressors, the expression of transgenes is distilled in overlapping areas to either activate or inhibit expression in discrete subsets of cells (Potter et al. 2010). For example, for GAL4 lines that drive expression in both neurons and glial cells, the GAL4 activation in neurons is selectively suppressed with neuron-specific expression of LexA- or Q- driven GAL80 (i.e. op-GAL80 or QUAS-GAL80).
MOSAIC ANALYSIS WITH THE MARCM SYSTEM
Conventional MARCM system
As mentioned above, the MARCM system is a very powerful method for labeling and/or manipulating a small subset of GAL4-positive cells. The MARCM system allows one to generate labeled homozygous mutant clones in an otherwise unlabeled and heterozygous background (Lee and Luo 1999). Thus, even if a gene mutation causes embryonic lethality in the homozygote, the effect of mutant clones in particular cells can be analyzed at later developmental stages using MARCM. Taking advantage of this system, the effect of loss-of-function of gcm on post-embryonic gliogenesis has been analyzed (Awasaki et al. 2008). Using MARCM analysis, many genes having essential function in neural development have been identified through mutant screening (Lee et al. 2000a; Lee et al. 2000b; Wang et al. 2004; Zheng et al. 2003; Zhu et al. 2006). So far, this approach has not been used to identify genes that are functionally required for glial cell development, but by using glial subtype-specific GAL4 lines, it will be possible to isolate critical genes required for glial cell formation and function. MARCM analysis will be a powerful approach to complement loss-of-function mutant and cell type-specific gene silencing analysis to understand the molecular underpinnings of glial cell function.
The MARCM system is also a powerful method to analyze cell lineage and proliferation. Because of the enormous number of glial cells and long-distance migration that occurs during development, cell lineage analysis of glial cells is quite difficult in mammalian models. In contrast, in Drosophila, both neurons and glial cells in the CNS are derived from a limited number of progenitor cells (Doe 1992; Schmid et al. 1999; Schmidt et al. 1997; Urbach and Technau 2003). Therefore, glial lineage in Drosophila can be analyzed much more easily. Previously, focusing on the post-embryonic development of adult brain glia, proliferation patterns were analyzed using the MARCM system with a pan-glial GAL4 driver, repo-GAL4 (Awasaki et al. 2008). This study indicates that three out of five major subtypes of adult glia are extensively generated during the larval period. Further systematic MARCM analysis using various glial subtype-specific GAL4 drivers will enable a better understanding of the details of cell lineage and division patterns of diverse glial subtypes.
Variants of MARCM system
In addition to the conventional system, useful variants of MARCM have been established. The dual expression control MARCM system allows us to perform mosaic analysis with two different populations of cells, labeled with GAL4/UAS and LexA::GAD/lexAop (Lai and Lee 2006). By using this system, it is possible to determine whether two different subtypes of glial cells are derived from same progenitor or not. In fact, with this strategy, it was revealed that two different subtypes of adult neuropile glia -- ensheathing glia and astrocyte-like glia -- are generated from different precursors in post-embryonic development (Awasaki et al. 2008).
In the conventional MARCM system, while one set of sister clones derived from dividing cells is labeled by expression of a reporter, the other set remains unlabeled. New versions of MARCM system, including Twin-spot MARCM system and coupled MARCM system, allow us to visualize both sets of sister clones with different reporters, such as GFP and RFP (Potter et al. 2010; Yu et al. 2009). These systems will give more detailed insight into cell lineage and division patterns of glial cells. When dividing larval glial precursors are labeled with the twin-spot MARCM system in embryos, twin-spot clones of larval glia are labeled side-by-side (Fig. 3), suggesting that these twin glial cells or clones are generated by symmetric cell-division. These methods can also be used to evaluate the effects of gene mutations: when mutant and wild-type chromosomes are linked with different reporters, the homozygous mutant and wild-type sister clones can be visualized with different colors (e.g. GFP and RFP) and the consequence of the mutation is easily evaluated.
Fig. 3.
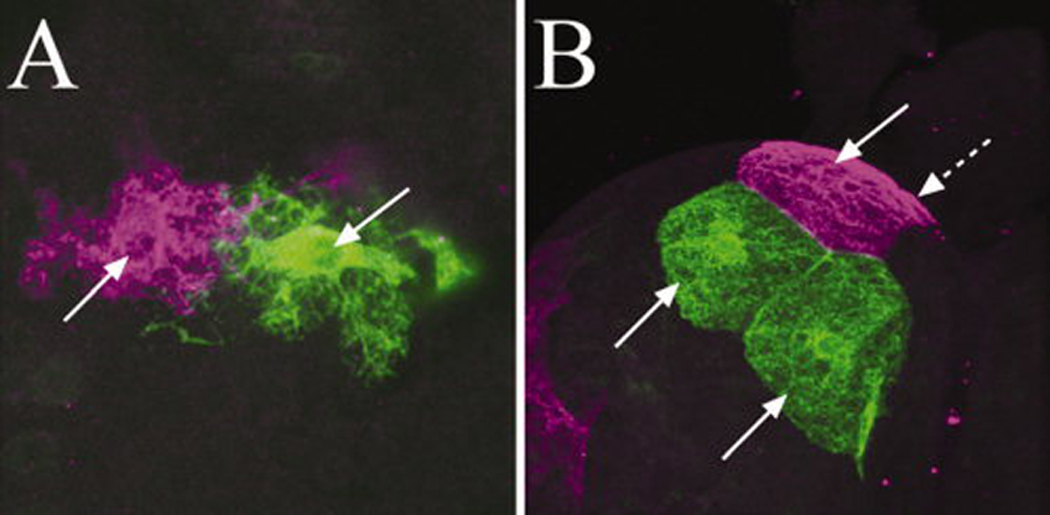
Glial cells labeled with twin-spot MARCM. Twin-spot clones of larval astrocyte-like glia in the neuropile (A) and larval subperineurial glia on the brain surface (B). Note in [B], two green and two magenta (one is shown by arrow with a broken line) subperinurial glial cells were labeled in cluster.
CONCLUSIONS
Drosophila studies have contributed significantly to the field of molecular and cellular neurobiology and to our understanding of the fundamental mechanisms underlying neural development and function (e.g. cell fate specification, neurite genesis, synapse formation, learning and memory and innate behavior). As shown in other review articles in this issue, Drosophila also provides a valuable model system to study glial cell biology. Of note, most of these studies yield cellular and molecular insights into the in vivo development and function of glia. As additional powerful genetic tools become available, Drosophila will be an outstanding model system for making rapid advances in our understanding of glial cell biology. These advances will uncover the important roles these cells play in brain formation and function.
Table 1.
List of useful genetic tools for glial biology of Drosophila.
| Tools | Description | References |
|---|---|---|
| GAL4/UAS system | ||
| GAL4 | Drives transgenes down stream of UAS | Brand and Perrimon 1993 |
| UAS | Allows express transgenes placed in downstream by the control of GAL4 | Brand and Perrimon 1993 |
| GAL80 | Suppresses of GAL4 transcriptional activity | Lee and Luo 1999 |
| GAL4/UAS related systems | ||
| FLP-out system | Labels a subset of GAL4 positive cells by the FLP-FRT recombination in the FLP-out cassette. Two types; GAL4-FLP-out and UAS-FLP-out | Ito et al. 1997; Struhl and Basler 1993 |
| GeneSwitch | Controls expression of UAS-genes by the hormone, RU486 (mifepristone) | Osterwalder et al. 2001 |
| TARGET system | Temporal control of GAL4/UAS system with GAL80ts (temperature sensitive GAL4 repressor). | McGuire et al. 2003 |
| Other binary systems | ||
| LexA/lexAop | LexA::VP16 or LexA::GAD (drivers) drives genes under the lexAop (reporter/effector). LexA::GAD is suppressed by GAL80 | Lai and Lee 2006 |
| Q system | QF (driver) drives expression of genes under QUAS (reporter/effector). QF is suppressed by QS (repressor) | Potter et al. 2010 |
| MARCM analysis | ||
| MARCM (conventional) | GAL4 drives UAS-genes in the GAL80 minus sister clone(s) induced by somatic recombination | Lee and Luo 1999 |
| Dual Expression Controlled MARCM | GAL4 and LexA::GAD drive UAS-genes and lexAop-genes, respectively, in the GAL80 minus sister clones | Lai and Lee 2006 |
| Twin Spot MARCM | Different reporters of GAL4/UAS system label twin spot clones derived from mitotic recombination, respectively. | Yu et al. 2009 |
| Coupled MARCM | Reporters of GAL4/UAS and Q system label twin spot clones derived from mitotic recombination, respectively. | Potter et al. 2010 |
ACKNOWLEDGMENTS
We thank T. Wolff, M. A. Logan and C.-F. Kao for critical reading of the manuscript. We thank National Institutes of Health and Howard Hughes Medical Institute for grant support.
REFERENCES
- Altenhein B, Becker A, Busold C, Beckmann B, Hoheisel JD, Technau GM. Expression profiling of glial genes during Drosophila embryogenesis. Dev Biol. 2006;296(2):545–560. doi: 10.1016/j.ydbio.2006.04.460. [DOI] [PubMed] [Google Scholar]
- Awasaki T, Ito K. Engulfing action of glial cells is required for programmed axon pruning during Drosophila metamorphosis. Curr Biol. 2004;14(8):668–677. doi: 10.1016/j.cub.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Awasaki T, Lai SL, Ito K, Lee T. Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J Neurosci. 2008;28(51):13742–13753. doi: 10.1523/JNEUROSCI.4844-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasaki T, Tatsumi R, Takahashi K, Arai K, Nakanishi Y, Ueda R, Ito K. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron. 2006;50(6):855–867. doi: 10.1016/j.neuron.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Bainton RJ, Tsai LT, Schwabe T, DeSalvo M, Gaul U, Heberlein U. moody encodes two GPCRs that regulate cocaine behaviors and blood-brain barrier permeability in Drosophila. Cell. 2005;123(1):145–156. doi: 10.1016/j.cell.2005.07.029. [DOI] [PubMed] [Google Scholar]
- Beckervordersandforth RM, Rickert C, Altenhein B, Technau GM. Subtypes of glial cells in the Drosophila embryonic ventral nerve cord as related to lineage and gene expression. Mech Dev. 2008;125(5–6):542–557. doi: 10.1016/j.mod.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Bellaiche Y, Gho M, Kaltschmidt JA, Brand AH, Schweisguth F. Frizzled regulates localization of cell-fate determinants and mitotic spindle rotation during asymmetric cell division. Nat Cell Biol. 2001;3(1):50–57. doi: 10.1038/35050558. [DOI] [PubMed] [Google Scholar]
- Bergmann A, Tugentman M, Shilo BZ, Steller H. Regulation of cell number by MAPK-dependent control of apoptosis: a mechanism for trophic survival signaling. Dev Cell. 2002;2(2):159–170. doi: 10.1016/s1534-5807(02)00116-8. [DOI] [PubMed] [Google Scholar]
- Booth GE, Kinrade EF, Hidalgo A. Glia maintain follower neuron survival during Drosophila CNS development. Development. 2000;127(2):237–244. doi: 10.1242/dev.127.2.237. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Buchanan RL, Benzer S. Defective glia in the Drosophila brain degeneration mutant drop-dead. Neuron. 1993;10(5):839–850. doi: 10.1016/0896-6273(93)90200-b. [DOI] [PubMed] [Google Scholar]
- Chen CH, Huang H, Ward CM, Su JT, Schaeffer LV, Guo M, Hay BA. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science. 2007;316(5824):597–600. doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]
- Chen P, Nordstrom W, Gish B, Abrams JM. grim, a novel cell death gene in Drosophila. Genes Dev. 1996;10(14):1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- Chotard C, Leung W, Salecker I. glial cells missing and gcm2 cell autonomously regulate both glial and neuronal development in the visual system of Drosophila. Neuron. 2005;48(2):237–251. doi: 10.1016/j.neuron.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Cox RT, Spradling AC. A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development. 2003;130(8):1579–1590. doi: 10.1242/dev.00365. [DOI] [PubMed] [Google Scholar]
- Daneman R, Barres BA. The blood-brain barrier--lessons from moody flies. Cell. 2005;123(1):9–12. doi: 10.1016/j.cell.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448(7150):151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116(4):855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- Doherty J, Logan MA, Tasdemir OE, Freeman MR. Ensheathing glia function as phagocytes in the adult Drosophila brain. J Neurosci. 2009;29(15):4768–4781. doi: 10.1523/JNEUROSCI.5951-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Leemans R, Loop T, Kammermeier L, Fan Y, Radimerski T, Strahm MC, Certa U, Reichert H. Gliogenesis in Drosophila: genome-wide analysis of downstream genes of glial cells missing in the embryonic nervous system. Development. 2002;129(14):3295–3309. doi: 10.1242/dev.129.14.3295. [DOI] [PubMed] [Google Scholar]
- Ewer J, Frisch B, Hamblen-Coyle MJ, Rosbash M, Hall JC. Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells' influence on circadian behavioral rhythms. J Neurosci. 1992;12(9):3321–3349. doi: 10.1523/JNEUROSCI.12-09-03321.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichelson P, Gho M. The glial cell undergoes apoptosis in the microchaete lineage of Drosophila. Development. 2003;130(1):123–133. doi: 10.1242/dev.00198. [DOI] [PubMed] [Google Scholar]
- Fortier E, Belote JM. Temperature-dependent gene silencing by an expressed inverted repeat in Drosophila. Genesis. 2000;26(4):240–244. doi: 10.1002/(sici)1526-968x(200004)26:4<240::aid-gene40>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Franzdottir SR, Engelen D, Yuva-Aydemir Y, Schmidt I, Aho A, Klambt C. Switch in FGF signalling initiates glial differentiation in the Drosophila eye. Nature. 2009;460(7256):758–761. doi: 10.1038/nature08167. [DOI] [PubMed] [Google Scholar]
- Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ. Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron. 2003;38(4):567–580. doi: 10.1016/s0896-6273(03)00289-7. [DOI] [PubMed] [Google Scholar]
- Golic KG. Site-specific recombination between homologous chromosomes in Drosophila. Science. 1991;252(5008):958–961. doi: 10.1126/science.2035025. [DOI] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9(14):1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Griffiths RL, Hidalgo A. Prospero maintains the mitotic potential of glial precursors enabling them to respond to neurons. EMBO J. 2004;23(12):2440–2450. doi: 10.1038/sj.emboj.7600258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean Y, Grillet M, Augustin H, Ferveur JF, Featherstone DE. A glial amino-acid transporter controls synapse strength and courtship in Drosophila. Nat Neurosci. 2008;11(1):54–61. doi: 10.1038/nn2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halassa MM, Fellin T, Haydon PG. The tripartite synapse: roles for gliotransmission in health and disease. Trends Mol Med. 2007;13(2):54–63. doi: 10.1016/j.molmed.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Haley B, Hendrix D, Trang V, Levine M. A simplified miRNA-based gene silencing method for Drosophila melanogaster. Dev Biol. 2008;321(2):482–490. doi: 10.1016/j.ydbio.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbar S, Fernandes JJ. Glial remodeling during metamorphosis influences the stabilization of motor neuron branches in Drosophila. Dev Biol. 2010;340(2):344–354. doi: 10.1016/j.ydbio.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Hidalgo A, Booth GE. Glia dictate pioneer axon trajectories in the Drosophila embryonic CNS. Development. 2000;127(2):393–402. doi: 10.1242/dev.127.2.393. [DOI] [PubMed] [Google Scholar]
- Hidalgo A, Kinrade EF, Georgiou M. The Drosophila neuregulin vein maintains glial survival during axon guidance in the CNS. Dev Cell. 2001;1(5):679–690. doi: 10.1016/s1534-5807(01)00074-0. [DOI] [PubMed] [Google Scholar]
- Hidalgo A, Urban J, Brand AH. Targeted ablation of glia disrupts axon tract formation in the Drosophila CNS. Development. 1995;121(11):3703–3712. doi: 10.1242/dev.121.11.3703. [DOI] [PubMed] [Google Scholar]
- Hosoya T, Takizawa K, Nitta K, Hotta Y. glial cells missing: a binary switch between neuronal and glial determination in Drosophila. Cell. 1995;82(6):1025–1036. doi: 10.1016/0092-8674(95)90281-3. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124(4):761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Ito K, Urban J, Technau G. Distribution, classification, and development of Drosophila glial cells in the late embryonic and early larval ventral nerve cord. Development Genes and Evolution. 1995;204(5):284–307. doi: 10.1007/BF02179499. [DOI] [PubMed] [Google Scholar]
- Jacobs JR, Goodman CS. Embryonic development of axon pathways in the Drosophila CNS. I. A glial scaffold appears before the first growth cones. J Neurosci. 1989;9(7):2402–2411. doi: 10.1523/JNEUROSCI.09-07-02402.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JR, Hiromi Y, Patel NH, Goodman CS. Lineage, migration, and morphogenesis of longitudinal glia in the Drosophila CNS as revealed by a molecular lineage marker. Neuron. 1989;2(6):1625–1631. doi: 10.1016/0896-6273(89)90051-2. [DOI] [PubMed] [Google Scholar]
- Jones BW, Fetter RD, Tear G, Goodman CS. glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell. 1995;82(6):1013–1023. doi: 10.1016/0092-8674(95)90280-5. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol. 2000;18(8):896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- Kidd T, Bland KS, Goodman CS. Slit is the midline repellent for the robo receptor in Drosophila. Cell. 1999;96(6):785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- Kinrade EF, Brates T, Tear G, Hidalgo A. Roundabout signalling, cell contact and trophic support confine longitudinal glia and axons in the Drosophila CNS. Development. 2001;128(2):207–216. doi: 10.1242/dev.128.2.207. [DOI] [PubMed] [Google Scholar]
- Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol. 2001;47(2):81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- Kitamoto T. Conditional disruption of synaptic transmission induces male-male courtship behavior in Drosophila. Proc Natl Acad Sci U S A. 2002;99(20):13232–13237. doi: 10.1073/pnas.202489099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klambt C, Goodman CS. The diversity and pattern of glia during axon pathway formation in the Drosophila embryo. Glia. 1991;4(2):205–213. doi: 10.1002/glia.440040212. [DOI] [PubMed] [Google Scholar]
- Kretzschmar D, Hasan G, Sharma S, Heisenberg M, Benzer S. The swiss cheese mutant causes glial hyperwrapping and brain degeneration in Drosophila. J Neurosci. 1997;17(19):7425–7432. doi: 10.1523/JNEUROSCI.17-19-07425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006;9(5):703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- Lam G, Thummel CS. Inducible expression of double-stranded RNA directs specific genetic interference in Drosophila. Curr Biol. 2000;10(16):957–963. doi: 10.1016/s0960-9822(00)00631-x. [DOI] [PubMed] [Google Scholar]
- Learte AR, Forero MG, Hidalgo A. Gliatrophic and gliatropic roles of PVF/PVR signaling during axon guidance. Glia. 2008;56(2):164–176. doi: 10.1002/glia.20601. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22(3):451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lee T, Marticke S, Sung C, Robinow S, Luo L. Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron. 2000a;28(3):807–818. doi: 10.1016/s0896-6273(00)00155-0. [DOI] [PubMed] [Google Scholar]
- Lee T, Winter C, Marticke SS, Lee A, Luo L. Essential roles of Drosophila RhoA in the regulation of neuroblast proliferation and dendritic but not axonal morphogenesis. Neuron. 2000b;25(2):307–316. doi: 10.1016/s0896-6273(00)80896-x. [DOI] [PubMed] [Google Scholar]
- Lin DM, Auld VJ, Goodman CS. Targeted neuronal cell ablation in the Drosophila embryo: pathfinding by follower growth cones in the absence of pioneers. Neuron. 1995;14(4):707–715. doi: 10.1016/0896-6273(95)90215-5. [DOI] [PubMed] [Google Scholar]
- Livet J. The brain in color: transgenic "Brainbow" mice for visualizing neuronal circuits. Med Sci (Paris) 2007;23(12):1173–1176. doi: 10.1051/medsci/200723121173. [DOI] [PubMed] [Google Scholar]
- MacDonald JM, Beach MG, Porpiglia E, Sheehan AE, Watts RJ, Freeman MR. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron. 2006;50(6):869–881. doi: 10.1016/j.neuron.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Martinek S, Young MW. Specific genetic interference with behavioral rhythms in Drosophila by expression of inverted repeats. Genetics. 2000;156(4):1717–1725. doi: 10.1093/genetics/156.4.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer F, Mayer N, Chinn L, Pinsonneault RL, Kroetz D, Bainton RJ. Evolutionary conservation of vertebrate blood-brain barrier chemoprotective mechanisms in Drosophila. J Neurosci. 2009;29(11):3538–3550. doi: 10.1523/JNEUROSCI.5564-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302(5651):1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Moffat KG, Gould JH, Smith HK, O'Kane CJ. Inducible cell ablation in Drosophila by cold-sensitive ricin A chain. Development. 1992;114(3):681–687. doi: 10.1242/dev.114.3.681. [DOI] [PubMed] [Google Scholar]
- Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H, et al. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 2009;182(4):1089–1100. doi: 10.1534/genetics.109.103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson L, Singh GK, Osterwalder T, Roman GW, Davis RL, Keshishian H. Spatial and temporal control of gene expression in Drosophila using the inducible GeneSwitch GAL4 system. I. Screen for larval nervous system drivers. Genetics. 2008;178(1):215–234. doi: 10.1534/genetics.107.081968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A. 2001;98(22):12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paladi M, Tepass U. Function of Rho GTPases in embryonic blood cell migration in Drosophila. J Cell Sci. 2004;117(Pt 26):6313–6326. doi: 10.1242/jcs.01552. [DOI] [PubMed] [Google Scholar]
- Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141(3):536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RD, Cavenee WK, Furnari FB, Thomas JB. A drosophila model for EGFR-Ras and PI3K-dependent human glioma. PLoS Genet. 2009;5(2):e1000374. doi: 10.1371/journal.pgen.1000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzenthaler S, Suzuki E, Chiba A. Postsynaptic filopodia in muscle cells interact with innervating motoneuron axons. Nat Neurosci. 2000;3(10):1012–1017. doi: 10.1038/79833. [DOI] [PubMed] [Google Scholar]
- Rogulja-Ortmann A, Luer K, Seibert J, Rickert C, Technau GM. Programmed cell death in the embryonic central nervous system of Drosophila melanogaster. Development. 2007;134(1):105–116. doi: 10.1242/dev.02707. [DOI] [PubMed] [Google Scholar]
- Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7(2):179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development. 1999;126(21):4653–4689. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev Biol. 1997;189(2):186–204. doi: 10.1006/dbio.1997.8660. [DOI] [PubMed] [Google Scholar]
- Scholz H, Sadlowski E, Klaes A, Klambt C. Control of midline glia development in the embryonic Drosophila CNS. Mech Dev. 1997;62(1):79–91. doi: 10.1016/s0925-4773(96)00652-1. [DOI] [PubMed] [Google Scholar]
- Schwabe T, Bainton RJ, Fetter RD, Heberlein U, Gaul U. GPCR signaling is required for blood-brain barrier formation in drosophila. Cell. 2005;123(1):133–144. doi: 10.1016/j.cell.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Sepp KJ, Auld VJ. Conversion of lacZ enhancer trap lines to GAL4 lines using targeted transposition in Drosophila melanogaster. Genetics. 1999;151(3):1093–1101. doi: 10.1093/genetics/151.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepp KJ, Auld VJ. Reciprocal interactions between neurons and glia are required for Drosophila peripheral nervous system development. J Neurosci. 2003;23(23):8221–8230. doi: 10.1523/JNEUROSCI.23-23-08221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepp KJ, Schulte J, Auld VJ. Peripheral glia direct axon guidance across the CNS/PNS transition zone. Dev Biol. 2001;238(1):47–63. doi: 10.1006/dbio.2001.0411. [DOI] [PubMed] [Google Scholar]
- Shiga Y, Tanaka-Matakatsu M, Hayashi S. A nuclear GFP/ beta-galactosidase fusion protein as a marker for morphogenesis in living Drosophila. Dev Growth Differ. 1996;38(1):99–106. [Google Scholar]
- Shishido E, Ono N, Kojima T, Saigo K. Requirements of DFR1/Heartless, a mesoderm-specific Drosophila FGF-receptor, for the formation of heart, visceral and somatic muscles, and ensheathing of longitudinal axon tracts in CNS. Development. 1997;124(11):2119–2128. doi: 10.1242/dev.124.11.2119. [DOI] [PubMed] [Google Scholar]
- Sonnenfeld MJ, Jacobs JR. Apoptosis of the midline glia during Drosophila embryogenesis: a correlation with axon contact. Development. 1995;121(2):569–578. doi: 10.1242/dev.121.2.569. [DOI] [PubMed] [Google Scholar]
- Spindler SR, Ortiz I, Fung S, Takashima S, Hartenstein V. Drosophila cortex and neuropile glia influence secondary axon tract growth, pathfinding, and fasciculation in the developing larval brain. Dev Biol. 2009;334(2):355–368. doi: 10.1016/j.ydbio.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemerdink C, Jacobs JR. Argos and Spitz group genes function to regulate midline glial cell number in Drosophila embryos. Development. 1997;124(19):3787–3796. doi: 10.1242/dev.124.19.3787. [DOI] [PubMed] [Google Scholar]
- Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72(4):527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- Suh J, Jackson FR. Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron. 2007;55(3):435–447. doi: 10.1016/j.neuron.2007.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney NT, Brenman JE, Jan YN, Gao FB. The coiled-coil protein shrub controls neuronal morphogenesis in Drosophila. Curr Biol. 2006;16(10):1006–1011. doi: 10.1016/j.cub.2006.03.067. [DOI] [PubMed] [Google Scholar]
- Sweeney ST, Broadie K, Keane J, Niemann H, O'Kane CJ. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron. 1995;14(2):341–351. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development. 2003;130(16):3621–3637. doi: 10.1242/dev.00533. [DOI] [PubMed] [Google Scholar]
- Vincent S, Vonesch JL, Giangrande A. Glide directs glial fate commitment and cell fate switch between neurones and glia. Development. 1996;122(1):131–139. doi: 10.1242/dev.122.1.131. [DOI] [PubMed] [Google Scholar]
- Waddell S, Armstrong JD, Kitamoto T, Kaiser K, Quinn WG. The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell. 2000;103(5):805–813. doi: 10.1016/s0092-8674(00)00183-5. [DOI] [PubMed] [Google Scholar]
- Wang J, Ma X, Yang JS, Zheng X, Zugates CT, Lee CH, Lee T. Transmembrane/juxtamembrane domain-dependent Dscam distribution and function during mushroom body neuronal morphogenesis. Neuron. 2004;43(5):663–672. doi: 10.1016/j.neuron.2004.06.033. [DOI] [PubMed] [Google Scholar]
- White K, Tahaoglu E, Steller H. Cell killing by the Drosophila gene reaper. Science. 1996;271(5250):805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- Witte HT, Jeibmann A, Klambt C, Paulus W. Modeling glioma growth and invasion in Drosophila melanogaster. Neoplasia. 2009;11(9):882–888. doi: 10.1593/neo.09576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AM, Wang JW, Axel R. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell. 2002;109(2):229–241. doi: 10.1016/s0092-8674(02)00707-9. [DOI] [PubMed] [Google Scholar]
- Xiong WC, Okano H, Patel NH, Blendy JA, Montell C. repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev. 1994;8(8):981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117(4):1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yu HH, Chen CH, Shi L, Huang Y, Lee T. Twin-spot MARCM to reveal the developmental origin and identity of neurons. Nat Neurosci. 2009;12(7):947–953. doi: 10.1038/nn.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Wang J, Haerry TE, Wu AY, Martin J, O'Connor MB, Lee CH, Lee T. TGF-beta signaling activates steroid hormone receptor expression during neuronal remodeling in the Drosophila brain. Cell. 2003;112(3):303–315. doi: 10.1016/s0092-8674(03)00072-2. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wildonger J, Ye B, Zhang Y, Kita A, Younger SH, Zimmerman S, Jan LY, Jan YN. Dynein is required for polarized dendritic transport and uniform microtubule orientation in axons. Nat Cell Biol. 2008;10(10):1172–1180. doi: 10.1038/ncb1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Lin S, Kao CF, Awasaki T, Chiang AS, Lee T. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 2006;127(2):409–422. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]
- Ziegenfuss JS, Biswas R, Avery MA, Hong K, Sheehan AE, Yeung YG, Stanley ER, Freeman MR. Draper-dependent glial phagocytic activity is mediated by Src and Syk family kinase signalling. Nature. 2008;453(7197):935–939. doi: 10.1038/nature06901. [DOI] [PMC free article] [PubMed] [Google Scholar]