Abstract
Substrate discrimination in the ubiquitin–proteasome system is believed to be dictated by specific combinations of ubiquitin–protein ligases (E3s) and ubiquitin-conjugating enzymes (E2s). Here we identify Doa10/Ssm4 as a yeast E3 that is embedded in the endoplasmic reticulum (ER)/nuclear envelope yet can target the soluble transcription factor Matα2. Doa10 contains an unusual RING finger, which has ubiquitin-ligase activity in vitro and is essential in vivo for degradation of α2 via its Deg1 degradation signal. Doa10 functions with two E2s, Ubc6 and Ubc7, to ubiquitinate Deg1-bearing substrates, and it is also required for the degradation of at least one ER membrane protein. Interestingly, different short-lived ER proteins show distinct requirements for Doa10 and another ER-localized E3, Hrd1. Nevertheless, the two E3s overlap in function: A doa10Δ hrd1Δ mutant is far more sensitive to cadmium relative to either single mutant and displays strong constitutive induction of the unfolded protein response; this suggests a role for both E3s in eliminating aberrant ER proteins. The likely human ortholog of DOA10 is in the cri-du-chat syndrome critical region on chromosome 5p, suggesting that defective ubiquitin ligation might contribute to this common genetic disorder.
Keywords: Ubiquitin, ERAD, proteasome, protein degradation, UPR
Selective protein degradation plays an essential role in a diverse array of biological processes. The most common mechanism for degrading intracellular proteins in eukaryotes uses the ubiquitin–proteasome system. In this system, polymers of ubiquitin, a 76-residue protein, are conjugated to a substrate protein, resulting in recognition and destruction of the substrate by the 26S proteasome (Hochstrasser 1996; Pickart 2001; Weissman 2001). For ubiquitin–protein conjugation, the C-terminal carboxyl group of ubiquitin is first activated in an energy-dependent reaction by the ubiquitin-activating enzyme (E1), followed by transfer of the ubiquitin to a ubiquitin-conjugating enzyme (E2) via transthiolation. The E2, together with a third factor called a ubiquitin–protein ligase or E3, transfers ubiquitin to a lysine side-chain(s) of a target protein. E3s are factors that stimulate the E2-dependent ubiquitination of substrates (Reiss et al. 1989).
The E3 proteins are thought to be largely responsible for the high degree of specificity in protein ubiquitination. For instance, the Rsp5 E3 enzyme ubiquitinates the large subunit of RNA polymerase II, and WW domains in Rsp5 interact directly with a repeated proline-rich motif (PxY) in the polymerase subunit (Chang et al. 2000). Whereas E1 and E2 components of the ubiquitin conjugation machinery can be readily identified by their signature sequence motifs, E3s have not been as easily categorized. However, known E3s divide into two heterogeneous families (Weissman 2001). E3s of the HECT domain family have a ∼350-residue domain that includes a conserved Cys residue, which attacks the ubiquitin–E2 to form another thioester intermediate before ubiquitin transfer to substrate. The second group of E3s comprise the RING finger family. RING fingers bind two zinc atoms in a characteristic “cross-brace” arrangement of coordinating Cys and His residues and can bind directly to the E2 (Zheng et al. 2000).
Surprisingly, the ubiquitin–proteasome system is also responsible for the degradation of membrane and lumenal proteins of the endoplasmic reticulum (ER) (for review, see Plemper and Wolf 1999). This ER-associated degradation (ERAD) requires retrotranslocation of proteins or protein segments back into the cytoplasm, a process often mediated by the same Sec61 translocon responsible for anterograde translocation of ER proteins. Ubc1, Ubc6, and Ubc7 are the primary E2s responsible for ERAD, whereas Hrd1/Der3, a RING finger protein, is the only known E3 that participates in this process, although additional ERAD E3s are likely to exist (Plemper and Wolf 1999; Friedlander et al. 2000; Hill and Cooper 2000; Wilhovsky et al. 2000; Bays et al. 2001). Ubc6, Ubc7, and Hrd1 all localize to the ER/nuclear envelope.
Our studies of the ubiquitin system have focused on the short-lived transcriptional repressor Matα2 in the yeast Saccharomyces cerevisiae (Hochstrasser et al. 1999). Previously, we identified two distinct ubiquitination pathways that are both required for normal rates of α2 degradation (Chen et al. 1993). The first of these involves the closely related E2s Ubc4 and Ubc5 and recognizes an as-yet-undefined degradation signal in α2. The second pathway uses Ubc6 and Ubc7; this pathway recognizes a degradation signal within the N-terminal 62 residues of α2, which we named Deg1. A key determinant of the Deg1 signal is the exposed hydrophobic face of an amphipathic helix, which we had suggested could be a recognition site for an E3 or E2/E3 complex (Johnson et al. 1998). Despite several previous screens for doa (degradation of alpha2) mutants, no E3 that ubiquitinates α2 had been identified (Hochstrasser et al. 1999; Swanson and Hochstrasser 2000). Here we describe a new genetic selection that has led to the identification of Doa10, a novel RING finger E3 that promotes the ubiquitination of Deg1-containing proteins. Doa10 is also shown to be required for the degradation of an integral ER membrane protein. The Doa10 ubiquitin ligase has an overlapping role with Hrd1/Der3 in promoting ER-associated degradation and negatively regulating the ER unfolded protein response.
Results
Genetic selection for Deg1–protein degradation defects
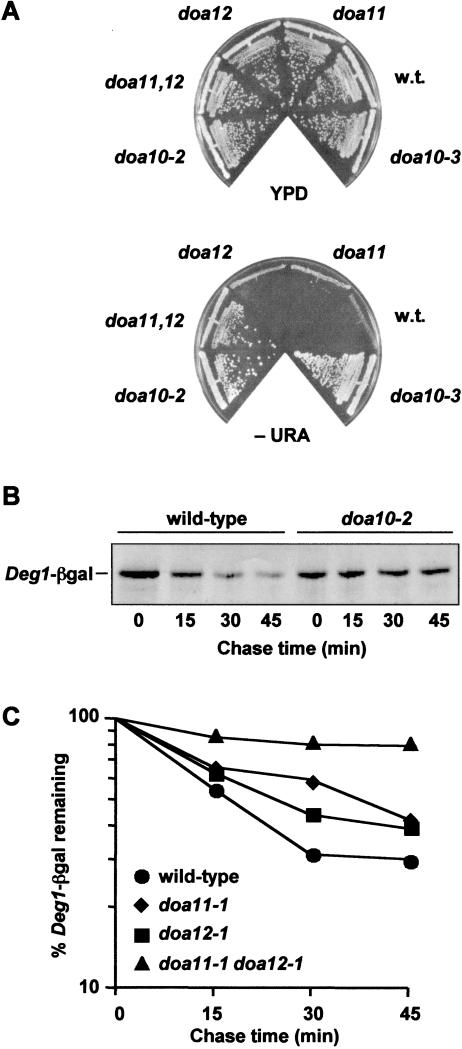
We devised a mutant selection strategy that took advantage of the fact that fusion of the Deg1 degradation signal to Ura3, an enzyme required for uracil biosynthesis, creates a short-lived fusion product. Rapid degradation of Deg1–Ura3 severely impairs growth of wild-type cells on media lacking uracil when this protein is the only source of Ura3 activity (Chen et al. 1993). A selection for mutants in which Deg1–Ura3 was long lived offered two principal advantages in a search for an E3. First, the positive growth selection allowed a large number of mutants to be examined. Second, the selection is biased against proteasome mutants and mutants involved in general ubiquitin metabolism because most of these genes are essential. A strain with one of these genes mutated may express a more stable Deg1–Ura3 fusion but would not be isolated because of its concomitant poor growth. In contrast, yeast cells lacking Ubc6 and Ubc7, the E2s that act through Deg1, are not growth compromised, so we hypothesized that the E3 for Deg1 also would not be necessary for rapid growth.
Deg1–URA3 ura3 cells were mutagenized and plated on minimal medium lacking uracil (SD-ura). We recovered 960 mutants that grew rapidly. To avoid characterizing mutations in genes already known to act in Deg1-mediated degradation, the mutants were tested for their ability to complement the following mutants: ubc6Δ ubc7Δ; cue1Δ, which lacks an accessory factor for Ubc7 (Biederer et al. 1997); doa4Δ, which is missing a deubiquitinating enzyme required for ubiquitin homeostasis (Swaminathan et al. 1999); and rpn4, which is defective for a nonessential regulator of the proteasome (Mannhaupt et al. 1999; Swanson and Hochstrasser 2000). Of the isolated mutants, 602 were mutated in UBC6 and/or UBC7 (Table 1, group I), and none was defective for any of the other tested genes. Analysis of the remaining mutants revealed three additional complementation groups. The group II mutants carried a recessive mutation in a single gene, which we named DOA10. These mutants had no discernable growth defect on rich media and showed robust growth on SD-ura (Fig. 1A). Mutant doa10 cells were not hypersensitive to high or low temperatures, to the amino-acid analog canavanine, or to cadmium (see Fig. 7A, below; data not shown); such defects are common among mutants with a global disruption of ubiquitin-mediated proteolysis. To confirm that the enhanced growth of doa10 cells on SD-ura was caused by stabilization of Deg1-containing substrates, the degradation of a Deg1–β-galactosidase protein (Deg1–βgal) was examined by pulse-chase analysis (Fig. 1B). In wild-type cells, Deg1–βgal has a half-life of <15 min, but very little was degraded in doa10 cells during the 45-min chase. Therefore, doa10 cells do not display aberrations associated with global defects in the ubiquitin–proteasome system, but they have a striking defect in the degradation of Deg1-containing substrates.
Table 1.
Mutant doa complementation groups
| Complementation
| ||
|---|---|---|
| Group
|
No. isolates
|
Mutant
|
| I | 602 | ubc6/doa2 and/or ubc7 |
| II | 356 | doa10 |
| III | 1 | doa11-1 doa12-1 |
| IV | 1 | DOA13-1 |
Figure 1.
Characterization of new doa mutants. (A) Wild-type and doa mutant strains were grown on rich medium (YPD) and minimal plates lacking uracil (−URA). (B) Pulse-chase analysis of Deg1–βgal in wild-type and doa10-2 strains. (C) Degradation kinetics of Deg1-βgal in wild-type, doa11-1, doa12-1, and doa11-1 doa12-1 strains. Proteins were precipitated with antibodies to βgal for B and C.
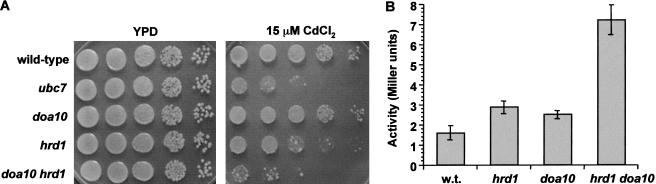
Figure 7.
Overlap in function of the ER-localized Doa10 and Hrd1 ubiquitin ligases. (A) The doa10Δ hrd1Δ double mutant is hypersensitive to cadmium. Congenic cells of the indicated genotypes were spotted in 10-fold dilutions on minimal medium containing CdCl2 or on YPD and incubated for 4 d and 2 d at 30°C, respectively. (B) Induction of the unfolded protein response in a hrd1Δ doa10Δ mutant. Congenic cells carried the plasmid pSZ1, which contains a UPRE–lacZ reporter (Friedlander et al. 2000).
The group III mutant phenotype was also recessive but resulted from mutations in two unlinked genes, which we called DOA11 and DOA12. Whereas the doa11-1 doa12-1 double mutant grew well on SD-ura, the doa11-1 and doa12-1 strains grew poorly (Fig. 1A). Consistent with these growth characteristics, mutations in both DOA11 and DOA12 were needed for strong perturbation of Deg1-mediated degradation (Fig. 1C). The doa11-1 mutant, but not doa12-1, was also somewhat hypersensitive to cadmium but displayed no other obvious abnormalities (data not shown). Finally, the single group IV mutant carried a dominant mutation in a gene we named DOA13. DOA13-1 cells were hypersensitive to various stress conditions, which suggested that this mutant had a widespread defect in ubiquitin-mediated degradation (data not shown).
Because mutations in DOA10 were isolated repeatedly and had such a strong effect on Deg1-containing substrates, DOA10 was chosen for further study.
Identification of the DOA10 gene
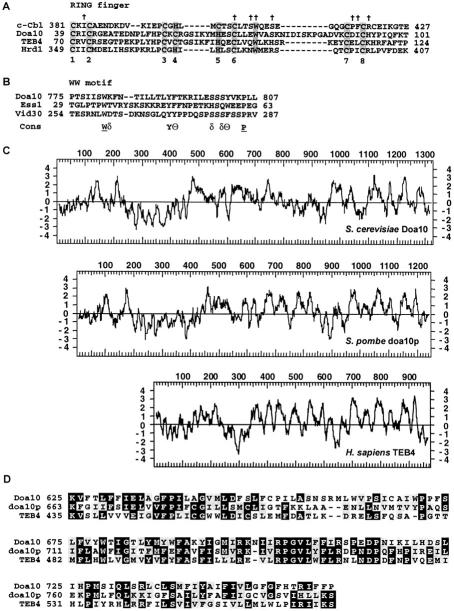
Attempts to clone DOA10 through complementation of the recessive doa10 degradation defect using multiple genomic yeast DNA libraries were unsuccessful. We resorted to localizing the DOA10 gene by genetic mapping (see Materials and Methods). Following assignment of DOA10 to chromosome IX, the doa10-2 allele was fine-mapped by meiotic mapping. The doa10-2 strain was mated to a strain in which the ULP2 gene on chromosome IX was replaced with HIS3 (Li and Hochstrasser 2000). After sporulation of the resulting diploid, 96 complete tetrads were dissected. Surprisingly, no recombination between the doa10-2 and ulp2Δ∷HIS3 alleles was detected, indicating very tight linkage of the two loci (<0.5 cM). One of the genes flanking ULP2 is SSM4/YIL030c. SSM4 had been identified by a mutation that suppressed the temperature-sensitive growth of an rna14-1 mutant, but the biochemical function of Ssm4 was unknown (Mandart et al. 1994). We regarded this gene as a likely candidate for DOA10 for two reasons. First, SSM4 had been shown to be toxic in Escherichia coli, which could explain our inability to clone DOA10, inasmuch as the yeast DNA libraries were maintained in E. coli hosts. Second, the predicted protein had a putative RING finger (Fig. 2A), which would be consistent with it being a ubiquitin ligase.
Figure 2.
Doa10 structural features. (A) Alignment of the closely related yeast Doa10 and human TEB4 RING fingers with examples of a RING-HC finger (human c-Cbl) and a RING-H2 finger (yeast Hrd1). The metal-coordinating His and Cys residues are highlighted in gray, as is the Trp residue commonly found in RING finger ubiquitin ligases. An 11-residue segment after residue 359 of Hrd1 was removed for clarity. (†) Residues in the c-Cbl RING that contact the E2 in the c-Cbl–UbcH7 structure (Zheng et al. 2000). (B) The Doa10 WW domain compared to two other yeast non-type-I WW motifs. The consensus sequence is from Kasanov et al. (2001). (δ) hydrophilic; (Θ) aromatic. (C) Similar membrane topology predicted from hydropathy plots of S. cerevisiae Doa10 and its likely orthologs in S. pombe (SPBC14F5.07) and humans (TEB4; GenBank KIAA0597). The predicted human protein lacks several internal, poorly conserved segments shared by the two yeast proteins. (D) The TEB4–Doa10 (TD) domain. The same proteins as in C are compared.
To confirm that SSM4 was DOA10, the SSM4 coding sequence was replaced by HIS3. Degradation of Deg1–βgal in the ssm4Δ::HIS3 strain was severely inhibited (data not shown). Furthermore, the ssm4Δ strain failed to complement the Deg1–Ura3 proteolytic defect when mated to several different doa10 strains isolated in our selection. These complementation and mapping results show that SSM4 and DOA10 are the same gene.
Doa10 structural features
DOA10 is predicted to encode a 151-kD protein that includes segments bearing similarity to several known protein motifs. As noted above, one such motif is a RING finger, which resides in the N-terminal 100 residues of Doa10 (Fig. 2A). The classical RING finger (RING-HC) has a histidine at the fourth coordinating position and a cysteine at the fifth. In the RING-H2 variant, both the fourth and fifth positions are occupied by histidines. The Doa10 RING, which is very similar to the RING finger in the human TEB4 protein (Fig. 2A), differs from both of these variants in that it has a Cys residue in the fourth position and a His in the fifth. We suggest calling this widespread variant a RING-CH finger. Another difference between Doa10 and the common RING variants is a somewhat longer peptide segment between the fourth and fifth zinc-coordinating residues.
A second motif found in Doa10 is a WW domain at residues 775 to 807 (Fig. 2B). The WW domain is a motif of ∼32 to 40 residues (Kasanov et al. 2001). It has been found in several HECT-class E3s, in which it can function as either a substrate- or cofactor-binding site. WW motifs fall into several sequence subclasses with characteristic peptide binding preferences. Many type I WW domains, for example, bind to proline-rich PPxY motifs in target proteins. Although the Doa10 WW domain cannot be placed easily in any of the known subclasses, it is intriguing that Ubc6 is unique among the yeast E2s in that it bears a PPxY motif (PPPY). Motif-searching algorithms also predict that Doa10 contains 10 to 14 transmembrane segments, suggesting that it is an integral membrane protein (Fig. 2C).
Doa10 is related to predicted proteins in Schizosaccharomyces pombe, Arabidopsis thaliana, Drosophila melanogaster, Caenorhabditis elegans, and Homo sapiens. Similarity among these proteins, none of which had a previously described function, is concentrated in two segments. The N-terminal regions that span the RING finger are 29% to 43% identical to Doa10 (see Fig. 2A). A number of shorter viral proteins also have RING domains closely related to the Doa10 finger. Both human Kaposi's sarcoma-associated herpesvirus IE1 proteins and swinepox C7 protein have an N-terminal RING-CH as well as several predicted transmembrane segments (Nicholas et al. 1997), suggesting that these viruses can redirect the cellular ubiquitination machinery to facilitate virus reproduction. The second region of similarity is an internal block of ∼130 residues that shows 27% to 37% identity to Doa10 (Fig. 2D); we call this region the TEB4–Doa10 (TD) domain. The TD domain appears to be unique to the Doa10-homologous proteins, which also all contain N-terminal RING-CH fingers. Finally, comparisons of hydropathy profiles for the predicted orthologs strongly suggest that they also all share similar membrane topologies; this is particularly clear in their C-terminal regions (Fig. 2C).
Doa10 is an integral membrane protein that localizes to the ER
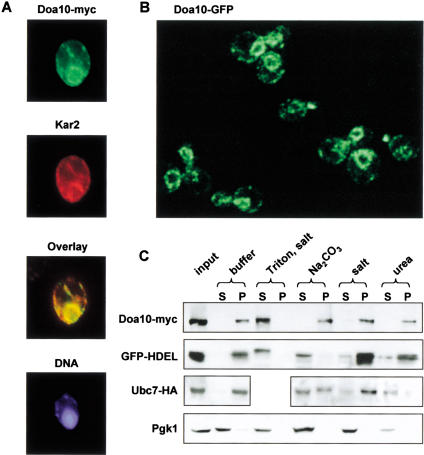
The predicted transmembrane disposition of Doa10 led us to examine its cellular localization using both microscopy and cell fractionation. Immunofluorescent staining of a functional Doa10 derivative tagged at its C terminus with nine myc epitopes (Doa10–myc) revealed bright perinuclear staining as well as more peripheral stained structures (Fig. 3A). The pattern was reminiscent of ER staining. This was confirmed in double-labeling experiments, which showed nearly identical staining by antibodies directed against Doa10–myc and against the ER chaperone Kar2. The same ER staining pattern was observed in live cells expressing a functional fusion between Doa10 and GFP (Fig. 3B).
Figure 3.
Doa10 is an integral membrane protein of the ER/nuclear envelope. (A) MHY1657 cells expressing myc9-tagged Doa10 were stained with an antibody to the myc epitope, an antibody to Kar2, and the Hoechst 33258 dye. (B) GFP fluorescence visualized by confocal microscopy in MHY1658 cells expressing a fusion between Doa10 and GFP. (C) Subcellular fractionation of MHY1690 cells carrying pHA-UBC7. Cell lysates were divided into microsomal pellet and supernatant fractions, which were examined by immunoblotting. Lysates were treated with buffer alone or buffer containing 1% Triton X-100 and 0.5 M NaCl (Triton, salt), 0.1 M Na2CO3 at pH 11.5, 0.5 M NaCl (salt), or 2.5 M urea. (P) pellet; (S) supernatant.
To verify that Doa10 is an integral membrane protein, subcellular fractionation was performed. Cells were gently lysed and then separated by high-speed centrifugation into pellet and supernatant fractions after various treatments (Fig. 3C). Doa10–myc could be extracted from the membrane pellet with detergent plus salt, a condition that solubilizes membranes. In contrast, salt or urea treatments that strip peripheral membrane proteins could not extract Doa10–myc nor could sodium carbonate (pH 11.5), which strips peripheral membrane proteins and releases lumenal ER proteins such as GFP-HDEL, a model lumenal ER protein (Rossanese et al. 2001). Ubc7 was used as a peripheral membrane protein control. It was substantially solubilized by sodium carbonate or urea but not by salt. Pgk1 (phospho-glycerate kinase) fractionated as expected for a cytosolic protein. Therefore, by multiple biochemical and cell biological criteria, Doa10 is an integral membrane protein that localizes primarily to the ER/nuclear envelope, the same localization as the Ubc6 and Ubc7 enzymes.
Substrate specificity of Doa10
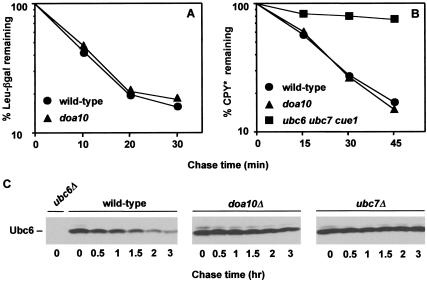
E3s are expected to show a high degree of substrate specificity. To gauge the range of ubiquitin pathway substrates that require Doa10, additional substrates were examined. Neither degradation of Leu-βgal, a substrate for the N-end rule pathway, nor that of Ub-Pro-βgal, a UFD pathway substrate (Varshavsky 1997), was affected by deletion of DOA10 (Fig. 4A; data not shown). We also tested an ERAD substrate, CPY*, the degradation of which requires two of the same E2s, Ubc6 and Ubc7, that act in the DOA pathway (Chen et al. 1993; Hiller et al. 1996). CPY* is a mutant version of the vacuolar carboxypeptidase Y. CPY* is retained in the ER, ejected into the cytoplasm, and degraded by the proteasome (Hiller et al. 1996). In wild-type cells, CPY* had a half-life of ∼15 min, whereas in ubc6 ubc7 cue1 cells, its degradation was severely compromised (Fig. 4B). In contrast, CPY* degradation kinetics were identical in wild-type and doa10Δ cells. These data also indicate that Ubc6, Ubc7, and Cue1 are still active in doa10Δ cells.
Figure 4.
Doa10 shows high substrate specificity in vivo. (A) Degradation kinetics of the N-end rule substrate Leu–βgal in wild-type (MHY501) and doa10Δ strains (MHY1631) carrying the plasmid p415GPD-Ub-L-lacZ. Radiolabeled proteins were precipitated with an antibody to βgal. (B) Degradation kinetics of CPY* in wild-type and mutant strains. Radiolabeled proteins were precipitated with an antibody to CPY. (C) Degradation of Ubc6. At time zero, cycloheximide was added, and disappearance of Ubc6 was followed by anti-Ubc6 immunoblotting.
Very recently, it has been shown that the transmembrane Ubc6 protein is itself a relatively short-lived protein with a constitutive degradation that depends on Ubc7, Cue1, and the proteasome (but not on Hrd1 or Sec61; Walter et al. 2001). Moreover, a catalytically inactive Ubc6 mutant is not degraded, even in cells that also express a wild-type version of the E2. The requirement for both Ubc6 activity in cis and for Ubc7 in Ubc6 ubiquitination suggests that these E2s assemble into a complex, as had been proposed previously (Chen et al. 1993). If Doa10 were also part of this complex, one would expect doa10 mutants to be defective for Ubc6 degradation as well. This was precisely what was observed (Fig. 4C). Ubc6 was not detectably degraded during the 3-h chase period, and its steady state levels were increased relative to wild-type. Therefore, Doa10 is capable of targeting not only soluble proteins for degradation but also integral ER membrane proteins.
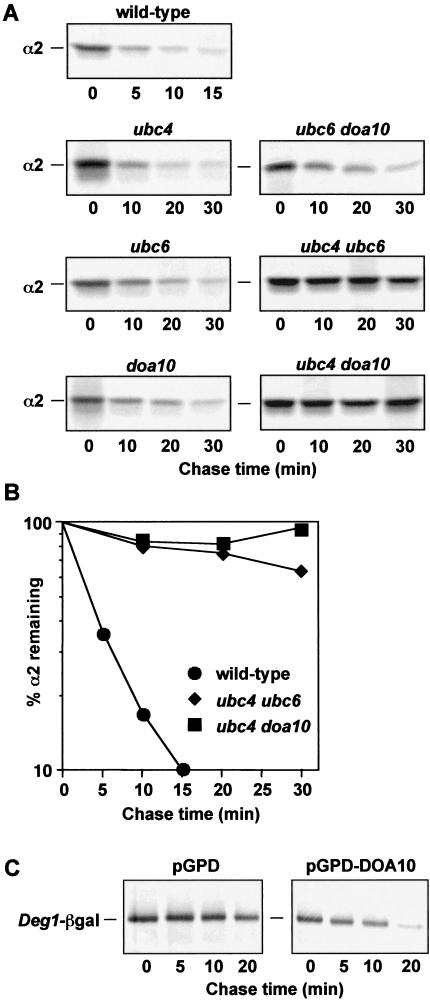
As noted above, the α2 protein is degraded by at least two distinct pathways. If either the Ubc4/Ubc5 or the Ubc6/Ubc7 pathway alone is disrupted, α2 degradation is slowed only two- to threefold, but if both are blocked, a synergistic inhibition is seen, with α2 half-life increasing 10- to 15-fold (Fig. 5A,B; Chen et al. 1993). We tested whether Doa10 functioned only with Ubc6 and Ubc7, which target the Deg1 signal of α2, or with both Ubc4/5 and Ubc6/7. Degradation of α2 was inhibited ∼threefold in doa10Δ cells (Fig. 5A), consistent with Doa10 acting in only one of the two pathways. Doa10 was confirmed to be in the Ubc6/Ubc7 pathway based on the finding that in a ubc6Δ doa10Δ double mutant, the half-life of α2 was no higher than that in the single mutants (Fig. 5A). In contrast, simultaneous deletion of UBC4 and DOA10 led to a striking inhibition of α2 degradation, indicating that both the Ubc4/5 and Ubc6/7 α2 ubiquitination pathways were blocked in this mutant (Fig. 5A,B).
Figure 5.
Matα2 degradation in mutant cells. (A) Pulse-chase analysis of α2 in wild-type cells and congenic deletion mutants. Proteins were precipitated with an antibody to α2. (B) Quantitation of the pulse-chase data in A for MHY501, MHY503, and MHY1648. (C) Deg1–βgal degradation in wild-type cells (MHY501) carrying the plasmid YEp13–Deg1–lacZ and either full-length DOA10 under the control of a strong promoter on a 2-μm plasmid (pGPD–DOA10) or the empty vector (pGPD). Proteins were precipitated with an antibody to βgal.
Hence, Doa10 has a substrate specificity even more restricted than that of the E2s that act in α2 turnover, and it functions specifically in the Deg1-mediated Ubc6/Ubc7 pathway of α2 degradation.
Doa10 is required for Deg1-mediated ubiquitination
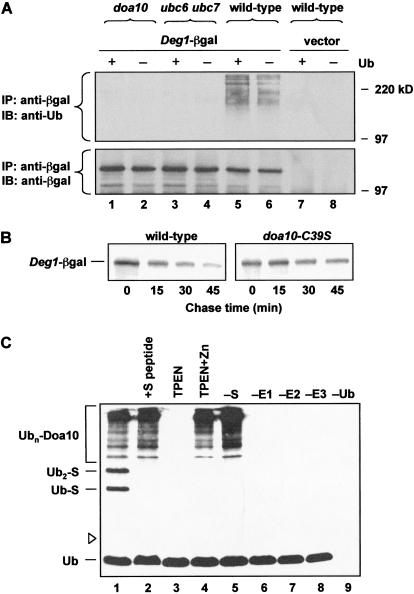
In light of the data described above, the most likely hypothesis for the mechanism of Doa10 action is that it stimulates the ubiquitination of Deg1-containing substrates. To test this, the levels of ubiquitin-Deg1–βgal conjugates were monitored (Fig. 6A). Deg1–βgal was immunoprecipitated from yeast cell extracts and then analyzed by anti-ubiquitin immunoblotting. In wild-type cells, polyubiquitinated Deg1–βgal species were detected, and the levels of these species were enhanced by ubiquitin overexpression (Fig. 6A, lanes 5,6). These conjugates were not detected in cells that did not express Deg1–βgal (Fig. 6A, lanes 7,8) or expressed a mutant Deg1–βgal with an inactivating Deg1 point mutation (Johnson et al. 1998; J. Laney and M. Hochstrasser, unpubl.). No ubiquitinated Deg1–βgal was detected in either ubc6Δ ubc7Δ or doa10Δ cells (Fig. 6A, lanes 1–4). Therefore, Doa10 is required in vivo for the ubiquitination of Deg1 substrates.
Figure 6.
Function of the Doa10 RING finger in vivo and in vitro. (A) Anti-ubiquitin immunoblot analysis of immunoprecipitated Deg1–βgal from cells carrying YEplac195–Deg1–lacZ or empty vector. Strains also contained a plasmid for overexpression of ubiquitin (+Ub) or an empty vector. Proteins were immunoprecipitated with antibodies to βgal. (Lower panel) The same blot reprobed with antibodies against βgal. (B) Degradation of Deg1–βgal in wild-type and doa10–C39S cells. Proteins were precipitated with an antibody to βgal. (C) In vitro ubiquitin ligase activity of the Doa10 RING domain. (Lanes 1,2) Complete reactions, including S protein substrate. In the reaction with S peptide, 1.5 μg of competitor peptide was added (lane 2). Ubiquitin–S protein conjugates (Ubn-S) are indicated. Omitted components are indicated above lanes 5–9, and no S protein was added in the zinc chelation experiment shown in lanes 3 and 4. Ubiquitinated proteins were detected by anti-ubiquitin immunoblotting. Open arrowhead indicates position of unmodified S protein.
A number of E3s in yeast are known to be present at limiting levels in vivo, and overproduction can enhance rates of substrate proteolysis (Bartel et al. 1990; Bays et al. 2001). Similarly, overexpression of DOA10 resulted in a reproducible two- to threefold increase in the rate of Deg1–βgal degradation (Fig. 5C). We also observed an increase in levels of ubiquitinated Deg1–βgal when Doa10 was overexpressed (data not shown), suggesting that enhanced ubiquitination of the substrate accounts for its accelerated degradation under these conditions.
The Doa10 RING finger is required in vivo and has ubiquitin ligase activity in vitro
The RING fingers found in many E3s are essential for their ability to catalyze substrate ubiquitination and degradation (Weissman 2001). To investigate whether the putative RING finger in Doa10 plays a role in Deg1-mediated degradation, the first cysteine in the RING consensus sequence was mutated to a serine (C39S). Epitope-tagged Doa10 derivatives from both DOA10 and doa10-C39S strains were expressed at similar levels (data not shown). However, degradation of Deg1–βgal was strongly impaired in doa10-C39S cells (Fig. 6B). Therefore, the RING-CH finger of Doa10 is crucial for Doa10-dependent proteolysis in vivo.
These in vivo data strongly suggest that Doa10 is the E3 ubiquitin ligase for the DOA pathway. To examine directly the ability of the Doa10 RING to catalyze the transfer of ubiquitin to another protein, a GST-S peptide–Doa10RING fusion was purified from E. coli and used in an in vitro ubiquitination assay with purified recombinant E1, E2, ubiquitin, and ATP. The 15-residue S peptide segment allows binding of GST-S–Doa10RING to S protein, which was added as substrate to the in vitro reaction (Bays et al. 2001). GST-S–Doa10RING catalyzed efficient ubiquitination of S protein (Fig. 6C, lane 1). Ubiquitination was prevented by omission of any single component of the reaction (Fig. 6C, lanes 5–9) or by adding an excess of competitor S peptide (Fig. 6C, lane 2), suggesting that the RING protein must bind the substrate to catalyze E2-dependent ubiquitin transfer. The RING fingers of many E3s catalyze polyubiquitination of themselves in vitro (Lorick et al. 1999), and high molecular mass ubiquitinated species also accumulated in reactions with the Doa10 RING finger (Fig. 6C). Based on our ability to reisolate a substantial fraction of these species on glutathione-Sepharose (data not shown) and their presence even in the absence of added S protein (Fig. 6C, lanes 4,5), we expect that most are polyubiquitinated GST-S–Doa10RING. Removal of zinc from GST-S–Doa10RING with the zinc chelator TPEN, which will disrupt the RING finger, destroyed its ligase activity, but activity was restored by reintroduction of zinc ions (Fig. 6C, lanes 3,4; no S protein added). We conclude that Doa10 is a ubiquitin–protein ligase.
Functional overlap of Doa10 and Hrd1
Doa10 and Hrd1 are the only known E3s that reside in the ER. The two proteins clearly have distinct substrate specificities in vivo (Fig. 4). However, their similar localization and common cofactors led us to investigate whether there might nevertheless be some overlap in their function. We created a hrd1Δ doa10Δ double mutant and asked whether any phenotypic abnormalities could be detected in this strain that were not seen in the single mutants. The double mutant grew normally at 30°C and 37°C, but when plated on medium containing cadmium, a pronounced growth defect was observed (Fig. 7A). The hrd1Δ single mutant is slightly hypersensitive to cadmium, but the hrd1Δ doa10Δ strain grew at least as poorly as a ubc7Δ mutant, which is known to be extremely sensitive to the heavy metal. The mechanistic basis of cadmium sensitivity is not known, but these results indicate that Doa10 and Hrd1 both participate in maintaining normal resistance.
Loss of Hrd1-dependent ERAD leads to a modest increase in unfolded proteins in the ER, which in turn causes a mild constitutive activation of the unfolded protein response (UPR; Friedlander et al. 2000). If Doa10 also contributes to the degradation of proteins in the ER, its deletion, either alone or in combination with that of Hrd1, would also be predicted to activate the UPR. Using a plasmid construct in which transcription of the lacZ gene was controlled by the UPRE sequence from KAR2, a gene that is induced by the UPR (Friedlander et al. 2000), we tested these possibilities (Fig. 7B). Under the conditions used, deletion of either HRD1 or DOA10 alone caused only modest (<twofold) induction of the UPR. In striking contrast, if both genes were deleted, strong induction of the UPRE–lacZ reporter was observed. These findings suggest that Hrd1 and Doa10 both stimulate degradation of aberrant proteins in the ER. Together with the substrate specificity studies described earlier, the data also imply that the ER/nuclear envelope-localized Doa10 ligase can act on substrates in distinct cellular compartments: the nucleus/cytosol and the ER membrane.
Discussion
The genetic selection described here has led to the discovery of a novel ubiquitin–protein ligase, Doa10/Ssm4. Remarkably, Doa10 functions not only in the degradation of the Matα2 transcription factor, which concentrates in the nucleus, but also in the degradation of proteins of the endoplasmic reticulum. Doa10 is a polytopic ER membrane protein with an unusual RING finger motif that is essential both for Deg1-dependent proteolysis in vivo and for ubiquitin ligase activity in vitro. Furthermore, our results reveal that the two ER-localized ubiquitin ligases Hrd1 and Doa10 have distinguishable substrate specificities but overlap in their ability to stimulate degradation of ERAD substrates. Loss of both Hrd1 and Doa10 causes a pronounced induction of the unfolded protein response, thereby identifying Doa10 as a new component of this highly conserved homeostatic mechanism.
Identification of novel genes required for Matα2 degradation
Our lack of knowledge about the E3s that function in Matα2 ubiquitination has hampered efforts to understand the molecular basis of specificity in this pathway. The Deg1 degradation signal in α2 includes the hydrophobic face of an amphipathic helix, and we have argued that the recognition of such exposed hydrophobic surfaces is likely to be a very common mechanism of substrate discrimination by the ubiquitin system (Johnson et al. 1998; Laney and Hochstrasser 1999). The identification of Doa10 as an E3 for Deg1-mediated ubiquitination should now allow us to dissect how these hydrophobic signals are recognized.
DOA10/SSM4 was originally identified as a suppressor of the temperature-sensitive growth of an rna14-1 strain (Mandart et al. 1994). Rna14 is a component of pre-mRNA cleavage and polyadenlyation factor I and is required for viability. Disruption of SSM4 suppresses rna14 mutants in an allele-specific manner and does not suppress an RNA14 deletion (Rouillard et al. 2000). One explanation for this suppression, which has ample precedent, is that the mutant Rna14-1 protein or an associated polypeptide unfolds at the high temperature and is degraded; inactivation of Doa10/Ssm4 reduces this degradation, allowing a partial rescue of Rna14 function. Because the Doa10 pathway can recognize exposed hydrophobic substrate elements, as would occur with partial protein unfolding, this hypothesis is appealing, although it remains to be tested.
Doa10 is a ubiquitin–protein ligase
Multiple lines of experimental evidence lead us to conclude that Doa10 is a ubiquitin–protein ligase or E3. First, a mutation in an E3, a substrate specificity factor, would be expected to affect only a restricted set of substrates. In fact, Doa10 is required only for the Deg1-mediated Ubc6/Ubc7 pathway of α2 degradation, and it is even not necessary for the degradation of another substrate, CPY*, that also depends on these E2s. Second, Doa10 is absolutely required for Deg1-mediated ubiquitination in vivo. Third, Doa10 colocalizes in the cell with the E2 enzymes that are also necessary for Deg1-dependent ubiquitination. Fourth, as is true for other yeast E3s, Doa10 levels are at least partially limiting for the ubiquitination and degradation of its targets. Fifth, Doa10 contains an N-terminal RING finger, a motif that defines the larger of the two known classes of E3 ligases, and the RING finger is necessary for Deg1-dependent proteolysis. Finally and most directly, the Doa10 RING domain shows ubiquitin ligase activity in a purified in vitro system, and the structural integrity of the RING is required for this activity in vivo and in vitro. Based on peptide competition experiments (Fig. 6C), the Doa10 RING-containing protein is likely to bind directly to the substrate to catalyze transfer of ubiquitin from the E2.
We have not yet succeeded in reconstituting Deg1-mediated ubiquitination in a fully homologous in vitro system. Because it is a relatively large integral membrane protein, full-length Doa10 may be difficult to manipulate in vitro. However, if the substrate- and E2-binding domains of the E3 can be defined, it might be possible to construct a mini-Doa10 derivative capable of directing Deg1-dependent substrate ubiquitination in vitro. As described in the next section, several predicted structural motifs in Doa10 offer clues as to what these key domains might be.
Significance of Doa10 structural motifs
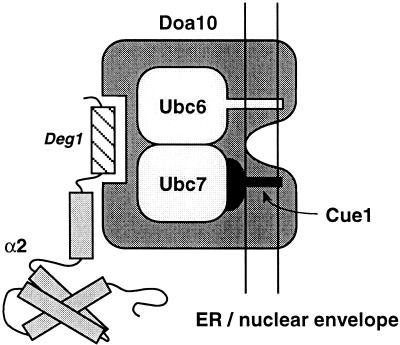
The RING finger ubiquitin ligases are thought to provide a platform for the simultaneous binding of E2 and substrate, thereby enhancing ubiquitin transfer. An unusual feature of the DOA pathway is that it requires two distinct E2 isozymes, Ubc6 and Ubc7. Yeast two-hybrid analysis had previously suggested a weak physical interaction (direct or indirect) between these two E2s (Chen et al. 1993). Certain structural features of Doa10 suggest how it might interact with both Ubc6 and Ubc7. It is tempting to speculate that the RING finger functions to recruit Ubc7. The Hrd1 E3 binds Ubc7 in vivo, and this interaction requires the intact Hrd1 RING finger (Bays et al. 2001). In the crystal structure of the UbcH7–c-Cbl complex, the c-Cbl RING makes direct contacts with the E2 (Fig. 2A; Zheng et al. 2000). Doa10 also contains a putative WW motif. The ligand specificity of this motif has been extensively characterized. The major class of ligands that can bind WW motifs is the proline-rich sequence PPxY, and Ubc6 is one of only 18 proteins (and the only E2) predicted from the yeast genome to contain a PPxY motif (Chang et al. 2000). We suggest that a noncanonical E3–E2 interaction between Doa10 and Ubc6, together with Doa10 RING association with Ubc7, allows simultaneous binding of Ubc6 and Ubc7 to the E3 in the ER membrane, as depicted schematically in Figure 8. Our data and those of others demonstrate that the E2 Ubc7 functions with two different E3s, Doa10 and Hrd1. Moreover, Ubc7 works both with Ubc6 to ubiquitinate Deg1-containing substrates and without Ubc6 to ubiquitinate Hrd1 substrates. These findings reinforce the idea, proposed earlier (Chen et al. 1993), that formation of different E2 and E2/E3 combinations would greatly expand the range of substrate specificities in the ubiquitin system.
Figure 8.
Model for Doa10 function. See text for details.
If Doa10 is indeed principally responsible for substrate discrimination, how might it recognize the Deg1 signal? As noted, the key determinant in Deg1 is an amphipathic segment with the potential to form a coiled-coil structure. It is intriguing in this regard that Doa10 has at its very N terminus a short stretch that is predicted to form a coiled coil (Lupas et al. 1991). Potentially, this could function as a sensor domain in Doa10 that allows it to interact with hydrophobic degradation signals, particularly those that are part of amphipathic helices, although other Doa10 elements may function in substrate recognition as well.
Many of the ER targets of the Hrd1 ubiquitin ligase appear to be retrotranslocated out of the ER via the Sec61 translocon (Plemper and Wolf 1999). Although it will be important to determine whether any ER substrates of Doa10 are also extracted via Sec61, Doa10-dependent degradation of the integral ER membrane protein Ubc6 has been shown to be independent of Sec61 (Walter et al. 2001). Doa10 is a large protein that appears to have at least ten transmembrane segments, and this organization is highly conserved (Fig. 2C). This suggests that the transmembrane topology has an important function(s) that goes beyond simply tethering the enzyme to the membrane. An attractive idea is that transmembrane segments in Doa10 contribute to a protein exit channel that allows retrotranslocation of substrates such as Ubc6 from the ER. It is also plausible that multiple transmembrane domains are required to coordinate the binding and proper orientation of E2s and substrate.
Overlapping and unique functions of two ER ubiquitin–protein ligases
Hrd1 and Doa10 are both RING finger E3s that localize to the ER/nuclear envelope. Hrd1 is important for the degradation of a number of ERAD substrates, but there are exceptions (Hill and Cooper 2000; Wilhovsky et al. 2000), which suggested that other ER ubiquitin ligases might exist. Indeed, we have now found that Doa10 is essential for the degradation of at least one Hrd1-independent ERAD substrate, Ubc6, and there are undoubtedly additional ER targets for this E3. Defects in ERAD have recently been linked to induction of the UPR (Friedlander et al. 2000; Travers et al. 2000). When Friedlander et al. (2000) compared UPR induction in hrd1Δ and ubc1Δ ubc7Δ cells, a much smaller effect was seen in the former strain. Loss of the E2s Ubc1 and Ubc7 eliminates all known ER-specific protein ubiquitination pathways, so this result implied that an ERAD ubiquitin ligase other than Hrd1 was needed for full UPR induction. Our data strongly suggest that Doa10 is such an E3. Consistent with this, UPR induction in doa10Δ hrd1Δ was nearly as great as that seen when wild-type cells were treated with 4 mM dithiothreitol, an inducer of the UPR (Friedlander et al. 2000; data not shown). Notably, transcriptional up-regulation of DOA10 in wild-type cells on UPR induction is comparable to that of UBC7 (Travers et al. 2000), and Ubc7 levels are known to go up as part of the UPR (Friedlander et al. 2000).
Because Doa10 and Hrd1 localize similarly, it is not yet clear how specific substrates are targeted to one E3 or the other. The two E3s (or E3/E2 complexes) might recognize distinct structural features of substrates, or they might localize to distinct microdomains within the ER/nuclear envelope. The ER localization of Doa10 and its cognate E2s raises the interesting question of how a substrate such as Matα2, which is primarily nuclear, gains access to the Doa10 ubiquitination complex. Three general models, which are not mutually exclusive, can be proposed. First, α2 may only be subject to Doa10-mediated ubiquitination between the time of its synthesis and its import into the nucleus. We do not favor this idea because the degradation kinetics of the steady-state pool of α2 are comparable to those of newly synthesized repressor (J. Laney and M. Hochstrasser, unpubl.), and most of α2 at steady state is in the nucleus. In another model, Doa10 (and Ubc6 and Cue1/Ubc7) would diffuse from the outer to the inner nuclear membrane (the two fuse at the nuclear pores), where the ubiquitination complex would have direct access to nuclear-localized α2. Such movement of membrane proteins to the inner membrane occurs for a number of proteins, but a size limit of ∼60 kD for the cytoplasmic domains of such proteins appears to exist, presumably because of the restricted space within the lateral channels of nuclear pore complexes (Worman and Courvalin 2000). We do not know the precise topology of Doa10, but transmembrane prediction algorithms suggest a fairly large cytoplasmically disposed component, which might restrict movement to the inner membrane. Finally, substrates such as α2 might be transported out of the nucleus to access the ER-embedded Doa10 complex. Other proteins, such as p53, have been shown to require nuclear export for efficient degradation (Liang and Clarke 2001). Experiments to distinguish among these possibilities are underway.
The human ortholog of DOA10 maps to the cri-du-chat critical region
At present there are no published reports on any of the Doa10-like proteins predicted from other genomes. However, the human TEB4 gene, the likely homolog of DOA10 (see Fig. 2) maps to the cri-du-chat critical region on chromosome 5p15.2. Segmental aneuploidy of this region causes a relatively common genetic disorder (at least one in 50,000 live births) that is associated with severe mental retardation and developmental delay (Mainardi et al. 2001). Deficiencies for multiple genes in this region appear to contribute to the disease phenotype, so it has been difficult to evaluate the contribution of individual genes. It is intriguing that another disease characterized by severe mental retardation and some of the same craniofacial abnormalities, Angelman syndrome, has also been associated with a defective ubiquitin ligase, the HECT E3 E6-AP (Kishino et al. 1997). Our work on Doa10 leads to the hypothesis that defective protein ubiquitination at the ER/nuclear envelope is important for the etiology of cri-du-chat syndrome, although the precise contribution of TEB4 to this disease remains to be firmly established. Interestingly, the mouse protein axotrophin has a RING-CH domain similar to that of Doa10 and is required for normal brain development: Disruption of the axotrophin gene results in callosal agenesis and neural degeneration (see GenBank NP_065600).
Several other neurodegenerative disorders have been linked recently to defective ubiquitin/proteasome-dependent proteolysis, including ERAD. Parkin, a RING finger E3 that is mutated in an inherited form of Parkinson's disease, was shown to interact with the human orthologs of Ubc6 and Ubc7 and to suppress cell death induced by unfolded protein accumulation in the ER (Imai et al. 2001). Aggregation of aberrant huntingtin protein, which is associated with Huntington's disease, has been correlated with a block to the degradation of a Ubc6/Ubc7-dependent proteolytic test substrate in vivo (Bence et al. 2001). The degradation signal of this substrate was initially defined in yeast (Gilon et al. 1998). We have found that yeast proteins bearing this class of signals are targeted by the Doa10 ubiquitin ligase (data not shown). It will therefore be of interest to determine whether defects in the human Doa10 ortholog are linked to neurodegenerative diseases such as Huntington's.
Materials and methods
Bacterial and yeast methods
Rich (YPD) and minimal (SD) media were prepared as described (Ausubel et al. 1989). Standard methods were used for genetic analysis of yeast. The E. coli strains used in this study were JM101, MC1061, BL21(DE3), and DH10B, and bacterial methods and media were as described (Ausubel et al. 1989).
Construction of yeast strains
For a complete list of strains, see Table 2. The wild-type strain MHY1410 was prepared by integrating pRS303 (Sikorski and Hieter 1989) into the his3 locus of JY112, and MHY1411 was prepared by integrating pRS304 (Sikorski and Hieter 1989) into the trp1 locus of JY113. MHY1412 was a segregant from a cross between MHY551 and JY112. MHY1690 was derived from a cross between MHY551 and MHY1657; MHY1651, from a cross between MHY1631 and MHY1366; MHY1648, from a cross between MHY1631 and MHY513; and MHY1670, from a cross between MHY1631 and MHY496.
Table 2.
Strains used in present study
| Strain
|
Genotype
|
|---|---|
| JY112a | α his3-Δ200 leu2-Δ1∷LEU2-Deg1-lacZ ura3-52 lys2-801∷LYS2-Deg1-URA3 trp1-Δ63 |
| JY113a | a his3-200 leu2Δ1∷LEU2-Deg1-lacZ ura3-52 lys2∷LYS2-Deg1-URA3 trp1-Δ63 |
| MHY496b | MHY501 with ubc6-Δ1∷HIS3 |
| MHY498b | MHY501 with ubc4-Δ1∷HIS3 |
| MHY500b | a his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 |
| MHY501b | α his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 |
| MHY503b | MHY501 with ubc4-Δ1∷HIS3 ubc6-Δ1Δ∷HIS3 |
| MHY513b | MHY500 with ubc4-Δ1∷HIS3 |
| MHY551b | MHY500 with ubc7∷LEU2 |
| MHY552b | MHY501 with ubc6-Δ1∷HIS3 ubc7∷LEU2 |
| MHY884c | MHY500 with doa4Δ∷leu2∷HIS3 |
| MHY1364d | MHY501 with cue1Δ∷HIS3 |
| MHY1366 | MHY500 with prc1-1 |
| MHY1367 | MHY500 with cue1Δ∷HIS3 ubc6Δ∷TRP1 ubc7∷LEU2 prc1-1 |
| MHY1410 | α his3-200∷pRS303(HIS3) leu2-Δ1∷LEU2-Deg1-lacZ ura3-52 lys2-801∷LYS2-Deg1-URA3 trp1-Δ63 |
| MHY1411 | a his3-200 leu2-Δ1∷LEU2-Deg1-lacZ ura3-52 lys2-801∷LYS2-Deg1-URA3 trp1-Δ63∷pRS304(TRP1) |
| MHY1412 | α his3-Δ200 leu2-Δ1∷LEU2-Deg1-lacZ ura3-52 lys2-801∷LYS2-Deg1-URA3 trp1 ubc7∷LEU2 |
| MHY1631 | α his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 doa10Δ∷HIS3 |
| MHY1648 | α his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 doa10Δ∷HIS3 ubc4∷HIS3 |
| MHY1651 | α his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 doa10Δ∷HIS3 prc1-1 |
| MHY1657 | α his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 DOA10-myc9∷his5+ |
| MHY1658 | α his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 DOA10-GFP∷HIS3 |
| MHY1669e | MHY501 with hrd1Δ∷LEU2 |
| MHY1670 | α his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 doa10Δ∷HIS3 ubc6Δ∷HIS3 |
| MHY1690 | α his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 ubc7∷LEU2 DOA10-myc9∷his5+ |
| MHY1703 | MHY501 with hrd1Δ∷LEU2 doa10Δ∷HIS3 |
| MHY1740 | α his3-200 leu2-Δ1∷LEU2-Deg1-lacZ ura3-52 lys2∷LYS2-Deg1-URA3 trp1-Δ63 doa11-1 doa12-1 |
| MHY1745 | α his3-200 leu2-Δ1∷LEU2-Deg1-lacZ ura3-52 lys2∷LYS2-Deg1-URA3 trp1-Δ63 doa11-1 |
| MHY1747 | a his3-200 leu2-Δ1∷LEU2-Deg1-lacZ ura3-52 lys2-801∷LYS2-Deg1-URA3 trp1-Δ63∷pRS304(TRP1) doa12-1 |
| MHY2091 | α his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 doa10-C39S-myc9∷his5+ |
| MHY2093f | a leu2-Δ1∷LEU2-Deg1-lacZ ura3-52 lys2-801 rpn4∷URA3(mTn3) |
| MHY2094 | α his3-200 leu2-Δ1∷LEU2-Deg1-lacZ ura3-52 lys2-801∷LYS2-Deg1-URA3 trp1-1 doa10-2 |
| MHY2095 | α his3-200 leu2-Δ1∷LEU2-Deg1-lacZ ura3-52 lys2-801∷LYS2-Deg1-URA3 trp1-1 doa10-3 |
J. Laney and M. Hochstrasser, unpubl.
M. Hochstrasser, unpubl.
MHY1631 was generated by disrupting the entire DOA10 open reading frame (ORF) with the HIS3 gene via polymerase chain reaction (PCR)-mediated homologous recombination in MHY501 (Baudin et al. 1993). Primer sequences are available on request. Proper integration of the HIS3 fragment was determined by colony PCR, and the strain was backcrossed to MHY500 to show single site integration. MHY1657 was made using a C-terminal myc-tagging cassette (a gift of R. Verma, Caltech, Pasadena, CA). For this, a PCR product was generated that encoded the C terminus of Doa10 in frame with a sequence encoding nine copies of a c-myc epitope followed by the S. pombe his5+ gene, which functionally complements the S. cerevisiae HIS3 gene. This DNA fragment was integrated into MHY501, and correct integration was verified as above. MHY1657 was shown to complement the doa10 phenotype in crosses to several doa10 strains isolated in the screen. MHY1658 was made using a C-terminal GFP-tagging cassette (Longtine et al. 1998). A PCR product that contained the 3′ end of the DOA10 ORF in frame with the GFP ORF followed by the S. kluyveri HIS3 gene was generated from the plasmid pFA6a-GFP(S65)HIS3MX6. The DOA10-GFP allele fully complemented the doa10 degradation defect.
To construct the RING finger mutation in MHY2091, a two-part PCR method was used (Ausubel et al. 1989). Primers were used to amplify a 5′ segment of the DOA10 locus beginning 393 bp upstream of the translational start site and extending through the Glu45 codon, with the downstream primer introducing the Ser 39 mutation. A second pair of primers was used to amplify an internal fragment of DOA10, encoding Asp 32 to Arg 400, with the upstream primer introducing the Ser 39 mutation. The two fragments were combined by a second PCR step and subcloned into YIplac211, and the mutation was confirmed by DNA sequencing. The linearized plasmid was transformed into MHY1657, and uracil prototrophs were then plated on 5-fluoroorotic acid, which is toxic to cells expressing URA3, to select for plasmid excision. Strains were screened for the mutation by amplification of the 5′ region of the DOA10 ORF followed by DNA sequencing to identify the doa10–C39S–myc9 allele.
Plasmid constructions
DOA10/SSM4 is toxic to E. coli (Mandart et al. 1994), so full-length DOA10 plasmids were constructed in yeast. pGPD–DOA10 was constructed by gap repair from the chromosomal copy of DOA10. The 5′ end of the DOA10 ORF from the start codon to Pro 167 was PCR amplified, as was the 3′ end of the ORF from Arg 1250 to the stop codon. These two fragments were inserted into separate pGEM-T/EASY vectors (Promega). The 5′ DOA10 fragment was first subcloned into the p426GPD vector (Mumberg et al. 1995) using BamHI and HindIII sites, and the 3′ DOA10 fragment was then subcloned into the modified plasmid using HindIII and SalI sites. The resulting plasmid, which lacks sequences encoding Doa10 residues 168 to 1249, was linearized with HindIII and transformed into MHY501, with selection for uracil prototrophy. Gap repair was verified by colony PCR.
To express the Doa10 RING domain in E. coli BL21(DE3) as a GST-Speptide fusion, a segment of DOA10 encoding the first 112 residues of Doa10 was amplified by PCR using primers that allowed subsequent in-frame fusion with the sequence encoding GST-Speptide in pET-42b(+) (Novagen). The entire insert was sequenced to verify the absence of errors.
YCplac22-HA-UBC7 was constructed by a two-step PCR procedure similar to that described above. The plasmid insert contained 200 bp upstream of the UBC7 translational start site, the sequence encoding the HA epitope fused to the UBC7 ORF, and 150 bp of sequence downstream of the UBC7 stop codon. The fragment was initially ligated into the vector pGEM-T/Easy and was subsequently excised as an EcoRI fragment and subcloned into YCplac22 (Gietz and Sugino 1988). pHA-UBC7 was shown to reverse the cadmium hypersensitivity and rapid growth on medium lacking uracil, which characterized the ubc7Δ strain MHY1412.
Isolation of doa mutants
MHY1410, MHY1411, and MHY1412 strains (the last as a control that was not subjected to mutagenesis) were grown overnight in YPD and then mutagenized with ethyl methanesulfonate (Kodak) to ∼28% survival (∼3 × 106 survivors). Half the cells were plated on uracil drop-out plates immediately, and half were resuspended in SD complete medium, grown for 4 h at 30°C, and then plated on plates lacking uracil. Cells were grown for 1 to 2 d at 30°C, and the first 960 colonies that appeared were picked into 96-well microtiter plates. Mutants were retested by streaking on plates lacking uracil to confirm the Ura+ phenotype.
The mutants were first tested for their ability to complement the following known mutants of the Deg1-mediated degradation pathway: MHY552 (ubc6Δ ubc7Δ), MHY884 (doa4Δ), MHY1364 (cue1Δ), and MHY2093 (rpn4). Mutants of opposite mating type to the tester strain were spotted into fresh 96-well plates containing YPD. The tester strain was then spotted into the same plates, and mating was allowed to occur overnight at 30°C without agitation. Cells were pelleted and resuspended in cold sterile water. After incubating for 2 to 3 h at 4°C to slow cell growth, cells were spotted onto drop-out plates that were selected for diploid prototrophs. Plates were incubated overnight at 30°C, and the diploids were tested for complementation of the Deg1–Ura3 degradation defect by streaking onto minimal plates lacking both uracil and the nutrients required to maintain the selection for diploid cells. Complementation of MHY2093 were performed on 1 μg/mL canavanine sulfate. Mutants that did not fall into one of the tested groups were then crossed to one another and tested in the same manner as above for placement into new groups. Representative mutants were backcrossed to determine the number of unlinked mutations responsible for the degradation defect and to check whether they were recessive or dominant.
Identification of the DOA10 gene
The DOA10 gene was localized by genetic mapping of a representative doa10 mutant allele. First, the doa10-2 allele was localized to a specific chromosome by the 2-μm mapping method. After mating doa10-2 to the 2-μm tester strains, the resulting diploids were assayed quantitatively for βgal activity using O-nitrophenyl β-D-galactopyranoside (ONPG) as the substrate (Ausubel et al. 1989). Only diploids from the cross to the strain with the 2-μm plasmid in chromosome IX showed the recessive doa10 trait of increased Deg1–βgal activity (6 out of 19 tests, 31%). Subsequently, the doa10-2 Deg1-URA3 mutant was crossed to a strain with an integrated Deg1-URA3 marker and the ulp2Δ∷HIS3 allele on chromosome IX (Li and Hochstrasser 2000). After sporulation, 96 full tetrads (and >60 incomplete tetrads) were tested for marker segregation. No recombination between the two markers was seen, indicating very tight linkage of doa10-2 and ulp2Δ∷HIS3. A strain with a deletion of the nearby SSM4 gene was constructed and mated to mutants doa10-1 through doa10-5; the resulting diploids all grew rapidly on uracil drop-out plates.
Yeast subcellular fractionation
Yeast were grown to mid-log phase, and five OD600 equivalents were harvested and washed in spheroplast buffer (1 M sorbitol, 20 mM sodium phosphate at pH 7.5, and 75 mM NaCl). Cells were incubated for 10 min at 30°C in 1 mL of spheroplast buffer and 30 mM dithiothreitol (DTT) and then were spun down and resuspended in spheroplast buffer and 2 mM DTT. After addition of 70 μg of zymolase 100T (ICN), the cells was incubated for 20 min at 30°C, washed in spheroplast buffer, and lysed by glass-bead shearing for 1 min in 1 mL of fractionation buffer (200 mM D-mannitol, 75 mM NaCl, 20 mM sodium phosphate at pH 7.5, and 1 mM MgCl2) plus protease inhibitors. Unlysed cells and cell debris were pelleted at 600g for 6 min, and the supernatant was divided into separate tubes that were subjected to one of the following treatments for 1 h on ice: (1) 2.5 M urea, 0.1 M Na2CO3 (pH 11.5), 1% Triton X-100, and 0.5 M NaCl or (2) 0.5 M NaCl. Samples were then separated into pellet and supernatant fractions by centrifugation at 100,000 rpm for 1 h at 4°C. Pellets were washed once with fractionation buffer containing the original additions, as appropriate, and resuspended in fractionation buffer. Proteins were precipitated in 10% trichloroacetic acid for 30 min on ice, pelleted, washed in ethanol, resuspended in gel loading buffer, and evaluated by immunoblotting.
Pulse-chase and immunoblot analyses
Pulse-chase analyses were performed as described (Chen et al. 1993). Deg1–βgal and Leu–βgal fusions were immunoprecipitated with anti-βgal (Organon Teknika); Matα2, with anti-α2 (Chen et al. 1993); and CPY*, with anti-CPY antibodies (gift from N. Segev, University of Chicago). Immunoblot analysis was performed as described (Swaminathan et al. 1999), with antibody binding detected with enhanced chemiluminescence reagents (Amersham). Doa10–myc was visualized with an anti-myc monoclonal antibody (9E10, Roche); Pgk1, with anti-PGK (Molecular Probes); Kar2, with an anti-Kar2 antiserum (gift of J. Broach, Princeton University, NJ); Ubc7-HA, with an anti-HA antibody (16B12, Berkeley Antibody); and Ubc6, with anti-Ubc6 (gift of T. Sommer, Max Delbrück Center, Berlin, Germany). For Deg1–βgal immunoprecipitation/anti-ubiquitin immunoblotting, 20 mM N-ethylmaleimide was included in the lysis buffer. Precipitated proteins were resolved by SDS-PAGE and subjected to immunoblotting with anti-ubiquitin (hybridoma P4G7-H11; gift of D. Gottschling, Fred Hutchinson Cancer Research Center, Seattle, WA).
In vitro analysis of Doa10RING-mediated protein ubiquitination
Following the procedure of Bays et al. (2001), a typical 20 μL reaction contained the following purified components: 0.5 μg yeast Uba1 (Boston Biochem); 0.5 μg human Ubc4-His6 (Bays et al. 2001), which was purified from an E. coli BL21(DE3) lysate with a Talon affinity resin; 2.5 μg of bovine ubiquitin (Sigma); 10 mM ATP; 1.5 μg of S-protein (Sigma); and 1.5 μg of GST-S–Doa10RING, which was purified on glutathione-Sepharose and then dialyzed. Reactions were performed in 50 mM Tris-HCl (pH 7.5), 2.5 mM MgCl2, and 0.5 mM DTT for 4 h at 30°C and were stopped by addition of SDS gel loading buffer and heating for 3 min at 100°C. Proteins were resolved on 16.5% Tricine-SDS–polyacrylamide gels and visualized by anti-ubiquitin immunoblotting.
Fluorescent staining of yeast cells
Immunofluorescent staining was performed as described previously (Rossanese et al. 2001). Samples were visualized on an Axioplan microscope (Carl Zeiss) equipped with a 100× Plan-Apo 1.4 NA objective lens and with band-pass filters for visualizing Hoechst 33258, fluorescein, and Texas red fluorescence. Secondary antibodies (Molecular Probes) for primary antibody detection were goat anti-rabbit IgG coupled to Texas Red and goat anti-mouse IgG coupled to Alexa 488. For Doa10–GFP localization, MHY1658 cells were grown to mid-log phase and then washed twice and resuspended in sterile water; 5 μL were spotted on a slide and spread with a coverslip. The GFP chromophore was excited with a 488-nm laser and visualized with a 505- to 550-nm bandpass filter on an Zeiss LSM 510 confocal microscope.
Acknowledgments
We thank Jeff Laney, Randy Hampton, Dieter Wolf, Thomas Sommer, Rati Verma, and Ben Glick for plasmids or yeast strains; Ben Glick for advice on subcellular fractionation; Nate Bays for advice on in vitro ubiquitination; and Jeff Laney for comments on the manuscript. This work was supported by grant GM46904 from the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mark.hochstrasser@yale.edu; FAX (203) 432-5175.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.933301.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: Wiley; 1989. [Google Scholar]
- Bartel B, Wunning I, Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nuc Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/ Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin–proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- Chang A, Cheang S, Espanel X, Sudol M. Rsp5 WW domains interact directly with the carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2000;275:20562–20571. doi: 10.1074/jbc.M002479200. [DOI] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol. 2000;2:379–384. doi: 10.1038/35017001. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gilon T, Chomsky O, Kulka RG. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 1998;17:2759–2766. doi: 10.1093/emboj/17.10.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Cooper AA. Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J. 2000;19:550–561. doi: 10.1093/emboj/19.4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin–proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Ann Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M, Arendt CS, Swaminathan S, Johnson PR, Amerik AY, Li S-J, Swanson R, Laney J, Pals-Rylaarsdam R, Nowak J, et al. The Saccharomyces cerevisiae ubiquitin–proteasome system. Philos Trans R Soc Lond B Biol Sci. 1999;354:1513–1522. doi: 10.1098/rstb.1999.0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of parkin. Cell. 2001;105:891–902. doi: 10.1016/s0092-8674(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Johnson PR, Swanson R, Rakhilina L, Hochstrasser M. Degradation signal masking by heterodimerization of MATα2 and MATα1 blocks their mutual destruction by the ubiquitin–proteasome pathway. Cell. 1998;94:217–227. doi: 10.1016/s0092-8674(00)81421-x. [DOI] [PubMed] [Google Scholar]
- Kasanov J, Pirozzi G, Uveges AJ, Kay BK. Characterizing class I WW domains defines key specificity determinants and generates mutant domains with novel specificities. Chem Biol. 2001;8:231–241. doi: 10.1016/s1074-5521(01)00005-9. [DOI] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- Laney JD, Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol. 2000;20:2367–2377. doi: 10.1128/mcb.20.7.2367-2377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang SH, Clarke MF. Regulation of p53 localization. Eur J Biochem. 2001;268:2779–2783. doi: 10.1046/j.1432-1327.2001.02227.x. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mainardi PC, Perfumo C, Cali A, Coucourde G, Pastore G, Cavani S, Zara F, Overhauser J, Pierluigi M, Bricarelli FD. Clinical and molecular characterisation of 80 patients with 5p deletion: Genotype-phenotype correlation. J Med Genet. 2001;38:151–158. doi: 10.1136/jmg.38.3.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandart E, Dufour ME, Lacroute F. Inactivation of SSM4, a new Saccharomyces cerevisiae gene, suppresses mRNA instability due to rna14 mutations. Mol Gen Genet. 1994;245:323–333. doi: 10.1007/BF00290112. [DOI] [PubMed] [Google Scholar]
- Mannhaupt G, Schnall R, Karpov V, Vetter I, Feldmann H. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 1999;450:27–34. doi: 10.1016/s0014-5793(99)00467-6. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nicholas J, Ruvolo V, Zong J, Ciufo D, Guo HG, Reitz MS, Hayward GS. A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J Virol. 1997;71:1963–1974. doi: 10.1128/jvi.71.3.1963-1974.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Plemper RK, Wolf DH. Retrograde protein translocation: ERADication of secretory proteins in health and disease. Trends Biochem Sci. 1999;24:266–270. doi: 10.1016/s0968-0004(99)01420-6. [DOI] [PubMed] [Google Scholar]
- Reiss Y, Heller H, Hershko A. Binding sites of ubiquitin–protein ligase: Binding of ubiquitin–protein conjugates and of ubiquitin-carrier protein. J Biol Chem. 1989;264:10378–10383. [PubMed] [Google Scholar]
- Rossanese OW, Reinke CA, Bevis BJ, Hammond AT, Sears IB, O'Connor J, Glick BS. A role for actin, Cdc1p, and Myo2p in the inheritance of late Golgi elements in Saccharomyces cerevisiae. J Cell Biol. 2001;153:47–62. doi: 10.1083/jcb.153.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillard JM, Brendolise C, Lacroute F. Rna14p, a component of the yeast nuclear cleavage/polyadenylation factor I, is also localised in mitochondria. Mol Gen Genet. 2000;262:1103–1112. doi: 10.1007/pl00008653. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Amerik AY, Hochstrasser M. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol Biol Cell. 1999;10:2583–2594. doi: 10.1091/mbc.10.8.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R, Hochstrasser M. A viable ubiquitin-activating enzyme mutant for evaluating ubiquitin system function in Saccharomyces cerevisiae. FEBS Lett. 2000;477:193–198. doi: 10.1016/s0014-5793(00)01802-0. [DOI] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- Walter J, Urban J, Volkwein C, Sommer T. Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p. EMBO J. 2001;20:3124–3131. doi: 10.1093/emboj/20.12.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- Wilhovsky S, Gardner R, Hampton R. HRD gene dependence of endoplasmic reticulum-associated degradation. Mol Biol Cell. 2000;11:1697–1708. doi: 10.1091/mbc.11.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Courvalin JC. The inner nuclear membrane. J Membr Biol. 2000;177:1–11. doi: 10.1007/s002320001096. [DOI] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl–UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]