Abstract
We have designed a real-time computer vision system, the Multi-Worm Tracker (MWT), that can simultaneously quantify the behavior of dozens of Caenorhabditis elegans on a traditional petri plate at video rates. Three traditional behavioral paradigms are examined using this system: spontaneous movement on food, where the behavior changes over tens of minutes; chemotaxis, where turning events must be detected accurately to determine strategy; and habituation of response to tap, where the response is stochastic and changes over time. In each case, manual analysis or automated single-worm tracking would be tedious and time-consuming, but the MWT system allows rapid quantification of behavior with minimal human effort. Thus, this system will enablelarge scale forward and reverse genetic screens for complex behaviors.
Since Caenorhabditis elegans was introduced as a model organism, a large number of mutants in genes that modulate easily-scored behaviors have been isolated in genetic screens. However, it has been challenging to manually screen for behaviors that unfold over long periods of time, that are difficult to score quickly and accurately by eye, or that manifest stochastically. Thus, there is a need for high-throughput automated techniques that can be used on such behaviors.
Individual actions performed by worms, such as reversals and turns, are relatively easy to detect. The key computational difficulty is that to accurately monitor a long-lasting behavior consisting of many individual actions, a huge amount of image data needs to be acquired and processed: each animal needs to cover enough pixels to allow accurate quantification and frame rates need to behigh enough to maintain identity and resolve fast behaviors.
A variety of computer vision systems have previously been applied to C. elegans behavior. Systems that use motorized stages to follow individual worms at high resolution allow precise quantification of behavior1–3, but screening for stochastic behaviors is still highly labor- and time-intensive. Systems that monitor many worms simultaneously on a single plate4–6 use a two-step process: first, an experiment is run and a video is stored; then, image analysis software extracts behavioral parameters from the animals in view. Unfortunately, since storing high-speed high-resolution video is impractical, these systems use resolutions (~75 μm/pixel) and frame rates (3–5 frames per second) that make it difficult to accurately analyze a wide range of behaviors.
We therefore designed and built a tracking system that analyzes image data in real time to monitor many worms on a single plate, using commodity computer hardware and a high-speed high-resolution camera. This allows automated behavioral analysis of C. elegans on an unprecedented scale, and should enable genetic screening for defects in complex worm behaviors.
Results
Accuracy and speed of automated tracking
The Multi-Worm Tracker (MWT) package consists of real-time image analysis software, the MWT, and offline behavioral parameter measurement software, Choreography, that together analyze the behavior of multiple worms on a single petri plate (Fig. 1a,b). The MWT provides low-level features including the position and outline of the animal. Choreography selects appropriate objects to analyze and computes additional features, such as direction of motion, that benefit from statistics that span the experimental session. Organism- or condition-specific behavioral parameters are computed using plugins for Choreography.
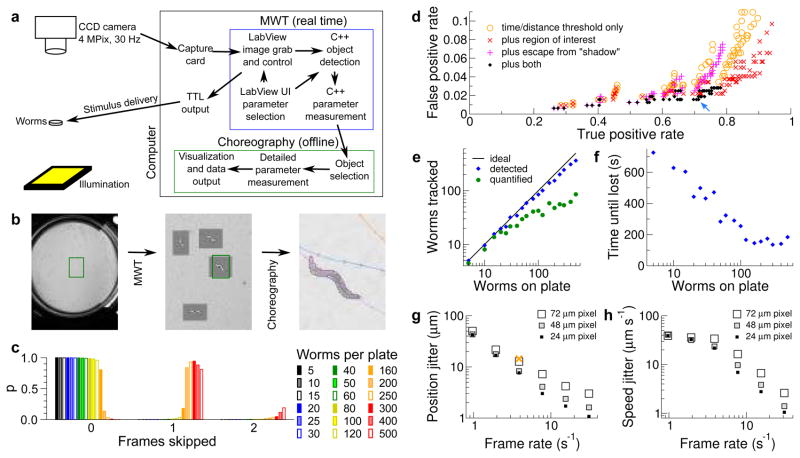
Figure 1.
Accuracy and performance of the Multi-Worm Tracker. (a) The major components of the Multi-Worm Tracker system. (b) Examples of full-field images, the regions selected for analysis by the MWT, and post-capture visualization in Choreography. (c) The plot shows the probability that a given number of frames will be dropped for each frame processed, with increasing number of worms. (d) Accuracy of detection and selection of worms for analysis. A false positive is either an object which is not a worm, or a worm scored by hand as partially obscured. Each dot corresponds to a different choice for time and distance threshold. Orange circle: no additional criteria. Red X: animals must be in a region of interest away from the edges of the plate. Magenta +: animals must move one body length before being quantified to avoid image processing artifacts. Black diamond: both additional criteria. Blue arrow: our typical choice of parameters. (e) Effect of crowding on fraction of animals detected (blue diamonds) or selected for analysis (green dots, using parameters chosen in d). (f) Effect of crowding on duration one worm can be followed (identity is lost upon collision). (g, h) Effect of pixel size and frame rate on estimate of animal position (g; the orange X indicates typical parameters for a non-real-time tracker, Parallel Worm Tracker) and of animal speed (h; movement averaged over 0.5 s).
To test the ability of the system to perform image processing in real time, we recorded from a series of plates with increasing numbers of worms. Using a 3 GHz Core 2 Duo processor with a 4 megapixel, 30 Hz camera, essentially all frames were processed when 5–120 worms were present; the processing load from greater numbers of worms caused frames to be dropped (Fig. 1c). Post-processing with Choreography is typically 10–30× faster than real time. Taken together, the MWT and Choreography process image data at approximately 110 megapixels per second, which is ~50 times faster than existing tools4,5 (~2 MPix/s; data not shown and G. Tsechpenakis, personal communication).
Although the MWT is designed to be used in real-time mode, we recorded 10 minutes of high-speed high-resolution video to disk as a reference data set that could be both scored manually and analyzed using the MWT. We used a plate that might be typical for high-throughput screening (imperfect staging, some small worms present, imperfect lighting at edges, etc.), such that not all worms were suitable for analysis. Manual scoring yielded an average of 76 visible worms per frame, of which 20 were at least partially obscured by shadows at the edge of the plate, four were touching another animal, and 52 were unobstructed. The MWT found and tracked worms in all categories as well as a small number of shadows and other non-worm artifacts. We used Choreography to select worms that were suitable for analysis; ideally, it would pick the unobstructed animals and reject all others. We tested different combinations of time and distance thresholds with several selection strategies, and found parameters that accepted a majority of animals manually annotated as suited for analysis, while rejecting all non-worms, all touching worms, and almost all worms obscured by shadows (Fig. 1d). We selected a combination of parameters (Fig. 1d, and Online Methods) that yielded 97.5% of selected worms suitable for analysis and used these for all future experiments.
To estimate the pixel-level error remaining after this selection process, a human expert segmented 37 worms by hand and found the total error rate to be 2.8% (1.0% of in-worm pixels were classified as background, and 1.8% extra background pixels were added to the true worms). These parameters allowed nearly all animals to be quantified at low density; at 100 worms per plate the system quantified approximately half of the animals (Fig. 1e). Larger numbers of animals also increased the number of collisions, causing animal identity to be lost more quickly (Fig. 1f).
To estimate measurement noise, we assumed that worm behavior is predominantly slowly-varying and thus high-order derivatives of parameters with respect to time are dominated by stochastic noise (Table 1). To see if reducing computational load was feasible, we subsampled our dataset in time and space to emulate slower and lower resolution cameras. Both pixel size and frame rate had major effects on apparent noise in the position and speed of animals (Fig. 1g,h). We therefore continued using our system at full speed and resolution.
Table 1.
Estimates of measurement noise in selected behavioral parameters.
| Quantity | Mean | Noise |
|---|---|---|
| Area | 0.145 mm2 | 1.7% |
| Bearing | --- | 4.1° |
| Position | --- | 1.2 μm |
| headposition | --- | 23 μm |
| spine length | 1.11 mm | 2.5% |
| spine curvature | 260° mm−1 | 8.0% |
| speed (over 0.5 s) | 0.186 mm s−1 | 0.55% |
| angular speed (over 0.5 s) | 5.8° s−1 | 2.4% |
Analysis of movement speed
C. elegans modulates its speed in response to a variety of stimuli including both acute insults and chronic environmental conditions such as presence of food. We first monitored basal movement rates of adult wild-type hermaphrodites on food. The process of putting a plate on the tracker involves temporarily removing the lid, and presumably changes temperature and humidity, induces mechanical vibration, and increases light levels. We observed, both by eye and with the MWT, that this process increased the worms’ movement speed, which then decended to a basal level after 10 minutes (Fig. 2a).
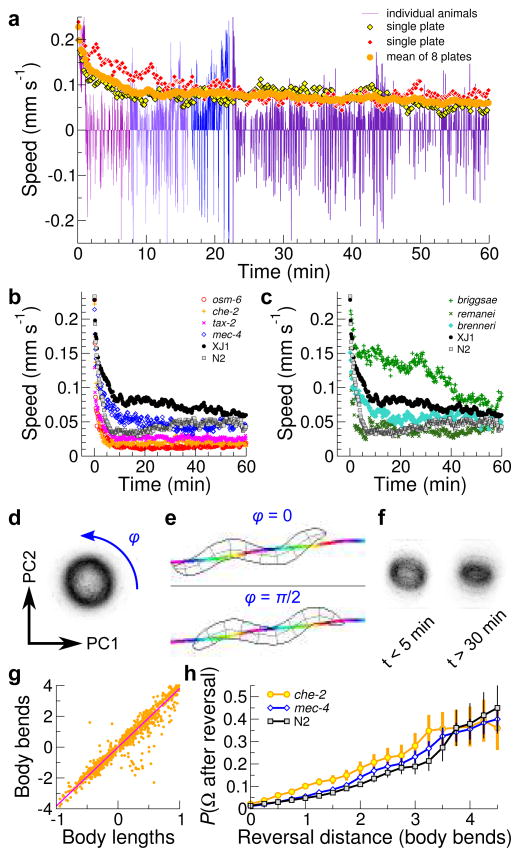
Figure 2.
Worm movement on food. (a) The plot shows worm speeds for selected individual plates (red or yellow diamonds) or as a mean for all plates (orange dots) over time (n ≈ 30 tracked animals per plate). (b, c) The plots show the return to steady-state speeds in the indicated strains after being placed on the tracker (b; mean of four plates, n ≥ 15 worms per plate) and in different Caenorhabditis species (c; mean of four plates, n ≥ 20 worms per plate). (d) Intensity map of first two principal components of worm shape computed from the reference data set; darker colors indicate more observations. The ring shape of the map corresponds to the worm’s sinusoidal movement cycle; the blue arrow indicates forward movement or an increase in phase. (e) Example of phase progression. Snapshots correspond to sine-like and cosine-like postures; rainbow line is the animal’s path colored by phase. (f) Intensity map as in d at the indicated time points after worms are placed on the tracker. n ≈ 20 tracked worms on one plate, strain XJ1. (g) Worm centroid movement (in body lengths) is plotted against estimated number of body bends (phase advance). Individual movements (orange dots) and best linear fit (magenta line) are shown. (h) The probability of omega turn initiation is plotted against reversal distance(number of body bends) for the indicated strains. n ≈ 400 omega turns per strain.
To assess the reproducibility of these results, we recorded from seven additional plates of wild-type worms (Fig. 2a) with an average of 27 worms tracked per timepoint per plate. Every plate showed approximately the same trend; monte carlo sampling of tracks suggested that plate-to-plate variability was approximately 60% larger than expected if all animals on a plate were independent, so we ran all experiments on several plates to average out this variability.
To explore which sensory modalities were responsible for the initial elevation in movement speed, we recorded from mec-4 mutants, which are defective in gentle touch response, and che-2 mutants which have defective sensory cilia and fail to chemotax. mec-4 responded similarly to wild-type; che-2 increased speed but returned to baseline much faster (Fig. 2b). Unexpectedly, a line of the wild-type strain (N2) recently obtained from the C. elegans Genomic Center also showed notable differences from the lab’s wild-type line (hereafter XJ1, to distinguish it). To determine whether the che-2 phenotype was a result of a chemosensory defect or was due to genetic drift in strains, we tested two additional chemosensory mutants, osm-6 and tax-2. These phenocopied che-2 (Fig. 2b), indicating that a major portion of the initial increase in speed requires properly functioning ciliated sensory neurons. We also investigated whether the difference between wild-type strains was typical of variation found in the wild. We examined an extreme case by tracking other Caenorhabditis species, namely briggsae, remanei, and brenneri. All three species showed similarly elevated movement rates, but briggsae had a markedly slower return to baseline (Fig. 2c). Collectively, these tests demonstrate that the MWT is broadly suited for studying baseline locomotion in Caenorhabditis.
Quantification of body posture
The MWT captures postural information in addition to position. However, the accuracy of this information is limited by the small number of pixels per animal. To determine whether the available postural information is useful, we sought to replicate work demonstrating that principal component analysis (PCA) of the worm skeleton yields two major components, analogous to sine and cosine in circular motion, and that these components cycle as the worm moves7. We added a PCA plugin to Choreography and found that in our reference data set, the top two components explained over 75% of the variance in the worm’s posture and the top three explained 90%. During movement, posture followed a phase advance along a ring in the space of the first two principal components (Fig. 2d,e). When we observed worms on food after movement had returned to baseline, we found that the same principal components still explained a similar portion of the variance. However, the relationship between these components changed: the ring partially collapsed as movement returned to basal levels (Fig. 2f), suggesting that mechanisms that control worm posture during rapid locomotion are relaxed during grazing.
Manual annotation of C. elegans motion often uses body bends as an easily-quantified parameter; a body bend corresponds to an advance of π in phase. We compared distances and phase advance for ~1,300 short forward or backward motions detected by Choreography in our reference data set and found that wild-type worms needed 3.8 body bends to advance one body length (Fig. 2g). Omega turns (very deep bends) are also frequently quantified manually; we used postural information to obtain automatic detection with ~5% error rates. Omega turns occur prominently after reversals during taxis and escape behaviors, but the relationship on food is less well characterized. To detect reversals, we classified movement direction based on the propensity of worms to travel forwards; out of 6.2 worm-hours of tracks hand-annotated for movement direction, 1.7% was classified by Choreography as unknown and 0.20% was classified incorrectly. We then looked for omega turns that followed reversals and found a nearly linear dependence between reversal size and turn probability that was consistent across chemosensory and mechanosensory mutants (Fig. 2h). This suggests that the relationship may be generated by the animal’s motor program, not driven by sensory input.
Chemotaxis
C. elegans uses two strategies to navigate in chemical gradients: a random tumble strategy (klinokinesis), where worms pirouette (reorient) when a favorable chemical decreases8, and a weathervane strategy (klinotaxis) where worms traveling perpendicular to the gradient turn up the gradient9. To assay chemotaxis, we seeded 1 μl of LB medium as a control and 1 μl of OP50 bacteria on either side of a plateand placed 5–10 worms in the center.
Within approximately three minutes, wild-type worms reliably reached the food spot (Fig. 3a); without food, no spatial preference was observed (data not shown). Chemosensory mutants were also able to preferentially aggregate on food, perhaps because they, like wild-type, are able to slow down when they reach food (10 and data not shown). We quantified pirouettes and found that wild-type and mechanosensory mutants regulated reversals as a function of direction while chemosensory mutants che-2 and osm-6 did not (Fig. 3b). tax-2 was omitted from this assay because it retains considerable sensitivity to a subset of volatile odorants11.
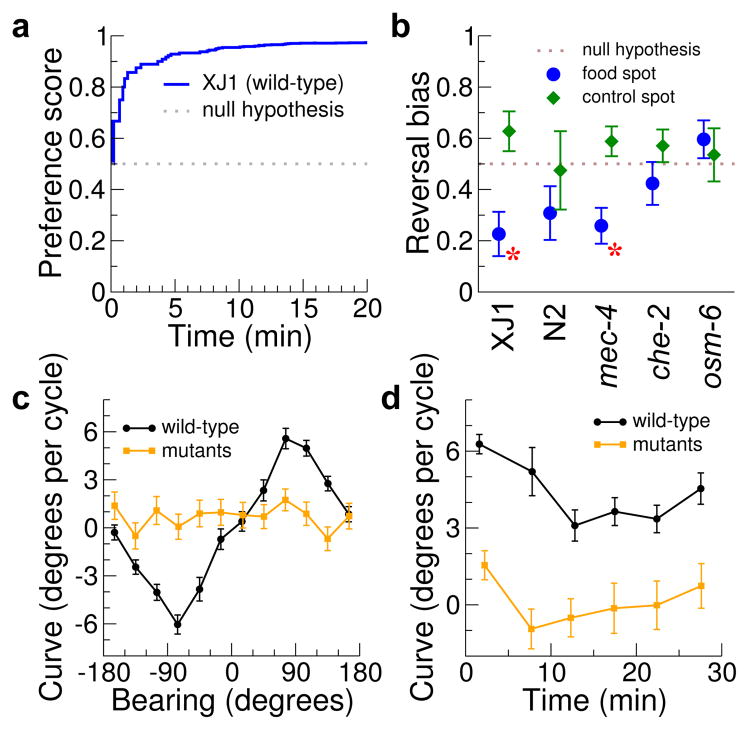
Figure 3.
Analysis of chemotaxis. (a) Chemotaxis effectiveness of wild type worms. Preference score is the Bayesian estimate of the probability that an animal will travel into the food spot instead of the control spot. n = 8 plates, ~8 worms tracked per plate. (b) Pirouette frequencyof the indicated strains near food. Reversal bias is fT/(fT+fA), where f is the frequency of reversals when moving towards (fT) or away (fA) from food (blue) or control spot (green). Error bars, s.e.m. from monte carlo simulation. Red asterisks: P < 0.05 that reversals are equally common when traveling towards vs. away from food (χ2 test). (c) Weathervane chemotaxis to NaCl in the absence of food. The mean curve of the animal’s path is plotted against its bearing relative to the gradient (see Online Methods for more detail); signs are chosen so that positive curve at positive bearing is a turn up the gradient. Black is wild-type (N2, 30 plates of ten worms each). Orange shows chemotaxis mutants (che-2, osm-6, and tax-2; five plates of ten worms of each, mutant data are pooled since we found no significant differences between the strains). Error bars, s.e.m.. (d) Decrease in strength of weathervane chemotaxis with time. Positive curve is up the gradient and results are averaged over quadrants around ±90° bearing. P < 0.001 (t-test) that the first 10 minutes and the next 20 are the same.
We also conducted a salt chemotaxis assay with NaCl at the center of the plate. As previously reported9, wild-type animals traveling perpendicular to the attractant followed a path that curved with a bias towards the attractant, while chemotaxis mutants did not regulate their turning (Fig. 3c). Animals also clustered less tightly at the end of longer recordings. Consistent with this, we found that curving bias decreases over time (Fig. 3d). Together, these results indicate that the MWT is suited for analysis of chemotaxis behavior.
Tap habituation
Worms respond to non-localized vibration by executing an escape response (reversal), and habituate to repeated stimuli by lowering the magnitude and probability of response12,13. To deliver tap stimuli automatically, we constructed a solenoid tapper that drove a metal rod into the side of the plate at intervals specified within the MWT, and wrote a reversal-detection plugin. Careful manual annotation disagreed with the plugin in 5% of cases where reversals were small; since the manual annotator had to revisit the data several times to catch their own errors, we concluded that automatic annotation is likely more reliable than routine manual annotation.
When we applied taps with a 10 s inter-stimulus interval, initially nearly all worms responded with large reversals, but with repeated taps the probability and size of responses decreased (Fig. 4a,b); these results were consistent across six independent wild-type plates (strain XJ1). As expected, the mechanosensory mutant mec-4 had low reversal probabilities (Fig. 4c) and small reversal sizes (Fig. 4d) that were not significantly different from reversals in the absence of tap. In contrast, all chemosensory mutants responded to tap (Fig. 4c,d) and showed a surprising range of phenotypes. che-2 worms showed a dramatically increased habituation rate, and none of the chemosensory mutants matched wild-type reversal distances; tax-2 had larger responses, while osm-6 responses were as small as basal reversals shown by mec-4. Sensory deprivation is reported to cause a smaller response to tap14. Consistent with this, osm-6 lacks functional sensory cilia15 and che-2 encodes a gene expressed in many ciliated sensory neurons but not in touch neurons16. However, this does not explain the wide diversity of phenotypes in the chemosensory mutants; further work is required to understand this.
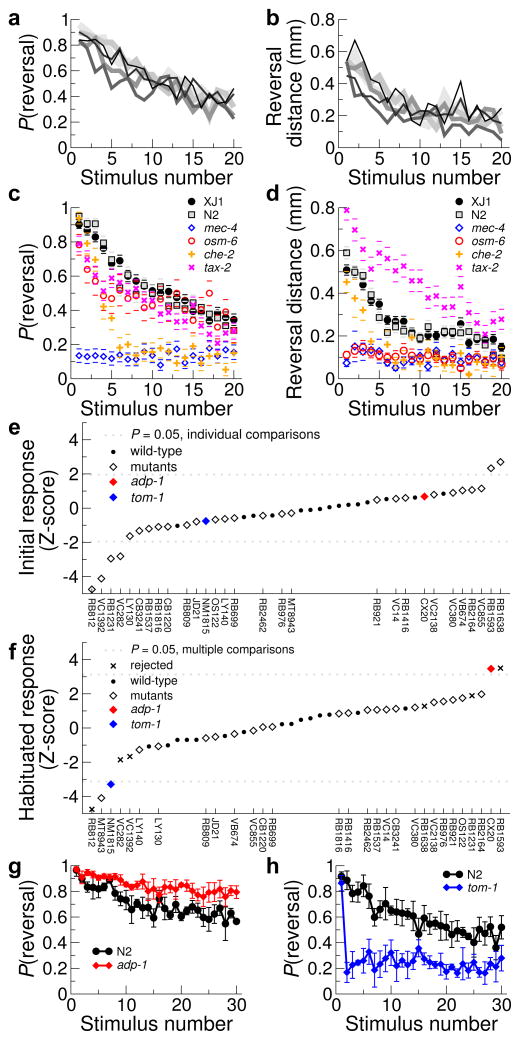
Figure 4.
Analysis of tap habituation. (a, b) Probability of reversing (a, Bayesian estimate) or reversal distance (b) after a tap are plotted against the number of tap stimuli. Six plates of wild-type (XJ1) animals are plotted in different shades of gray. n ≈ 30 animals per plate. (c, d) Reversal probability (c) and distance (d) for mechanosensory and chemosensory mutants are plotted against the number of tap stimuli. n = 3 plates for mutants and N2, 6 plates for XJ1, ≥ 10 animals per plate; error bars are s.e.m. (e) Probability of response to first tap of various mutants (diamonds) and wild-type controls (circles); Z-score is normalized by wild-type distribution. (f) Habituated response probabilities (at stimuli 28–30). X denotes mutants with abnormal initial response. (g, h) Probability of reversal after tap is plotted for the loss-of-habituation mutant adp-1 (g) and the hyper-habituation mutant tom-1 (h).
We then used the MWT to conduct a pilot screen for novel tap habituation mutants. 33 strains were chosen with mutations in genes with a diversity of predicted functions (Table 2). Although we quantified a variety of parameters, to simplify analysis we focused on only the probability of reversal on initial and final taps. We recorded from 3–4 plates of each strain, averaging 136 animals per tap, plus 6–8 plates of wild-type each day as a control. Of the 33 strains, four had movement defects that were too severe to allow proper scoring of response to tap, 20 were not significantly different from wild-type, six had defects in initial (non-habituated) response (Fig. 4e), and three were identified as specifically tap habituation defective (Fig. 4f). adp-1 was isolated during a screen for chemosensory and salt adaptation17; in our assay it showed a loss of tap habituation (Fig. 4g). MT8943 (bas-1; cat-4) showed unusually strong habituation; it is deficient in dopamine and serotonin, and other dopamine mutants have been shown to be strong habituators18,19. Finally, the tomosyn ortholog tom-1 had a striking phenotype: only its first response was normal; all following responses were at fully habituated levels (Fig. 4h). Interestingly, tom-1 mutants have excess neurotransmitter release20. How this relates to the habituation phenotype we observed is unclear. The MWT will be valuable in elucidating the roles of these and other genes in tap habituation as well as in other behaviors.
Table 2.
Tap habituation phenotypes.
| Strain | Gene | Description | Phenotype | |
|---|---|---|---|---|
| Init | Hab | |||
| CX20 | adp-1 | Unknown locus | wt | high |
| MT8943 | bas-1; cat-4 | Monoamine biosynthesis enzymes | wt | low |
| JD21 | cca-1 | T-type Ca2+ channel α subunit | wt | wt |
| OS122 | cfi-1 | DNA-binding protein | wt | wt |
| VC855 | cle-1 | Type XV/XVIII collagen homolog | wt | wt |
| CB3241 | clr-1 | Receptor tyrosine phosphatase | wt | wt |
| RB812 | fax-1 | Nuclear hormone receptor | low | low |
| RB1816 | gpa-16 | G protein α subunit | wt | wt |
| VC2138 | kcnl-2 | Ca2+ activated K+ channel | wt | wt |
| VC2209 | lgc-46 | Ligand-gated ion channel | n/a | n/a |
| RB1593 | klp-15 | Kinesin-like protein | high | high |
| RB1416 | nhr-83 | Nuclear hormone receptor family | wt | wt |
| RB921 | nhr-84 | Nuclear hormone receptor family | wt | wt |
| VB674 | nnt-1 | Nicotinamide nucleotide transhydrogenase | wt | wt |
| RB2462 | oct-1 | Organic cation transporter | wt | wt |
| RB2164 | pde-1 | cGMP phosphodiesterase | wt | wt |
| RB1231 | pde-4 | cAMP phosphodiesterase | low | wt |
| VC282 | pdl-1 | cGMP phosphodiesterase | low | wt |
| RB809 | ptl-1 | Microtubule-associated protein | wt | wt |
| RB1638 | rab-18 | small GTPase | high | wt |
| RB1537 | rab-19 | small GTPase | wt | wt |
| VC14 | rap-2 | RAS-like GTPase | wt | wt |
| RB699 | rgs-9 | Regulator of G protein signalling | wt | wt |
| RB976 | rhgf-1 | Rho-like GTPase | wt | wt |
| RB2027 | src-1 | Tyrosine kinase | n/a | n/a |
| VC380 | tag-68 | TGFβ receptor signalling family | wt | wt |
| RB1182 | tba-1 | α tubulin | n/a | n/a |
| NM1815 | tom-1 | Tomosyn ortholog | wt | low |
| LY130 | twk-20 | TWiK K+ channel | wt | wt |
| CB246 | unc-64 | Syntaxin | n/a | n/a |
| CB1220 | unc-82 | Serine/threonine kinase | wt | wt |
| VC1392 | zip-5 | bZip transcription factor | low | wt |
| LY140 | -- | Voltage-gated K+ channel | wt | wt |
Discussion
Within the restrictions we have imposed to enable real-time tracking, the MWT has been designed to be as general as possible. It can record animals with either bright- or dark-field illumination, provides triggers to activate up to four different stimulators at defined times, and we have verified that the system robustly tracks swimming worms and Drosophila larvae in addition to crawling worms (data not shown). Since the MWT is intended for high throughput use, we have opted to reliably detect problematic cases and omit that data, rather than track reliably but slowly in those cases; if there is any doubt about the quality of image processing, the experiment can be replicated.
Other methods for recording many worms simultaneously have recently been developed, both straighforward and with sophisticated techniques to handle noise and adjacent worms4–6,21,22. However, these all require stored video, and thus have yet to demonstrate the throughput and accuracy achieved by the MWT. Commercial software supports real-time tracking (for example, Noldus EthoVision), but it is targeted mostly towards tracking small numbers of rodents, and thus uses relatively low-resolution cameras and does not provide shape information. Efforts at high-throughput behavioral screening in C. elegans have focused on worms in 96 well plates with no23 or very minimal24 image processing. These methods can achieve much higher throughput than can the MWT, but are only able to measure aggregate motion of all animals in a well. In contrast, the strength of the MWT is its applicability to a wide range of behaviors.
High throughput behavioral analysis has many advantages. One can run many controls along with experiments and test many conditions instead of being restricted to one carefully-defined protocol without knowing which aspects are important and which are irrelevant. One can test large numbers of strains or screen for new mutants instead of relying upon previous research and intuition to select a small number of candidate genes for analysis. As an example, our pilot tap habituation screen examined 33 strains with only 44 hours of tracking, including numerous wild-type controls. In contrast, only 26 strains covering 17 genes have been reported in the literature (spanning the previous 11 years). As tools like the MWT become available, studies of C. elegans behavior will increasingly benefit from this data-rich regime.
Methods
Computer vision hardware
A Dalsa Falcon 4M30 camera (8 bits; 2352 × 1728 pixels, 31 Hz) was used with a Rodenstock 60 mm f/4.0 Rodagon lens to image a 5 cm plate of worms (resulting in an image with 24.3 μm per pixel) under brightfield illumination (backlit with 4″ × 4.9″ light plate, Schott A08925 with ACE I illuminator). All tracking was performed on computers with 3 GHz Intel Core 2 Duo processors and 4 GB of RAM. Images were captured using National Instruments’ PCIe-1427 CameraLink capture card.
Online image processing
Custom software written in LabView (National Instruments) presents an interface to set up image processing and experimental parameters; once tracking is started, the software captures frames from the camera via CameraLink. (The software also supports GigE, Firewire, and USB input, and can read saved video from AVIs.) Captured frames are passed, on-line, to a custom image analysis library written in C++. Collectively, this software is called the MWT (Multi-Worm Tracker) and is available under an open-source license. The version used in this study is available as Supplementary Software 1 and the most recent version can be found at http://sourceforge.net/projects/mwt. LabView is required to modify the MWT code but not to run it. However, a runtime license for the LabView Vision package is required in all cases.
To identify moving objects, the MWT searches for portions of the image that are darker (optionally, lighter) than background by some threshold T. Objects are segmented using flood-fill from dark pixels. To avoid segmenting single pixels due to camera noise, T is set stringently but objects are filled out to a less stringent threshold βTT (typical βT ≈ 0.8).
Objects that fall within user-specified size thresholds (with more stringent thresholds S−, S+ for initial capture and less stringent ones βSS− and βT−1S+ thereafter) are presumed to be worms, and are followed by rectangular subregions 10 pixels (by default) larger on each side than the bounding box of the object. In the next frame, the object is sought only in this subregion. Thus, only a small portion of each frame needs to be analyzed. In addition, new objects are sought in a moving band covering 1/16 (by default) of the full image.
The background estimate is updated with a decaying average: B(t + 1) = (1 − α)B(t) + αI(t) where B is the estimate of the background, I is the measured intensity of a certain pixel at a given time, and by default α = 2−5. This estimate is only used to update pixels within the band used to search for new objects, and it only updates those pixels that are not in any animal’s subregion. Permanently motionless animals are treated as part of the background.
Objects are considered to persist from one frame to the next if four criteria are satisfied: (1) the new object is within the (less stringent) size threshold; (2) the new object overlaps an old object with at least 50% of its pixels; (3) only one old object overlaps the new object; and (4) only one new object overlaps that old object. Objects failing the first two criteria are discarded. Objects failing the third criteria (plus several others) are considered “collided” and are tracked as a new joint object; objects failing (4) are considered “split” and are tracked as two new objects. Due to rules (3) and (4), collisions can be robustly detected and isolated from analysis; when animals separate they can be immediately tracked again (original identities are lost, as robust maintenance of object identity is too computationally intensive to solve in real time).
The centroid position, area, best-fit line (least squares), and exterior contour of each object is computed from the binary segmentation. A spine (branchless skeleton) is computed using a fast algorithm that assumes that the objects have two pointed ends. One end isthe sharpest angle:
where n is the number of points in the contour, c is a vector between contour points, ci,k = xi+k − xi, and xi is the ith contour point. The other end is the sharpest weighted by distance from the first:
where i is the index of the selected end and j is the index being tested. Eleven spine points are then placed at the midpoint of equal divisions along the outline from the endpoints.
All quantities are computed at each timepoint for each object and accumulated in memory. When an object is lost, all of its data is written to disk. On-line estimates of speed are computed and can be displayed during tracking as a diagnostic tool.
The image processing and analysis are single-threaded (although largely parallelizable in principle), and are not strictly real-time. In particular, if a heavy workload or operating system call causes enough delay so that several images arrive while the previous one is being processed, only the latest image is used while older ones are skipped.
By default, only the first image of an experiment is saved to disk as a reference, since disk I/O would be a limiting factor unless specialized hardware was used. Timelapse images can optionally be saved to disk; on our system, aframe every two or three seconds is tolerated well.
Offline analysis
To refine the data saved by the MWT and to quantify specific behavioral parameters, we created a second software package, Choreography. Choreography is written in Java and is available under an open source license as part of the MWT package at http://sourceforge.net/projects/mwt.
A variety of conditions can be set to select which animals are suitable for analysis; the primary selection criteria are a minimum duration of tracking (typically 20 seconds) and a minimum movement (here, three times the animal’s length).
Choreography computes a variety of metrics and can provide population-averaged or animal-by-animal output, as appropriate. Metrics include area covered by the animal, bearing of the animal as defined by a least-squares line fit to the segmented pixels, speed and angular speed computed using boxcar averaging with user-defined width, length measured along the spine, average curvature along the spine, changes in direction, and—assuming that the animal’s dominant movement direction is forward—direction of motion. Note that the resolution used does not permit direction of motion to be inferred from shape analysis; head and tail are largely indistinguishable.
For noise estimates, we assumed that high-order derivatives of behavioral parameters should tend towards zero and therefore any residual is caused by measurement noise. Specifically, we assumed Gaussian noise and thus
as n → ∞. In most cases, the estimate converges by n = 2 (second derivative). The estimate is conservative for stochastic camera noise alone since behavior probably contributes somewhat to high-order derivatives. However, it is imperfect: it does not take into account rare large changes due to aliasing along pixel boundaries.
Metrics that rely upon the spine, such as length and curvature, are typically computed by first low-pass filtering the outline using an exponentially decaying kernel (2−1/2 decrease per pixel) before applying a spine-finding algorithm similar to that used in the MWT. If the plate is particularly messy and the outlines are therefore not smooth, a heuristic is applied to the outline to detect T-shapes and remove the stem of the T (presumed to be a spurious projection).
Metrics that rely upon the geometry of the animal’s path are not computed from raw centroid positions. Instead, we perform a greedy geometric decomposition of the path defined by the centroid positions. First, we classify those portions statistically indistinguishable from random jitter. Since our noise-measurement routines do not test for normality, we view the tests not as a guarantee of statistical relevance, but rather as a principled heuristic to obtain a decomposition of the animal’s path that is sufficient for robust behavioral analysis. Remaining points are fit by straight lines that are extended until new points are statistically distinguishable. Straight lines are converted to circular arcs where doing so provides statistically significant improvement (in mean squared displacement of points from the line). To speed computation, circular arcs are fit using the “HyperFit” method with zero essential bias25. This method requires finding the eigenvalues of a 4 × 4 fitting matrix, which we compute byusing an analytic solution of the quartic equation.
A change of direction is considered to be either two adjacent line segments that meet with a negative dot product, or motion along a single line that backtracks a statistically significant distance (P < 0.05 over the entire track, assuming Gaussian noise). Movement direction is scored by first splitting the path into segments where ends can be unambiguously followed---by rejecting segments whose perimeter to area ratio is statistically consistent with an animal folded back on itself along at least 1/3 of the body---and then scoring direction changes and assuming that any sufficiently long path (by default, three times body length) is predominantly forward.
Averages of computed metrics can be saved to disk or viewed as graphs within Choreography. In addition, metrics can be saved on an animal-by-animal basis or can be viewed as a color-coding of the tracks of the animal on a 2D map. Along with the tracks, the animals’ body postures can be replayed at various zoom levels and speeds to allow manual validation of the results. The replay feature also allows the creation of multiple minimaps that provide an overview of different regions on the plate; by switching between these, one can rapidly move the main viewback and forth between different locations and scales.
Organism- and experiment-specific functionality is implemented as plugins for Choreography. Plugins are tightly integrated in that they have access to all internal data used by the software, and can intervene at various stages of analysis. Although Choreography is open source and therefore can be modified directly, users are encouraged to write plugins when extending the software to reduce the difficulty of moving to new versions.
Strains
All assays were conducted with the wild-type Bristol isolate of Caenorhabditis elegans (N2) unless otherwise stated. The mutant strains CB1033 (che-2), PR811 (osm-6), RB2464 (tax-2), and CB1611 (mec-4), as well as the strains of wild-type Caenorhabditis species N2 (C. elegans, Bristol isolate) HK104 (C. briggsae), PB4641 (C. remanei), and PB2801 (C. brenneri), were obtained from the Caenorhabditis Genomic Center (CGC). These strains were maintained unfrozen for less than 30 generations before testing. The strain XJ1 is descended from N2 but was maintained unfrozen for at least several years (hundreds of generations). The 33 strains tested for tap habituation defects (Table 2) were also obtained from the CGC.
Movement experiments
Baseline recordings on food were performed on 5 cm NGM plates seeded 24 hours earlier with 50 μl of OP50 bacteria. Worms were transferred to the plate at 80–90 hours of age and allowed to acclimate for 4 hours before being tracked for 60 minutes. Unless otherwise specified, all analysis was performed with the Choreography options --shadowless -M 3 -t 20 -S --plugin Reoutline::exp --plugin Respine (see MTW User Guide in Supplementary Software 1 or at http://sourceforge.net/projects/mwt).
To assess whether plate-to-plate differences in movement rates were a result of the statistics of behavior of individual worms, we pooled all worms from eight plates and randomly sampled (with replacement) to generate data for 1,000 virtual plates. The observed differences between real plates and their mean was 60% larger than the difference between virtual plates and their mean (20.7 μm s−1 vs. 12.9 μm s−1); five of eight real plates had difference scores that were statistical outliers (P < 0.01) of the virtual plates’ distribution. Thus, the observed variability between plates was not solely due to statistical sampling. Other likely causes include different environmental conditions on the plates, subtle perturbations during recording, or behavioral states that last longer than identity is maintained (205 s on average here). Fortunately, plate-to-plate variability does not obscure the advantage of multi-worm tracking: monte carlo generation of virtual plates with 10 tracked worms produced a data set with slightly larger plate-to-plate variability than was observed for real plates.
Omega turns were detected with a combination of three factors. A spine-length-to-perimeter metric flagged regions where the worm had folded back to touch itself. To detect tight turns without self-contact, we used both the third principal component of the spine shape (which correlates with turns), and a folding score
where vb,a is the vector from spine point a to b. Intuitively, this last metric is small when the worm’s spine is folded exactly in half (all dot products will be close to −1); constants are chosen so that one gets a minimum of 0.5 for each pair of spine points folded onto each other. Thresholds for these metrics were set by hand; a wide range of parameters gave results that agreed with human judgment in obvious cases. This method detected 74 of 76 hand-annotated omega turns in our reference data set (3% false negatives), and 98 of 100 automatically-scored omega turns were confirmed by hand in wild-type movement data on relatively clean plates (2% false positives, though the error rate is higher on a sub-optimal plate such as our reference data set). Thus, the overall error rate is ~5%.
Chemotaxis experiments
Food chemotaxis plates were prepared by placing a 50 μl drop of OP50 bacteria at x,y coordinates (r/2,0) where (0,0) represents the center of a plate of radius r. In practice, on the 50 mm inner diameter plates we used, the actual coordinates (measured by hand) were (12.4,0.2) mm with standard deviation (±0.5,±0.9); the diameter of the spot of food was 3.7 ± 0.3 mm. A control spot of LB growth medium was placed at (−r/2,0). The OP50 was grown for 48 hours. For testing, 6–12 worms were picked from food to an empty plate to reduce the amount of stray food, and then picked again from the empty plate to coordinate (0,0). Tracking was started within 60 s of placement and left to run for 20 minutes.
To quantify food chemotaxis, we counted crossings of the animals’ centroids along a ring 1 mm outside of the spot of food or the control spot. We computed Bayesian preference estimates P(t) and the associated standard error σP(t) for each spot size, based on a binomial distribution with uniform prior26:
where F(t) is the cumulative number of (signed) crossings into the food spot at time t and C(t) is the same for the control spot.
To quantify pirouettes, we monitored the animal at 0.5 s intervals and calculated the angle between the direction of movement and the direction to the food. We then pooled the quadrant heading towards the food (angles from −π/4 to π/4) and the quadrant heading away, and counted reversal frequency (probability per unit time of executing a reversal). We compared the behavior of worms in a 1 cm radius circle about the center of the food spot to those in a 1 cm radius circle about the center of the control spot. Within-strain statistical comparisons were performed using a χ2 test; P-values for hypotheses about the similarity of strains were computed using monte carlo sampling.
Salt chemotaxis plates were prepared following previous work9. In brief, we placed a 5 μl drop of 500 mM NaCl at the center of a low salt plate (1 mM CaCl2, 1 mM MgSO4, 5 mM potassium phosphate, pH 6, 2% agar) three hours before tracking. Worms were washed 30 minutes before tracking in low salt buffer (as plate, less agar) and rinsed in low salt buffer plus 0.05% glycerol. 8–12 worms were transferred from the rinse bath to a < 0.5 μl droplet held in a glass loop, which was then dotted onto the plate 12.5 mm from the center. The droplet dried within 60 s; tracking was started within 120 s. To quantify weathervane turning, we segmented the path into one-movement-cycle chunks (two body bends), calculated the angle between the animal’s movement and the direction to the food (“bearing”), and how that angle changed over one cycle (“curve”). We considered only animals between 2.5 mm and 12.5 mm from the location of the NaCl drop, as this was predicted to be where the gradient was steepest (using the method of 8) and provided some robustness to inaccurate placement of the drop.
Tap habituation experiments
Tap habituation assays were performed on food after a minimum of 6 hours’ recovery from transfer; animals were tracked for 10 minutes to allow them to approach steady-state behavior, and then were tapped 20 or 30 times (Fig. 4a–d and e–g respectively) at a 10 s inter-stimulus interval with a custom-built solenoid tapper that drives a small plunger into the side of the plate. A reversal was scored when a worm was still or moving forward at the time of the tap and moved backwards within one second of the impact; the reversal was considered to be complete when the worm began a pause or forward motion lasting for more than 0.2 s. Cases where the animal was already reversing were removed from analysis. For reversal probability computations, reversal distances of less than 30 μm were considered “no reversal”, and we converted counts of reversals and failures into probabilities using a Bayesian estimate of probability with uniform prior.
Validation was performed by comparing with manually annotated reversals. Out of 148 responses scored by hand as either yes, no, or already-reversing, the tracker disagreed with eight (5% error rate). All but one discrepancy was due to small disagreements over the timing or size of a response.
Putative tap habituation mutants were prepared using a higher-throughput method: five gravid adults were placed on a plate seeded with 50 μl E. coli 24 hours before. The adults were left to lay eggs and then removed from the plate 3 hours later, leaving approximately 60–80 eggs. The plate was tracked once the animals reached adulthood (80 ± 3 hours later, at 20 °C). The recording protocol consisted of 100 s of baseline followed by 30 taps applied to the side of the plate at a 10 s inter-stimulus interval. The data was analyzed with Choreography’s reversal detection plugin: --plugin MeasureReversal::tap::dt=1::collect=0.5. For each mutant strain and wild-type replicate, three or four plates were tested (except CB1220: n = 2); on average, 34 worms per plate were suitable for analysis. A wild-type distribution for all wild-type plates tested on the same day was created for the initial and habituated (28th–30th) responses. All strains and wild-type replicates were standardized to the distribution for the day they were tested. Monte Carlo sampling was used to generate the null hypothesis distribution given this scheme, and the resulting P-values were re-interpreted as an effective Z-score for plotting. Values outside ±3.13 were considered to be significantly different than the wild-type distribution (two-tailed, P < 0.05, with Bonferroni correction for multiple comparisons). CX20 and NM1815 were confirmed using an unpaired two-tailed t-test between the raw reversal probabilities of the last stimulus versus the wild-type replicate that was tested most closely chronologically.
Reference data
The reference data set used for most hand-annotation and for speed and accuracy tests was a tap habituation screen assay with two differences. First, the worms were given only 5 minutes to recover before tapping instead of 10. Second, the worms were allowed to grow up for 100 hours. These conditions provided an extra challenge for the tracking algorithms, as there were a number of L1 progeny on the plate by that time. To improve the outlines in this more-difficult case, we used the::despike option of the Reoutline plugin.
Since the MWT cannot save video at the full rate (7.2 GB/minute), we used a custom LabView program to stream video to a solid state drive so we had a complete record of the experiment. We then ranthe MWT on the saved video.
Supplementary Material
The Multi-Worm Tracker consists of real-time image processing software, MWT, and offline analysis software, Choreography.
Acknowledgments
We appreciate the assistance of P. Liu in collecting data for the tap habituation screen as part of a directed studies course in the Department of Psychology at the University of British Columbia, and of D. Hoffmann in the design of the multi-worm tracker illumination platform and solenoid for tap habituation. M. Zlatic provided valuable bug reports, suggestions for features, and feedback on the usability of the MWT outside of the authors’ labs. We also appreciate advice from and discussions with members of our labs and our colleagues at the Janelia Farm Research Campus (JFRC). Strains used in this study were acquired with speed and ease from the Caenorhabditis Genetics Center, for whose services we are grateful;many of these strains were generated by the C. elegans Gene Knockout Consortium. This work was supported by the Howard Hughes Medical Institute’s JFRC (N.A.S. and R.A.K.) and the JFRC visitor program (A.C.G. and C.H.R.), and by a National Science and Engineering Research Council of Canada (NSERC) operating grant (to C.H.R.) and post-graduate scholarship (to A.C.G.). Preliminary investigations into multi-worm tracking were supported by the Peter Wall Institute for Advanced Studies’ Visiting Junior Scholars Program and by the lab of W. Schafer.
Footnotes
Author Contributions
N.A.S. and R.A.K. designed the MWT system, built the hardware, and wrote the software. A.C.G., C.H.R., and R.A.K. designed the experiments. A.C.G. and R.A.K. conducted the experiments and analyzed data. R.A.K. wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Baek JH, Cosman P, Feng Z, Silver J, Schafer WR. Using machine vision to analyze and classify Caenorhabditis elegans behavioral phenotypes quantitatively. J Neurosci Methods. 2002;118:9–21. doi: 10.1016/s0165-0270(02)00117-6. [DOI] [PubMed] [Google Scholar]
- 2.Feng Z, Cronin CJ, Wittig JH, Sternberg PW, Schafer WR. An imaging system for standardized quantitative analysis of C. elegansbehavior. BMC Bioinformatics. 2004;5:115. doi: 10.1186/1471-2105-5-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cronin CJ, Feng Z, Schafer WR. Automated imaging of C. elegans behavior. Methods Mol Biol. 2006;351:241–251. doi: 10.1385/1-59745-151-7:241. [DOI] [PubMed] [Google Scholar]
- 4.Ramot D, Johnson BE, Berry TL, Carnell L, Goodman MB. The Parallel Worm Tracker: a platform for measuring average speed and drug-induced paralysis in nematodes. PLoS ONE. 2008;3:e2208. doi: 10.1371/journal.pone.0002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsechpenakis G, Bianchi L, Metaxas D, Driscoll M. A novel computational approach for simultaneous tracking and feature extraction of C. elegans populations in fluid environments. IEEE Trans Biomed Eng. 2008;55:1539–1549. doi: 10.1109/TBME.2008.918582. [DOI] [PubMed] [Google Scholar]
- 6.Roussel N, Morton CA, Finger FP, Roysam B. A computational model for C. elegans locomotory behavior: application to multiworm tracking. IEEE Trans Biomed Eng. 2007;54:1786–1797. doi: 10.1109/TBME.2007.894981. [DOI] [PubMed] [Google Scholar]
- 7.Stephens GJ, Johnson-Kerner B, Bialek W, Ryu WS. Dimensionality and dynamics in the behavior of C. elegans. PLoS Comput Biol. 2008;4:e1000028. doi: 10.1371/journal.pcbi.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pierce-Shimomura JT, Morse TM, Lockery SR. The fundamental role of pirouettes in Caenorhabditis elegans chemotaxis. J Neurosci. 1999;19:9557–9569. doi: 10.1523/JNEUROSCI.19-21-09557.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iino Y, Yoshida K. Parallel use of two behavioral mechanisms for chemotaxis in Caenorhabditis elegans. J Neurosci. 2009;29:5370–5380. doi: 10.1523/JNEUROSCI.3633-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawin ER, Ranganathan R, Horvitz HRC. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26:619–631. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 11.Coburn CM, Bargmann CI. A putative cyclic nucleotide-gated channel is required for sensory development and function in C. elegans. Neuron. 1996;17:695–706. doi: 10.1016/s0896-6273(00)80201-9. [DOI] [PubMed] [Google Scholar]
- 12.Rankin CH, Beck CD, Chiba CM. Caenorhabditis elegans: a new model system for the study of learning and memory. Behav Brain Res. 1990;37:89–92. doi: 10.1016/0166-4328(90)90074-o. [DOI] [PubMed] [Google Scholar]
- 13.Rankin CH, Broster BS. Factors affecting habituation and recovery from habituation in the nematode Caenorhabditis elegans. Behav Neurosci. 1992;106:239–249. doi: 10.1037//0735-7044.106.2.239. [DOI] [PubMed] [Google Scholar]
- 14.Rose JK, Sangha S, Rai S, Norman KR, Rankin CH. Decreased sensory stimulation reduces behavioral responding, retards development, and alters neuronal connectivity in Caenorhabditis elegans. J Neurosci. 2005;25:7159–7168. doi: 10.1523/JNEUROSCI.1833-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- 16.Fujiwara M, Ishihara T, Katsura I. A novel WD40 protein, CHE-2, acts cell-autonomously in the formation of C. elegans sensorycilia. Development. 1999;126:4839–4848. doi: 10.1242/dev.126.21.4839. [DOI] [PubMed] [Google Scholar]
- 17.Colbert HA, Bargmann CI. Odorant-specific adaptation pathways generate olfactory plasticity in C. elegans. Neuron. 1995;14:803–812. doi: 10.1016/0896-6273(95)90224-4. [DOI] [PubMed] [Google Scholar]
- 18.Sanyal S, et al. Dopamine modulates the plasticity of mechanosensory responses in Caenorhabditis elegans. EMBO J. 2004;23:473–482. doi: 10.1038/sj.emboj.7600057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kindt KS, et al. Dopamine mediates context-dependent modulation of sensory plasticity in C. elegans. Neuron. 2007;55:662–676. doi: 10.1016/j.neuron.2007.07.023. [DOI] [PubMed] [Google Scholar]
- 20.Gracheva EO, et al. Tomosyn inhibits synaptic vesicle priming in Caenorhabditis elegans. PLoS Biol. 2006;4:e261. doi: 10.1371/journal.pbio.0040261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fontaine E, Burdick J, Barr A. Automated tracking of multiple C. Elegans. Conf Proc IEEE Eng Med Biol Soc; 2006. pp. 3716–3719. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Roysam B. Joint tracking and locomotion state recognition of C. elegans from time-lapse image sequences. IEEE International Symposium on Biomedical Imaging; 2010. pp. 540–543. [Google Scholar]
- 23.Simonetta SH, Golombek DA. An automated tracking system for Caenorhabditis elegans locomotor behavior and circadian studies application. J Neurosci Methods. 2007;161:273–280. doi: 10.1016/j.jneumeth.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 24.Buckingham SD, Sattelle DB. Fast, automated measurement of nematode swimming (thrashing) without morphometry. BMC Neurosci. 2009;10:84. doi: 10.1186/1471-2202-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Sharadqah A, Chernov N. Error analysis for circle fitting algorithms. Electronic Journal of Statistics. 2009;3:886–911. [Google Scholar]
- 26.Gelman A, Carlin JB, Stern HS, Rubin DB. Bayesian Data Analysis. Vol. 36. Chapman & Hall/CRC; 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Multi-Worm Tracker consists of real-time image processing software, MWT, and offline analysis software, Choreography.