Abstract
Background
LMO2 is highly expressed at the most immature stages of lymphopoiesis. In T-lymphocytes, aberrant LMO2 expression beyond those stages leads to T-cell acute lymphoblastic leukemia, while in B cells LMO2 is also expressed in germinal center lymphocytes and diffuse large B-cell lymphomas, where it predicts better clinical outcome. The implication of LMO2 in B-cell acute lymphoblastic leukemia must still be explored.
Design and Methods
We measured LMO2 expression by real time RT-PCR in 247 acute lymphoblastic leukemia patient samples with cytogenetic data (144 of them also with survival and immunophenotypical data) and in normal hematopoietic and lymphoid cells.
Results
B-cell acute lymphoblastic leukemia cases expressed variable levels of LMO2 depending on immunophenotypical and cytogenetic features. Thus, the most immature subtype, pro-B cells, displayed three-fold higher LMO2 expression than pre-B cells, common-CD10+ or mature subtypes. Additionally, cases with TEL-AML1 or MLL rearrangements exhibited two-fold higher LMO2 expression compared to cases with BCR-ABL rearrangements or hyperdyploid karyotype. Clinically, high LMO2 expression correlated with better overall survival in adult patients (5-year survival rate 64.8% (42.5%–87.1%) vs. 25.8% (10.9%–40.7%), P= 0.001) and constituted a favorable independent prognostic factor in B-ALL with normal karyotype: 5-year survival rate 80.3% (66.4%–94.2%) vs. 63.0% (46.1%–79.9%) (P= 0.043).
Conclusions
Our data indicate that LMO2 expression depends on the molecular features and the differentiation stage of B-cell acute lymphoblastic leukemia cells. Furthermore, assessment of LMO2 expression in adult patients with a normal karyotype, a group which lacks molecular prognostic factors, could be of clinical relevance.
Keywords: acute leukemia, B lymphocytes, gene expression, prognostic factor
Introduction
The Lim Domain Only 2 gene (LMO2, also termed RBTN2 and TTG2) encodes a cystein-rich LIM domain-containing transcription factor that is required for complete hematopoiesis in mice.1 While LMO2 is expressed at various levels in virtually every tissue in fetal and adult life, its expression in hematopoietic cells is tightly regulated and varies at different ontogeny stages of maturation.2 LMO2 was originally identified through its involvement in recurrent chromosomal translocations in T-cell acute lymphoblastic leukemia (T-ALL). Indeed, constitutive activation of LMO2 in T cells by juxtaposition with the T-cell receptor (TCR) gene loci through t(11;14)(p13;q11) or t(7;11)(q35;p13), or by the cryptic deletion del(11)(p12p13), is characteristic of T-ALL.3,4 In normal T-cell development, LMO2 is expressed in immature CD4/CD8 double negative thymocytes, but its expression is down-regulated during T-cell maturation and is absent in mature T cells.5 Ectopic expression of LMO2 in the T-cell lineage in transgenic mouse models leads to clonal expansion of T cells, eventually generating human-like T-ALL.6 Moreover, during gene therapy of patients with X-linked severe combined immunodeficiency (X-SCID), retroviral insertion in the proximity of the LMO2 gene resulted in the overexpression of this gene and development of T-ALL in 3 children.7 These data demonstrate that LMO2 functions as an oncogene in this leukemia.
In normal B-cell differentiation and maturation, LMO2 is expressed mainly at two stages: at early lymphopoiesis within the bone marrow and in germinal centers (GC) of secondary lymphoid organs. LMO2 expression is also found in B-cell lymphomas derived from GC lymphocytes, including follicular, Burkitt’s and diffuse large B-cell (DLBCL) lymphomas, as well as in lymphocyte-predominant Hodgkin’s lymphoma.2 Remarkably, LMO2 expression is an independent prognostic factor of survival in patients with DLBCL treated with anthracycline-based chemotherapy with and without rituximab,8 as well as in chronic myeloid leukemia9 and pancreatic cancer.10 While marked advances have been made in establishing the significance and function of LMO2 in T-ALL and B-cell lymphomas, its role in B-cell acute lymphoblastic leukemia (B-ALL) has not been investigated.
B-ALL comprises cytogenetically distinct subgroups defined by specific recurrent chromosomal translocations or by the presence of hyperdiploid and hypodiploid karyotypes. Clinically, the distinction between these genetic subgroups is important for prognosis and selection of optimal therapeutic options. In addition to cytogenetic analysis, the study of the immunophenotype of the blasts is also important in the diagnosis of B-ALL, and the distinction between T-, mature B- and precursor B-phenotypes affects therapeutic decision making.11
This report assesses LMO2 expression in a series of patients with B-ALL. Our data suggest that, in contrast to T-ALL, LMO2 expression reflects the developmental stage in which the blasts are arrested rather than being an oncogenic event in this disease. Furthermore, we show that LMO2 expression correlates with survival of B-ALL patients, being an independent prognostic factor in the subgroup with normal karyotype, particularly among patients over 15 years of age.
Design and Methods
Patients’ samples and cell lines
LMO2 expression was assessed in a cohort of 247 patients with acute lymphoblastic leukemia (ALL) who were treated according to the PETHEMA (Spanish Group for the Study and Therapy of Haematological Malignancies) protocols at the Departments of Haematology of the Clínica Universidad de Navarra (Pamplona), Hospital Reina Sofía (Cordoba) and other national institutions belonging to the PETHEMA group.12,13 Full clinical and laboratory data were available for 39 T-ALL and 145 B-ALL patients (one of them lacking survival data) with a median age of 11.5 years (range 0.5–87 years), including 100 children under 15 years of age (median 5.5 years, range 0.5–14) and 84 patients over 15 years of age (mean age 34.5, range 15–87). Sample collection was approved by the individual institutional review boards of the participating institutions and informed consent was obtained from all adult patients and children’s parents or legal guardians.
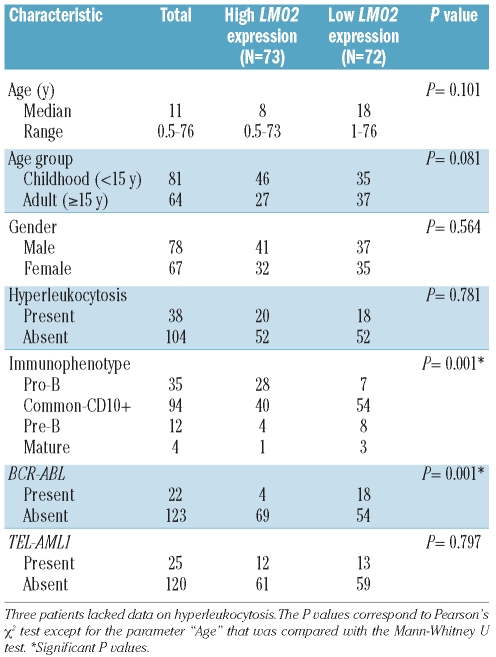
Cytogenetic and molecular analyses were performed for all samples (Online Supplementary Design and Methods). We found 32 cases (15%) with TEL-AML1 (ETV6-RUNX1) gene rearrangement, 41 cases (20%) with BCR-ABL rearrangement, 16 cases (8%) with rearrangements involving the MLL gene, 9 cases (4%) with hyperdiploid karyotype (more than 50 chromosomes), 21 cases (10%) with miscellaneous abnormalities and 89 cases (43%) with normal karyotype. Detailed information on the number of cases with cytogenetic, immunophenotypical and survival data available is provided in the Online Supplementary Table S1 and Figure S1. The clinical and immunophenotypical characteristics of the B-ALL patient samples are shown in Table 1. Methods for the sorting of the normal lymphocyte subpopulations are available in the Online Supplementary Appendix and Table S2, along with the cell lines included in this study.
Table 1.
Clinical parameters in B-ALL patients.
Quantitative real time PCR (Q-RT-PCR)
All patient samples were taken at initial diagnosis and come from bone marrow aspirates. Only samples with more than 80% blasts were included in this study. Total RNA from patients’ mononuclear bone marrow cells was extracted with Ultraspec RNA isolation system (Biotecx). RNA quantity and quality were assessed by spectrophotometric measurements with NanoDrop® ND-1000 Spectrophotometer. Reverse transcription and Q-PCR were performed as previously described14 using specific TaqMan probes and 10x LightCycler TaqMan Master (Roche Applied Science). LMO2 expression was normalized to ABL expression as recommended by the Europe Against Cancer group for leukemic samples.15 This group reported that ABL expression remained constant not only in normal blood/bone marrow samples but also in leukemic samples with different genetic alterations, including cases with BCR-ABL rearrangement. We confirmed that ABL expression remained constant among our samples with different genetic alterations and in the normal karyotype subgroup (P= 0.761) (Online Supplementary Figure S2). The following primers were used for PCR: forward LMO2 primer 5′-GGCGGCGCCTCTACTACA-3′, reverse LMO2 primer 5′-CCAAAAAGCCTGAGATAGTCTCT-3′, TaqMan LMO2 probe 5′-CTGGGCCGGAAGCTCTGCC-3′. Forward ABL primer 5′-TGGAGATAACACTCTAAGCATAACTAAAGGT-3′, reverse ABL primer 5′-GATGTAGTTGCTTGGGACCCA-3′, TaqMan ABL probe 5′-CCATTTTTGGTTTGGGCTTCACACCATT-3′.
Total RNA from cell lines and normal lymphocyte subpopulations was extracted using TRIzol reagent (Invitrogen). cDNA synthesis was performed with M-MLV reverse transcriptase (Invitrogen) and Q-PCR was performed in the 7300 Real Time PCR System with Universal PCR Master Mix (Applied Biosystems). Hs 00277106_m1 (Applied Biosystems) was the probe for LMO2 and Hs 99999906_m1 for PGK1, selected as reference gene due to its good performance in our experience with lymphoid samples and cell lines.16 LMO2 expression was also assessed using these probes in 54 B-ALL patients to compare them with normal lymphocytes and cell lines.
When using either ABL or PGK1 as housekeeping genes, LMO2 expression was calculated as a ratio between the amount of LMO2 referred to a standard curve of cDNA and the amount of the reference gene referred to a standard curve of the same cDNA. Correlation between the LMO2 expression results obtained using PGK1 or ABL as housekeeping genes was highly significant (P=0.003, Spearman’s Rho= 0.393).
Statistical analysis
The SPSS 15.0 software was used for statistical analyses. Kaplan-Meier curves and the log rank test were used for univariate survival analysis, for which LMO2 expression was categorized using its mean expression among all B-ALL patients as threshold value (2.29). Overall survival (OS) was defined as the time interval between the date of diagnosis and the date of death or last follow up. Event-free survival (EFS) was defined as the time interval from the date of initial diagnosis to the date of disease progression, death from any cause or last follow-up evaluation. The relevant clinical variables of these patients were also tested for their possible correlation with survival by the log rank test (Table 2), and a multivariate Cox’s regression assay was performed including all variables with P< 0.05. The possible association between nominal variables and categorical LMO2 expression was tested by Pearson’s χ2 test. LMO2 expression was used as a continuous variable for the remaining comparisons. Representative values are given as median and range. Due to the lack of fit to the normal distribution of LMO2 expression values, tested by Kolmogorov-Smirnov and Shapiro-Wilk tests, non-parametric comparisons were performed: Mann-Whitney U test for comparisons of two groups or Kruskal-Wallis test for comparisons of more than two groups. Holms-Bonferroni correction was applied in cases of multiple comparisons with the Mann-Whitney U test. Correlation between continuous variables was assessed by Spearman’s correlation test. Differences were considered significant when P< 0.05.
Table 2.
Factors associated with prolonged overall survival of B-ALL patients.
Results
LMO2 expression changes during B-cell lymphopoiesis and at distinct differentiation stages
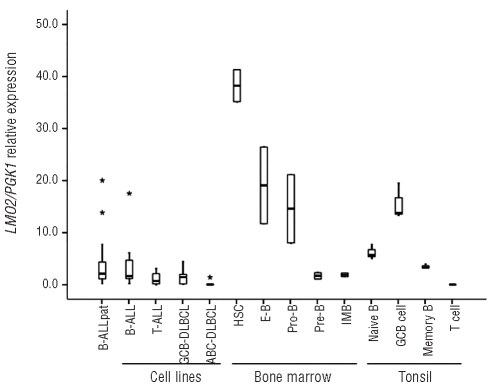
LMO2 expression was measured by Q-RT-PCR in different normal hematopoietic and B-cell subpopulations representing distinct differentiation stages. LMO2 expression was highest at the earliest stages of bone marrow hematopoiesis, decreasing gradually from hematopoietic stem cells (HSCs) to the pro-B cell stage: median 38.23 (range 35.16–41.30) in HSCs, 19.08 (11.72–26.44) in early B cells, and 14.59 (8.04–21.14) in pro-B cells. After the pro-B stage, a sharp approximately 8-fold decrease in LMO2 expression was observed: 1.71 (1.10–2.32) in pre-B cells and 1.90 (1.55–2.25) in immature B cells. In secondary lymphoid organs, LMO2 expression was increased at the GC B-cell stage [13.81 (13.29–19.54)] compared to naïve B cells [5.67 (5.06–7.78)] and memory B cells [3.26 (3.16–3.95)] (Figure 1).
Figure 1.
LMO2 expression during B-cell differentiation and in B-lymphoid malignancies. LMO2 expression was measured by Q-RT-PCR. The bar inside the boxes represents the median value, the limits of the box the interquartile range, the whiskers the range of expected values and (*) are outlier values. B-ALL: B-cell acute lymphoblastic leukemia, pat: patient samples, T-ALL: T-cell acute lymphoblastic leukemia, ABC-DLBCL: activated B-cell like diffuse large B-cell lymphoma, GCB-DLBCL: germinal center B-cell like diffuse large B-cell lymphoma, HSC: hematopoietic stem cell, E-B: early B cell, IMB: immature B cell, GCB=germinal center B cell.
Comparison of LMO2 expression in B-ALL, T-ALL, DLBCL and B-cell precursors
Similar levels of LMO2 expression were observed in primary B-ALL samples [2.13 (0.21–20.04)] and B-ALL cell lines [1.63 (0.21–17.54)] (P=0.986) as well as in the pre-B cell precursor subpopulation [1.71 (1.10–2.32)] (P=0.566) (Figure 1). Next, we compared LMO2 expression between cell lines of different origins. LMO2 expression tended to be lower in T-ALL [0.71 (0.00–3.11)] and GCB-DLBCL [1.45 (0.03–4.41)] cell lines compared to B-ALL cell lines but the difference did not reach statistical significance (P=0.151 and P=0.080, respectively) (Figure 1). As expected, LMO2 expression in ABC-DLBCL cell lines [0.02 (0.00–1.41)] was lower compared to B-ALL and GCB-DLBCL cell lines (P=0.018 and P=0.035, respectively).
Immunophenotypical and cytogenetic subgroups of B-ALL exhibit variable levels of LMO2 expression
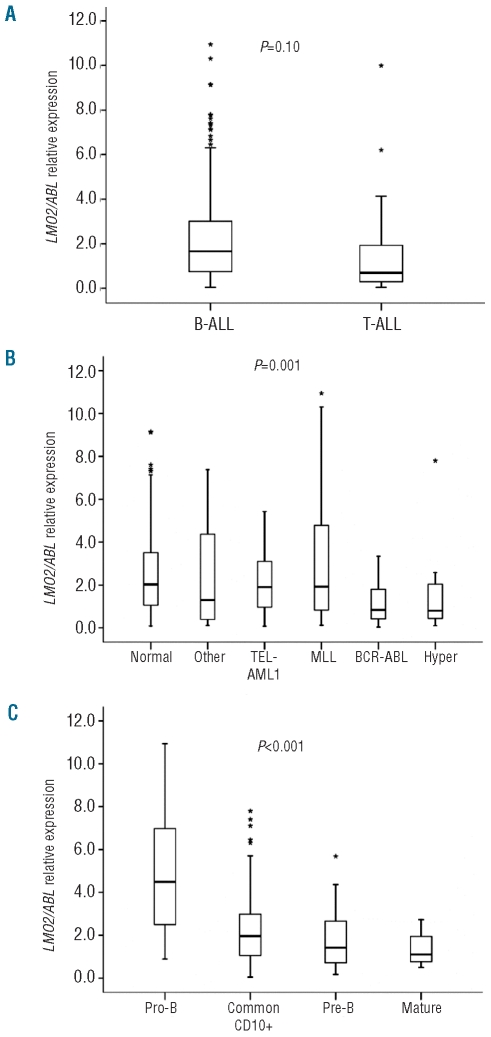
LMO2 expression was significantly higher in B-ALL patients than in T-ALL patients [1.66 (0.04–10.90) vs. 0.70 (0.04–10.00), P=0.010] (Figure 2A). Cytogenetic and molecular data on the B-ALL cases allowed the distinction of six specific genetic subgroups with hyperdiploidy, BCR-ABL rearrangement, TEL-AML1 rearrangement, translocations involving the MLL locus, normal karyotype and cases with miscellaneous alterations. B-ALL cases with BCR-ABL rearrangement or hyperdiploid karyotype showed the lowest LMO2 expression values [0.84 (0.00–3.30) and 0.80 (0.10–7.80) respectively], while all the remaining cases, including those with MLL translocations [1.92 (0.10–10.90)] or TEL-AML1 rearrangement [1.91 (0.10–5.40)] exhibited higher levels of LMO2 expression (P=0.001) (Figure 2B).
Figure 2.
LMO2 expression changes between B-ALL and T-ALL and among the main immunophenotypical and cytogenetic subtypes of B-ALL. LMO2 expression was measured by Q-RT-PCR. (A) B-ALL vs. TALL comparison was performed by the Mann Whitney U test. LMO2 expression among the different (B) cytogenetic and (C) immunophenotypical subgroups of B-ALL was compared by the Kruskal-Wallis test. The corresponding P values are shown within the graphs. BALL= B-cell acute lymphoblastic leukemia, T-ALL= T-cell acute lymphoblastic leukemia, Hyper= hyperdiploid karyotype (> 50 chromosomes), Normal: normal karyotype, MLL: translocations involving the MLL locus, Other: miscellaneous alterations. (*) depicts outlier values.
B-ALL patient samples were also subclassified in four different immunophenotypical subgroups (pro-B, common-CD10+, pre-B and mature). Among these subgroups, pro-B cell ALL showed the highest LMO2 expression [4.49 (0.90–10.90)] while B-ALL with common-CD10+ immunophenotype [1.96 (0.10–7.80)], pre-B cell ALL [1.42 (0.20–5.70)] and mature B-ALL [1.10 (0.50–2.70)] had lower expression (P<0.001) (Figure 2C). Notably, the difference in LMO2 expression between the pro-B and pre-B leukemia immunophenotypes resembled the decrease in LMO2 expression observed upon transition of pro-B to pre-B cells in normal bone marrow (Figure 1).
LMO2 expression is associated with survival in patients with B-ALL
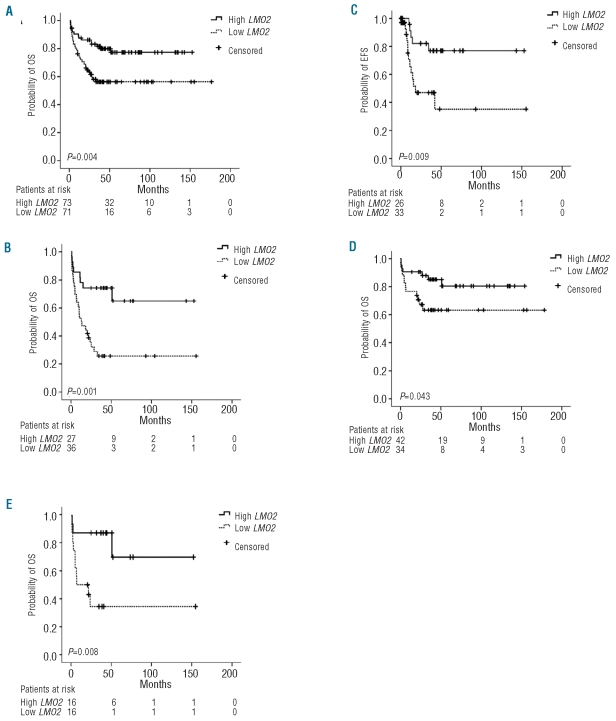
Previous studies demonstrated that increased LMO2 expression correlates with better survival in DLBCL, as well as in chronic myeloid leukemia and pancreatic cancer. Therefore, we examined the possible correlation between LMO2 RNA expression and patient clinical outcome in ALL. We divided the patients in two subgroups with high or low levels of LMO2 expression using the mean LMO2 expression value of the whole cohort of B-ALL patients (2.29) as cut-off value. The median overall survival (OS) was not reached in any of the two subgroups. The OS rate was significantly higher in B-ALL patients with high levels of LMO2 expression compared to patients with low levels of LMO2 expression: the 5-year cumulative survival rate with 95% CI was 77.2%(66.8%–87.6%) vs. 56.1%(44.1%–68.1%); P=0.004 (Figure 3A). To determine whether LMO2 expression is an independent predictor for OS in BALL, we initially examined whether other known clinical and laboratory prognostic factors also correlated with OS by log rank test. In univariate analysis, age under 15 years at diagnosis (childhood B-ALL) and the presence of TEL-AML1 gene rearrangement correlated with favorable prognosis, whereas the presence of hyperleukocytosis (leukocyte count > 50,000/mm2) or BCR-ABL gene fusion were associated with shorter OS (Table 2). Analyses of the possible correlation of MLL gene rearrangements or hyperdiploid karyotypes with survival in our cohort of patients were not performed because of the small number of cases in these subgroups, which precluded any robust statistical conclusion.
Figure 3.
LMO2 expression predicts overall survival in patients with B-ALL. B-ALL patient samples were analyzed for LMO2 expression by Q-RT-PCR. LMO2 expression was categorized using its mean expression value (2.29) as the cut-off value. Overall survival (OS) was compared between patients with high or low LMO2 expression (A) in the whole cohort, (B) in patients older than 15 years, (D) in the subgroup with normal karyotype, and (E) in patients older than 15 years and with normal karyotype. (C) Event-free survival (EFS) was compared between patients older than 15 years with high or low LMO2 expression. The patients remaining in the study (patients at risk) for each subgroup at each time point and the probability associated with the log rank test are displayed.
A multivariate Cox’s regression analysis including age group, hyperleukocytosis, presence of BCR-ABL or TEL-AML1 gene rearrangements and subgrouping based on LMO2 expression was performed. Only age over 15 years and the presence of BCR-ABL gene rearrangement were independently associated with worse OS in B-ALL (P=0.003 and P<0.001, respectively). In contrast, LMO2 expression was not independently correlated with OS, suggesting its association with age and/or the presence of BCR-ABL rearrangement. By using Pearson’s χ2 test, a highly significant association between the presence of BCR-ABL rearrangement and low LMO2 expression was found (P=0.001). As age group is one of the main prognostic factors in our cohort of patients, and the treatment protocols are more aggressive and effective in the age group under 15 years, we performed a separate study on the association between LMO2 expression and survival for each age group. We found that in adult B-ALL cases high LMO2 expression is a predictor of better outcome (5-year survival rate 64.8% (42.5–87.1%) vs. 25.8% (10.9–40.7%), P=0.001) (Figure 3B), while in patients under 15 years of age there is no difference in mortality between the patients with high or low LMO2 expression (P=0.673), probably due to the low number of events among patients in this age group (5-year survival rate 84.8% (76.4–93.2%) vs. 42.7% (28.8–56.6%) in adults). As far as event-free survival (EFS) is concerned, patients older than 15 years with high LMO2 expression displayed better survival rates than those with low LMO2 expression (5-year survival rate 76.9% (59.1–94.7%) vs. 35.2% (9.9–60.5%), P=0.009) (Figure 3C).
LMO2 expression is an independent prognostic factor among the B-ALL cases with normal karyotype
In our series, the subgroup with normal karyotype included the highest number of cases. The presence of a normal karyotype did not correlate with OS (P=0.101). Remarkably, in this subgroup of patients high LMO2 expression was associated with better OS: the 5-year survival rate was 80.3% (66.4–94.2%) vs. 63.0% (46.1–79.9%) (P=0.043) (Figure 3D). Moreover, when multivariate Cox’s regression analysis was applied, testing the variables LMO2 expression levels, age group and hyperleukocytosis, LMO2 expression and age group remained independent prognostic factors (P=0.045 and P=0.007, respectively).
When we performed a log rank test separately in the two age groups, we found that higher LMO2 expression remained a predictor of better outcome only in the patients over 15 years of age: 5-year survival rate 70.0% (36.7–103.3%) vs. 34.3% (9.4–59.2%), P=0.008 (Figure 3E). We did not find any statistical association of EFS with LMO2 expression in the group of patients with normal karyotype and over 15 years of age.
Discussion
The role of LMO2 as oncogene in T-ALL6 and, conversely, as favorable prognostic factor in DLBCL,8 CML9 and pancreatic cancer,10 led us to evaluate the practically unexplored role of LMO2 in B-ALL. We found that LMO2 is expressed in this disease, as previously proposed.2,17 Most B-ALL cases exhibited LMO2 expression levels similar to those measured in non-transformed pre-B cells, and even in the cases with the highest LMO2 expression these levels did not exceed those found in non-transformed pro-B cells. As bone marrow B-cell precursors (both pro-B and pre-B cells) are immunophenotypically very similar to the blasts of the majority of B-ALL cases, the range of LMO2 expression found in B-ALL probably reflects the stage at which these blasts undergo differentiation arrest. In this regard, measurement of the expression of LMO2 in a BALL sample might help to assess the grade of differentiation of the malignant cells. Furthermore, LMO2 expression was associated with different cytogenetic subtypes of B-ALL, being particularly low in BCR-ABL and hyper-diploid cases. Overall, LMO2 RNA expression levels in BALL seem to reflect the molecular and phenotypical characteristics of the leukemic cells.
Despite the clear oncogenic role of LMO2 in T-ALL, this is probably not the case in B-ALL. No translocations of this gene have been reported in this disease, and other genetic alterations involving the LMO2 locus are extremely rare among B-ALL cases. For instance, genome-wide analysis of 192 B-ALL samples using SNP arrays detected a deletion of 155 kb upstream of the LMO2 gene locus in only one case, in contrast to the 8% of T-ALL cases that showed this alteration.18 Furthermore, among 226 B-ALL cases analyzed in our institution by conventional cytogenetics, 154 presented chromosomal abnormalities, but none showed alterations in 11p13 (MJ Calasanz, unpublished data, 2010). Moreover, in 3 out of 10 X-SCID patients treated with retrovirus-mediated gamma(c) gene transfer into autologous CD34+ bone marrow cells, integration of the retrovirus vector in the LMO2 gene promoter induced aberrant LMO2 expression that led exclusively to T-ALL.19 Interestingly, while mature T cells repress LMO2 expression after their differentiation in the thymus,5 B cells greatly induce LMO2 expression during the immune response in the GC reaction, reaching levels similar to that of pro-B cells (Figure 1). This observation may suggest that elevated LMO2 expression in B cells is not necessarily an oncogenic event per se. Thus, all these data suggest that in contrast to T-ALL, LMO2 does not act as an oncogene in B-ALL cells. On the other hand, a recent report has described that in BALL cells with t(17;19) (found in only 1% of B-ALL patients20) the resulting fusion protein E2A/HLF induces the transcription of LMO2, and upon LMO2 silencing with shRNA the apoptotic rate increases by almost 10%.21 Nevertheless, this effect on apoptosis is modest and it is possible that the oncogenic potential of E2A/HLF depends on other transcription targets. Furthermore, mice transgenic for E2A/HLF fusion protein develop lymphoid malignancies with T-cell phenotype, and only very rarely with B-cell precursor phenotype.22,23
Remarkably, we have found that high LMO2 expression correlates with longer overall survival (OS) in patients with B-ALL, in agreement with reports in DLBCL, CML and pancreatic cancer. Part of this prognostic association in our cohort of B-ALL patients may be attributed to the low levels of LMO2 expression found in the subgroup of cases with BCR-ABL rearrangement, a well-known adverse prognostic factor.24,25 Interestingly, multivariate regression analysis identified high LMO2 expression as an independent favorable prognostic factor for OS in patients with normal karyotype, particularly in patients over 15 years of age, for which treatment protocols are not as intense and effective as those used in younger patients. Because this heterogeneous subgroup lacks known cytogenetic or molecular markers that may predict survival, evaluation of LMO2 expression in these patients may provide valuable additional information in terms of risk stratification and therapeutic tailoring.
In terms of event-free survival (EFS), high LMO2 expression correlated with better outcome in B-ALL patients over 15 years of age, agreeing with its usefulness as prognostic factor in this subgroup. We could not find any association between LMO2 expression and EFS in the subgroup of patients over 15 years of age and with normal karyotype, probably due to the low number of relapses in this subgroup (event-free survival data was only available in 5 of the 28 patients in this subgroup).
It was recently reported that LMO2 protein expression assessed by immunohistochemistry in 22 cases of B-ALL did not predict survival in that patient cohort.17 The low number of B-ALL cases enrolled in this study, as well as the lack of stratification by immunophenotypical or cytogenetic characteristics of the blasts are probably the reasons why this study failed to find statistical correlation between LMO2 expression and survival in B-ALL.
In summary, B-ALL cases express variable levels of LMO2 that are associated with the immunophenotypical and molecular features of this disease and correlate with OS in our cohort of patients. Measurement of LMO2 expression should be of particular interest in B-ALL adult cases with normal karyotype, in which high LMO2 expression is an independent favorable prognostic factor for OS that could have clinical and therapeutic relevance.
Acknowledgments
we thank Drs. Martin J. Dyer and Loraine Karran (Leicester, UK) for providing selected leukemia and lymphoma cell lines.
Footnotes
Funding: supported by grants from the Spanish Ministry of Science and Innovation (FIS, RTICC-RD06 and the Ramón y Cajal Program), the CITTIL Spanish-French EU Program, the UTE-CIMA project, the National Institutes of Health (NIH, Bethesda, MD) CA109335 and NIH CA 122105, Fidelity Foundation and Dwoskin Family Foundation. RM was supported by the AECC (Spanish Association Against Cancer) Scientific Foundation.
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Yamada Y, Warren AJ, Dobson C, Forster A, Pannell R, Rabbitts TH. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc Natl Acad Sci USA. 1998;95(7):3890–5. doi: 10.1073/pnas.95.7.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Natkunam Y, Zhao S, Mason DY, Chen J, Taidi B, Jones M, et al. The oncoprotein LMO2 is expressed in normal germinal-center B cells and in human B-cell lymphomas. Blood. 2007;109(4):1636–42. doi: 10.1182/blood-2006-08-039024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boehm T, Foroni L, Kaneko Y, Perutz MF, Rabbitts TH. The rhombotin family of cysteine-rich LIM-domain oncogenes: distinct members are involved in T-cell translocations to human chromosomes 11p15 and 11p13. Proc Natl Acad Sci USA. 1991;88 (10):4367–71. doi: 10.1073/pnas.88.10.4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Vlierberghe P, van Grotel M, Beverloo HB, Lee C, Helgason T, Buijs-Gladdines J, et al. The cryptic chromosomal deletion del(11)(p12p13) as a new activation mechanism of LMO2 in pediatric T-cell acute lymphoblastic leukemia. Blood. 2006;108(10):3520–9. doi: 10.1182/blood-2006-04-019927. [DOI] [PubMed] [Google Scholar]
- 5.Grutz GG, Bucher K, Lavenir I, Larson T, Larson R, Rabbitts TH. The oncogenic T cell LIM-protein Lmo2 forms part of a DNA-binding complex specifically in immature T cells. EMBO J. 1998;17(16):4594–605. doi: 10.1093/emboj/17.16.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisch P, Boehm T, Lavenir I, Larson T, Arno J, Forster A, et al. T-cell acute lymphoblastic lymphoma induced in transgenic mice by the RBTN1 and RBTN2 LIM-domain genes. Oncogene. 1992;7(12):2389–97. [PubMed] [Google Scholar]
- 7.McCormack MP, Rabbitts TH. Activation of the T-cell oncogene LMO2 after gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2004;350 (9):913–22. doi: 10.1056/NEJMra032207. [DOI] [PubMed] [Google Scholar]
- 8.Natkunam Y, Farinha P, Hsi ED, Hans CP, Tibshirani R, Sehn LH, et al. LMO2 protein expression predicts survival in patients with diffuse large B-cell lymphoma treated with anthracycline-based chemotherapy with and without rituximab. J Clin Oncol. 2008;26(3):447–54. doi: 10.1200/JCO.2007.13.0690. [DOI] [PubMed] [Google Scholar]
- 9.Sonmez M, Akagun T, Cobanoglu U, Topbas M, Erkut N, Yilmaz M, et al. Effect of LMO2 protein expression on survival in chronic myeloid leukemia patients treated with imatinib mesylate. Hematology. 2009;14(4):220–3. doi: 10.1179/102453309X426245. [DOI] [PubMed] [Google Scholar]
- 10.Nakata K, Ohuchida K, Nagai E, Hayashi A, Miyasaka Y, Kayashima T, et al. LMO2 is a novel predictive marker for a better prognosis in pancreatic cancer. Neoplasia. 2009;11(7):712–9. doi: 10.1593/neo.09418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371 (9617):1030–43. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 12.Ribera JM, Ortega JJ, Oriol A, Bastida P, Calvo C, Perez-Hurtado JM, et al. Comparison of intensive chemotherapy, allogeneic, or autologous stem-cell transplantation as postremission treatment for children with very high risk acute lymphoblastic leukemia: PETHEMA ALL-93 Trial. J Clin Oncol. 2007;25(1):16–24. doi: 10.1200/JCO.2006.06.8312. [DOI] [PubMed] [Google Scholar]
- 13.Ribera JM, Ortega JJ, Oriol A, Granada I, Hernandez-Rivas JM, Parody R, et al. Prognostic value of karyotypic analysis in children and adults with high-risk acute lymphoblastic leukemia included in the PETHEMA ALL-93 trial. Haematologica. 2002;87(2):154–66. [PubMed] [Google Scholar]
- 14.Agirre X, Roman-Gomez J, Jimenez-Velasco A, Garate L, Montiel-Duarte C, Navarro G, et al. ASPP1, a common activator of TP53, is inactivated by aberrant methylation of its promoter in acute lymphoblastic leukemia. Oncogene. 2006;25 (13):1862–70. doi: 10.1038/sj.onc.1209236. [DOI] [PubMed] [Google Scholar]
- 15.Beillard E, Pallisgaard N, van der Velden VH, Bi W, Dee R, van der Schoot E, et al. Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using ‘real-time’ quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR) - a Europe against cancer program. Leukemia. 2003;17(12):2474–86. doi: 10.1038/sj.leu.2403136. [DOI] [PubMed] [Google Scholar]
- 16.Lossos IS, Czerwinski DK, Wechser MA, Levy R. Optimization of quantitative real-time RT-PCR parameters for the study of lymphoid malignancies. Leukemia. 2003;17(4):789–95. doi: 10.1038/sj.leu.2402880. [DOI] [PubMed] [Google Scholar]
- 17.Cobanoglu U, Sonmez M, Ozbas HM, Erkut N, Can G. The expression of LMO2 protein in acute B-cell and myeloid leukemia. Hematology. 2010;15(3):132–4. doi: 10.1179/102453309X12583347113618. [DOI] [PubMed] [Google Scholar]
- 18.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758–64. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 19.Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118(9):3132–42. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278(5340):1059–64. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 21.Hirose K, Inukai T, Kikuchi J, Furukawa Y, Ikawa T, Kawamoto H, et al. Aberrant induction of LMO2 by the E2A-HLF chimeric transcription factor and its implication in leukemogenesis of B-precursor ALL with t(17;19) Blood. 2010;116(6):962–70. doi: 10.1182/blood-2009-09-244673. [DOI] [PubMed] [Google Scholar]
- 22.Honda H, Inaba T, Suzuki T, Oda H, Ebihara Y, Tsuiji K, et al. Expression of E2A-HLF chimeric protein induced T-cell apoptosis, B-cell maturation arrest, and development of acute lymphoblastic leukemia. Blood. 1999;93(9):2780–90. [PubMed] [Google Scholar]
- 23.Smith KS, Rhee JW, Naumovski L, Cleary ML. Disrupted differentiation and oncogenic transformation of lymphoid progenitors in E2A-HLF transgenic mice. Mol Cell Biol. 1999;19(6):4443–51. doi: 10.1128/mcb.19.6.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gleissner B, Gokbuget N, Bartram CR, Janssen B, Rieder H, Janssen JW, et al. Leading prognostic relevance of the BCR-ABL translocation in adult acute B-lineage lymphoblastic leukemia: a prospective study of the German Multicenter Trial Group and confirmed polymerase chain reaction analysis. Blood. 2002;99(5):1536–43. doi: 10.1182/blood.v99.5.1536. [DOI] [PubMed] [Google Scholar]
- 25.Vrooman LM, Silverman LB. Childhood acute lymphoblastic leukemia: update on prognostic factors. Curr Opin Pediatr. 2009;21(1):1–8. doi: 10.1097/MOP.0b013e32831f1f24. [DOI] [PubMed] [Google Scholar]