Abstract
Neural crest cells play a key role in craniofacial development. The endothelin family of secreted polypeptides regulates development of several neural crest sublineages, including the branchial arch neural crest. The basic helix–loop–helix transcription factor dHAND is also required for craniofacial development, and in endothelin-1 (ET-1) mutant embryos, dHAND expression in the branchial arches is down-regulated, implicating it as a transcriptional effector of ET-1 action. To determine the mechanism that links ET-1 signaling to dHAND transcription, we analyzed the dHAND gene for cis-regulatory elements that control transcription in the branchial arches. We describe an evolutionarily conserved dHAND enhancer that requires ET-1 signaling for activity. This enhancer contains four homeodomain binding sites that are required for branchial arch expression. By comparing protein binding to these sites in branchial arch extracts from endothelin receptor A (EdnrA) mutant and wild-type mouse embryos, we identified Dlx6, a member of the Distal-less family of homeodomain proteins, as an ET-1-dependent binding factor. Consistent with this conclusion, Dlx6 was down-regulated in branchial arches from EdnrA mutant mice. These results suggest that Dlx6 acts as an intermediary between ET-1 signaling and dHAND transcription during craniofacial morphogenesis.
Keywords: dHAND/HAND2, neural crest, branchial arches, endothelin-1, endothelin receptor A, Dlx6
The neural crest, which is induced at the interface between ectoderm and neurectoderm (for review, see García-Castro and Bronner-Fraser 1999), is a key vertebrate character. Neural crest cells emigrate from the neural tube or folds to many different sites in the embryo and differentiate into a large variety of cell types (for review, see Le Douarin and Kalcheim 1999), providing an important paradigm for studying mechanisms of cell differentiation. Heterotopic transplantation experiments have revealed that, although intrinsic differences in developmental potential between neural crest cell populations arising at different levels of the neural tube do exist, development of the neural crest is highly plastic, and signals from the cellular environment are paramount in determining its differentiation (see Le Douarin and Kalcheim 1999). It is not yet clear to what extent plasticity at the population level reflects a lack of commitment or respecification of cell fate at the cellular level (for review, see Dorsky et al. 2000 and references therein), although multipotent, putative stem cells have been isolated from neural crest (Stemple and Anderson 1992).
Insight into the signals involved in cell fate determination and differentiation has come mainly from studies involving trunk neural crest, which gives rise to sensory, sympathetic, and enteric neurons, glia, melanocytes, and adrenomedullary cells (Le Douarin and Kalcheim 1999). Cranial neural crest cells that migrate into the branchial arches yield three important cell types not normally formed by trunk neural crest: chondrocytes and osteocytes, which generate most of the craniofacial skeleton, and smooth muscle cells, which surround derivatives of the branchial arch arteries, including the great arteries in the thorax. Little is known about the signaling molecules involved in specification and differentiation of these three lineages, although in vitro experiments with trunk neural crest suggest that transforming growth factor-β may be a candidate inducer of smooth muscle differentiation (Shah et al. 1996).
Development of both craniofacial and vascular derivatives of the branchial arch neural crest critically depends on endothelin signaling, as shown by the effects of mutation of components of the endothelin signaling pathway (Kurihara et al. 1994, 1995; Clouthier et al. 1998; Yanagisawa et al. 1998a,b; Miller et al. 2000), and of pharmacological interference with endothelin signaling (e.g., Kempf et al. 1998). Neural crest cells migrating into the branchial arches express the endothelin receptor A (EdnrA) (Clouthier et al. 1998), whereas endothelin-1 (ET-1) is expressed in a complementary fashion, in the mesodermal core and the surface ectoderm of the arches, in the pharyngeal endoderm, and in the endothelium of the branchial arch arteries (Maemura et al. 1996; Clouthier et al. 1998).
As in other lineages, the basic domain helix–loop–helix (bHLH) family of transcription factors figures prominently in cell fate determination of the neural crest, and members of this family have been shown to control specification of the sensory neuron, autonomic neuron, and melanocyte sublineages (see Christiansen et al. 2000). ET-1 signaling has been shown to be essential for expression of the bHLH transcription factors dHAND/HAND2 and eHAND/HAND1, which are expressed in partially overlapping domains in the distal part of the branchial arches (Cserjesi et al. 1995; Thomas et al. 1998; Clouthier et al. 2000).
The early embryonic lethality caused by loss of function of dHAND (Srivastava et al. 1997), which is also expressed in extraembryonic tissues, heart, limb buds, sympathetic ganglia, and adrenomedullary cells (Srivastava et al. 1995; Charité et al. 2000; data not shown), has so far precluded assessment of its role in development of the vascular derivatives of the branchial arches. However, recent work from our laboratory has revealed that dHAND is essential for craniofacial development (H. Yanagisawa, J. Charité, D. McFadden, D. Clouthier, and E.N. Olson, in prep.). Furthermore, because both in vitro and in vivo experiments suggested that dHAND is involved in cell fate specification within the sympathetic neuron lineage (Howard et al. 1999, 2000), we set out to investigate the regulation of dHAND in the branchial arch neural crest, as a possible avenue for gaining insight into the specification of the chondrogenic and smooth muscle lineages.
Here, we describe the identification of an enhancer responsible for expression of dHAND in branchial arches 1 and 2. We show that enhancer function is dependent on the endothelin signaling pathway, and analyze trans-acting factors that bind to it. We provide evidence that the homeodomain transcription factor Dlx6 binds directly to this enhancer and is a key intermediate in transduction of the endothelin signal.
Results
An endothelin-dependent, dHAND branchial arch enhancer
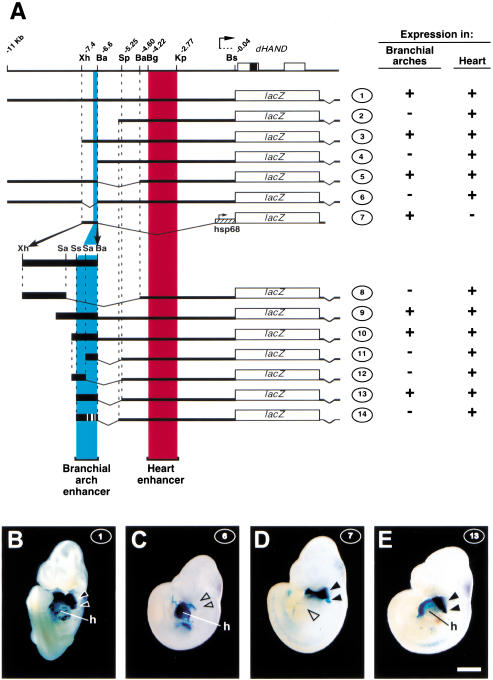
We have shown previously that a genomic fragment extending 11 kb upstream of the dHAND initiation codon (Fig. 1A, construct 1) drives expression in the heart and in the branchial arches at embryonic day 10.5 (E10.5; Fig. 1B; McFadden et al. 2000). The developmental expression pattern driven by this region was determined in transgenic lines carrying construct 1 randomly integrated in the genome. Four independent transgenic lines exhibited strong expression in heart and branchial arches, in addition to minor sites of ectopic expression, which were unique to each line and were therefore regarded as caused by elements present at the integration sites. β-Galactosidase expression in the first branchial arch was first observed around E9.25 (data not shown), and well-demarcated domains of expression in the distal part of arches 1 and 2, similar to those of the endogenous dHAND gene, were established by E9.5 (Fig. 2A). These domains persisted at E10.5 (Fig. 2B), at which stage more posterior branchial arches are well developed. These posterior arches did not express the transgene (Fig. 2C,D), although dHAND is expressed there. This suggests that a distinct enhancer not included in construct 1 is responsible for expression in arches 3 to 6.
Figure 1.
Identification of a branchial arch-specific enhancer. (A) Reporter constructs used in this study. The genomic organization of dHAND is depicted at the top; coding regions (boxes), relevant restriction sites, and their distance in kilobases from the start of the coding region are indicated. The lacZ reporter constructs used in this study are schematized underneath; thick black lines and boxes represent dHAND genomic sequences. Construct 2 extends to −5.49 kb. The Sau3A, SspI, and Sau3A sites used in generating constructs 8 and 11–14 are located 316, 203, and 121 bp, respectively, upstream of the BamHI site at −6.6 kb. The 5′ ends of constructs 9 and 10 are 413 and 196 bp, respectively, upstream of this BamHI site. Presence or absence of β-galactosidase expression driven by these constructs at E10.5 in branchial arches 1 and 2 and in the heart, respectively, is indicated on the right. (Ba) BamHI; (Bg) BglII; (Bs) BssHII; (Kp) KpnI; (Sa) Sau3A; (Sp) SpeI; (Ss) SspI; (Xh) XhoI. (B–E) Ventral/lateral views of E10.5, Xgal-stained F0 embryos transgenic for constructs 1, 6, 7, and 13, as indicated at upper right of panels. At least three F0 transgenic embryos were examined for each construct. B shows β-galactosidase activity in heart (h) and the first two branchial arches (black arrowheads). (C) Expression in heart but not in branchial arches (open arrowheads). (D) Expression in branchial arches (filled arrowheads) but not in heart (open arrowhead). (E) Expression in heart and branchial arches. Bar, 1 mm.
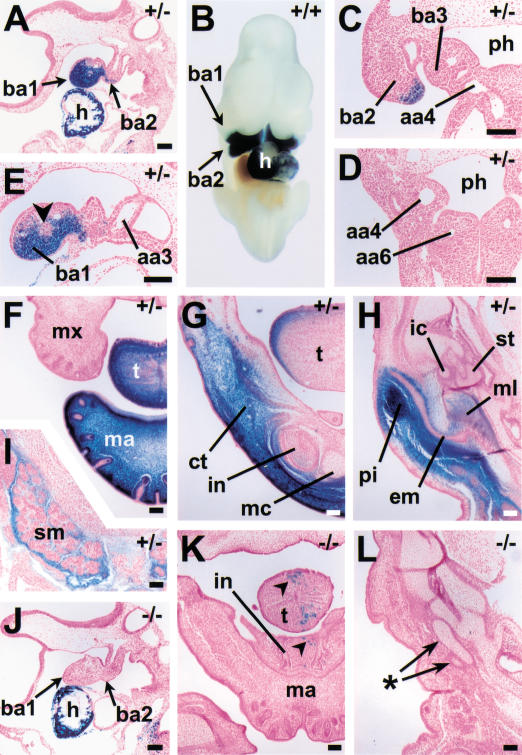
Figure 2.
Developmental expression pattern and EdnrA-dependence of the dHAND branchial arch enhancer. (A–I) β-Galactosidase expression driven by construct 1 in a wild-type or EdnrA+/− background. (J–L) β-Galactosidase expression driven by construct 1 in an EdnrA−/− background. Genotype with respect to the EdnrA locus is indicated in the top right corner of each panel. (A) Sagittal section through an E9.5, 27-somite embryo, showing expression in heart (h) and branchial arches 1 and 2 (ba1,2). (B) Ventral view of E10.5 embryo, posterior part removed, showing expression in branchial arches and heart. (C,D) Frontal/transverse sections through E10.5 embryo, showing expression in branchial arch 2 and lack of expression in branchial arches 3–6. (E) Sagittal section through the branchial arches of the embryo shown in A, showing lack of expression in the core (arrowhead) of the first branchial arch (ba1). (F–I) Frontal sections through the head of an E16.75 embryo. (F) Strong expression in the distal part of the mandible (ma) and tongue (t). (G) Section is cut at a more proximal level, showing expression dorsolaterally in the tongue, and in the ventral part of the mandible, in connective tissue (ct) and in the mesenchymal component of the incisor tooth bud (in), but not in Meckel's cartilage (mc). (H) Section is cut at the level of the middle ear, showing expression in the pinna (pi) and in part of the malleus (ml). (I) Another part of the section shown in H; β-galactosidase expression is observed in the mesenchyme of the submandibular gland (sm). (J–L) Sections through EdnrA−/− embryos. (J) Sagittal section, comparable to the one shown in A, through an E9.5, 27-somite embryo, showing strong expression in the heart but lack of expression in the branchial arches. (K) Frontal section through the head of an E16.75 littermate of the embryo shown in F–I, at approximately the same level as the section shown in G, showing few expressing cells (arrowheads) in the hypoplastic tongue (t) and mandible (ma). (L) Section corresponding to the one shown in H, showing lack of expression in cartilage elements possibly representing rudimentary middle ear ossicles (asterisk). (aa) aortic arch artery; (em) external acoustic meatus; (ic) incus; (mx) maxilla; (ph) pharynx; (st) stapes. In all panels except B, dorsal is toward the top; in A, E, and J, anterior is toward the left, in B, toward the top. Bars, 0.1 mm.
Histological sections of E9.5 embryos revealed that β-galactosidase activity was excluded from the center of the arches (Fig. 2E, arrowhead). This is consistent with expression being largely confined to neural crest cells, because at this stage mesoderm and neural crest cells are segregated to the core and periphery of the arch, respectively (Trainor and Tam 1995). However, lacking markers that identify each lineage with cellular resolution, we cannot exclude that some mesodermal cells express the transgene as well. At later stages, β-galactosidase expression was observed in the tip and the dorsolateral aspect of the tongue and in the mandible (Fig. 2F–H). Expression in the mandible appeared to be regional rather than tissue-specific, comprising much of the ventral connective tissue in the jaw (Fig. 2G) and the pinnae of the ear (Fig. 2H), but also mesoderm derivatives such as digastric muscle (data not shown) and submandibular glands (Fig. 2I). Expression in neural crest-derived cartilages was confined to part of the malleus (Fig. 2H), and a few cells in the proximal part of Meckel's cartilage (data not shown). Consistent with the absence of expression in arches 3 to 6 at E9.5–E10.5, no expression was observed at later stages in the vascular derivatives of the arch arteries of these branchial arches (data not shown).
To investigate the involvement of the endothelin signaling pathway in activation of the branchial arch enhancer, transgenic lines carrying construct 1 were crossed with EdnrA+/− mice (Clouthier et al. 1998), and embryos derived from intercrosses of transgenic, EdnrA+/− mice were stained for β-galactosidase activity. Transgene expression in the heart of E9.5 EdnrA−/− embryos appeared unaffected as compared with their EdnrA+/− littermates, but expression in the branchial arches was completely absent (Fig. 2, cf. J with A). At E16.75, a few expressing cells were present in the hypoplastic tongue and mandible of EdnrA−/− embryos (Fig. 2, cf. K with G). Aberrant cartilage elements located at the normal position of the ear ossicles did not contain β-galactosidase-positive cells (Fig. 2L). It should be noted that several structures, such as the pinnae and the submandibular glands, are missing in these embryos (Clouthier et al. 1998), making it impossible to establish whether gene expression in these structures is EdnrA-dependent. If we assume that the endothelin receptor A is expressed exclusively in neural crest cells, which is suggested by its expression pattern (Clouthier et al. 1998), we conclude that expression of dHAND in mesodermal structures at E16.75 reflects a distinct regulatory pathway, possibly acting through distinct regulatory elements. We have not attempted to identify these elements, but instead focused on identification of the early-acting, strictly EdnrA-dependent, branchial arch enhancer.
Deletion analysis defines a 208-bp enhancer
To delineate the cis-acting sequences responsible for driving expression in the branchial arches, serialdeletions of construct 1 were tested in E10.5 F0 embryos. Expression patterns generated by constructs 2–5 defined the XhoI and BamHI sites at −7.4 and −6.6 kb as the 5′ and 3′ boundaries, respectively, of a region necessary for branchial arch expression. Construct 6 showed that within construct 1 no sequences outside of the XhoI–BamHI region were capable of driving expression in the branchial arches (Fig. 1, cf. C with B), and construct 7 showed that the XhoI–BamHI fragment is sufficient to drive branchial arch expression from a heterologous promoter (Fig. 1D).
Further deletions of the XhoI–BamHI fragment were tested in conjunction with the cardiac enhancer identified previously (McFadden et al. 2000), which served as an internal control for integration sites being permissive to expression. Constructs 8–13 defined a minimally sufficient 208-bp branchial arch enhancer fragment (Fig. 1E), hereafter referred to as SspBam208; deletion of either the 5′ or the 3′ part of this fragment abolished branchial arch expression (constructs 11 and 12; data not shown).
Binding of EdnrA-dependent factors to SspBam208
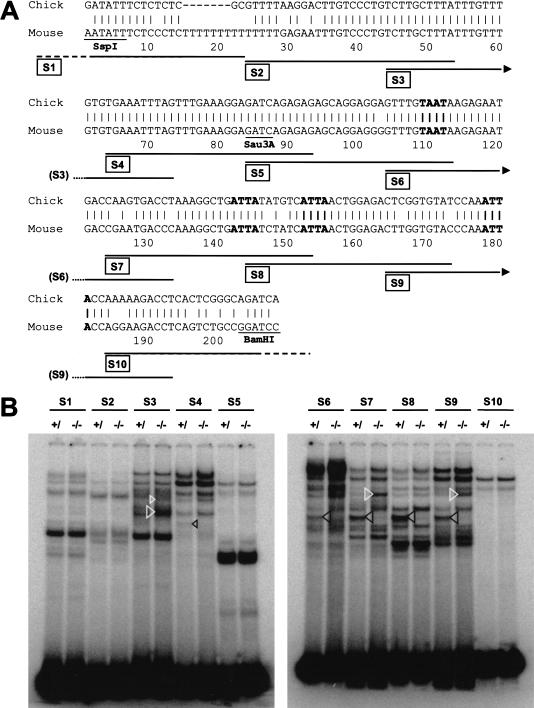
Comparison of the SspBam208 sequence with the corresponding region from chick revealed a high degree of homology (Fig. 3A), providing few clues with regard to putative transcription factor binding sites essential for enhancer function. Homology between the chick and mouse genomic sequences was confined to the neural crest enhancer and was not observed in surrounding genome regions. To identify trans-acting factors involved in mediating signaling from the EdnrA to SspBam208, 10 double-stranded oligonucleotides covering the SspBam208 sequence (see Fig. 3A) were used as probes in electrophoretic mobility shift assays (EMSAs) with protein extracts prepared from the first branchial arches of EdnrA−/− embryos and of wild-type or heterozygous littermates (Fig. 3B). No obvious differences between these extracts were observed with oligonucleotides S1, S2, S5, and S10, but the remaining oligonucleotides did reveal differential binding patterns. Differences included clear decreases as well as increases of certain complexes (indicated with black and white arrowheads, respectively, in Fig. 3B) upon removal of endothelin receptor function. Additional, less obvious differences were also apparent, but these could not be consistently quantitated. These observations suggested the existence of several potential, positive as well as negative regulatory proteins that could be involved in transducing the endothelin signal.
Figure 3.
Trans-acting factors binding to SspBam208. (A) Sequence comparison between SspBam208 and the corresponding region from chick. The SspI, Sau3AI, and BamHI sites and the position of oligonucleotides S1–S10 are indicated below the mouse sequence. (B) Electrophoretic mobility shift assays with oligonucleotides S1–S10 and whole cell extracts from the first branchial arch of EdnrA−/− (−/−), and a mixture of heterozygous and wild-type (+/) embryos. Complexes that are up-regulated or down-regulated in the EdnrA−/− background are indicated with white and black arrowheads, respectively.
Interestingly, oligonucleotides S6, S7, S8, and S9 yielded EdnrA-dependent complexes migrating at exactly the same level in the gel, and the complexes formed on S7 and S9 that were up-regulated in the EdnrA−/− background also comigrated. Because oligonucleotides S6–S9 all contain an ATTA motif, which forms the core binding site for many homeodomain-containing transcription factors, and because expression of several homeobox genes had been shown to be dependent on endothelin signaling (Clouthier et al. 1998, 2000), we investigated whether the ATTA motifs are important for enhancer function.
Homeodomain binding sites are essential for enhancer activity and binding of EdnrA-dependent factors
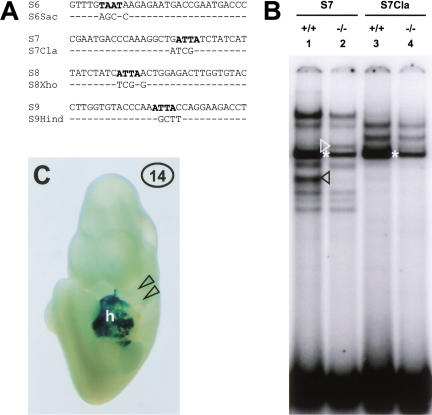
Oligonucleotides S6Sac, S7Cla, S8Xho, and S9Hind, containing mutations in the ATTA motifs (see Fig. 4A), were tested in EMSAs with branchial arch extracts. In each case, the ATTA mutation abolished formation of the endothelin-dependent complex (Fig. 4B, cf. lane 3 with lane 1, black arrowhead; data not shown). For S7 and S9, formation of the complex that was up-regulated in the EdnrA−/− background was abolished as well (Fig. 4B, cf. lane 4 with lane 2, white arrowhead; data not shown).
Figure 4.
Homeodomain binding sites are essential for enhancer activity and for binding of EdnrA-dependent factors. (A) Sequences of oligonucleotides S6–S9 and their mutant versions, containing several base changes, as indicated, destroying the ATTA motifs. (B) EMSA with wild-type and EdnrA−/− first arch extract and oligonucleotides S7 and S7cla. Both the EdnrA-dependent complex (lane 1, black arrowhead), and the complex up-regulated in the absence of endothelin signaling (lane 2, white arrowhead; note distance to the ubiquitous complex indicated by the asterisk) are not formed on S7cla (lanes 3 and 4, respectively). (C) E10.5 F0 embryo carrying construct 14, stained with Xgal. β-Galactosidase activity is observed in the heart (h), but not in the branchial arches (arrowheads).
Enhancer activity of SspBam208 containing all four ATTA mutations was tested, again using the cardiac enhancer as an internal control (construct 14, Fig. 1A). F0 embryos carrying construct 14 showed strong β-galactosidase expression in the heart, but lacked expression in the branchial arches (Fig. 4C), showing that one or more of the ATTA motifs is essential for enhancer function.
Identification of Dlx6 as an EdnrA-dependent enhancer binding factor
We sought to determine whether the EdnrA-dependent complexes identified in EMSAs represented any of several homeodomain proteins expressed in the branchial arches that were shown to depend on endothelin signaling (Clouthier et al. 1998, 2000). We initially focused on gsc and Dlx3, because the expression domains of these genes in the first branchial arch overlap at least in part with that of dHAND. However, expression of dHAND in the branchial arches of E10.5 gsc−/− embryos (a gift of R. Behringer, M.D. Anderson Cancer Center, Houston, TX) was not obviously different from that in wild-type littermates as determined by whole mount in situ hybridization (data not shown). Furthermore, in EMSAs, Dlx3 protein expressed in COS cells did not comigrate with the EdnrA-dependent complex (data not shown).
In the course of these experiments we did, however, find that formation of this complex could be abolished by an antibody directed against the homeodomain of Distal-less (Dll; a gift of G. Panganiban, University of Wisconsin, Madison; Fig. 5A, cf. lane 5 with lanes 3 and 4), indicating that Dlx proteins are present in this complex. Further experiments revealed that neither Dlx2 nor Dlx5 comigrated with the complex (data not shown), but that Dlx6 did migrate at exactly the same position in the gel (Fig. 5A, cf. lane 2 with lane 3). Binding of Dlx6 was abolished by anti-Dll (Fig. 5A, cf. lane 6 with lane 2) and by mutation of the ATTA motif (Fig. 5A, cf. lane 8 with lane 2). These data suggest that Dlx6 is the major EdnrA-dependent factor binding to SspBam208. We cannot formally exclude that the binding of Dlx6 to SspBam208 depends on an endothelin-dependent binding partner that is somehow expressed in COS cells as well. However, expression of Dlx6 was undetectable in the distal part of the branchial arches of EdnrA−/− embryos (Fig. 5, cf. E with C), whereas expression in the proximal part of the arch appeared to be independent of EdnrA signaling (Fig. 5E, closed arrowhead). Therefore, we favor the parsimonious conclusion that Dlx6 is the key EdnrA-dependent component of the EdnrA-dependent complex in the wild-type first arch.
Figure 5.
Involvement of Dlx6 in transduction of endothelin signaling to SspBam208. (A) Identification of the EdnrA-dependent complex. EMSA with oligonucleotides S7 and S7cla, and extracts from untransfected COS-1 cells (lanes 1,9), COS-1 cells transfected with pSG5-Dlx6 (lanes 2,6,8), or from branchial arches of wild-type (lanes 3,5,7) or EdnrA−/− (lane 4) embryos, showing that Dlx6 comigrates with the EdnrA-dependent complex (lanes 2 and 3, black arrowheads), that binding of both is dependent on the integrity of the ATTA motif (cf. lanes 7 and 8 with lanes 3 and 2), and is abolished by incubation with anti-Dll (cf. lanes 5 and 6 with lanes 3 and 2). (B–D) Dependence of Dlx6 expression on endothelin signaling. Brightfield (B,D) and darkfield (C,E) images of frontal sections through the head of E10.5 wild-type (B,C) and EdnrA−/− (D,E) embryos. Dlx6 is expressed in the first branchial arch (ba1) of wild-type embryos (arrowheads in B,C), but not in the distal part of the EdnrA−/− first arch (open arrowheads in D,E), although expression is still observed in the proximal region of the arch (filled arrowheads in D,E). Bar, 0.1 mm.
Discussion
A distinct developmental program for the mandibular and hyoid arch
We have described the identification and analysis of an enhancer responsible for expression of dHAND in branchial arches 1 and 2, the mandibular and hyoid arches. To our knowledge, this is the first description of an endothelin-responsive enhancer with a defined activity in vivo. Our finding of a distinct enhancer dedicated to driving dHAND expression in arches 1 and 2 versus the more posterior arches is surprising, but consistent with the distinct morphological development of these arches, which in more primitive gnathostomes form the jaw and its supportive apparatus, respectively. These structures are thought to have evolved at an early stage in gnathostome evolution, as specializations of the gill bars formed by the more posterior branchial arches. It remains to be established whether the presence of a dedicated arch 1/2 enhancer is related to different functional requirements of dHAND in these versus more posterior arches, for example, with respect to expression level, or exclusively reflects differences in upstream signaling pathways that may be contingent upon differences in development, including the degeneration of the branchial arch arteries and increased growth, of arch 1 and 2.
The existence of a distinct developmental program for the mandibular and hyoid arches is supported by the finding that mutations in several complementation groups uncovered by mutagenesis screens in zebrafish specifically affect skeletal elements derived from arches 1 and 2, leaving more posterior structures intact (Piotrowski et al. 1996). Interestingly, the most severe anterior arch phenotype is observed in the sucker mutant, caused by a missense mutation in the et-1 gene that abolishes ET-1 function (Miller et al. 2000). However, sucker not only affects development of arches 1 and 2, but also of arches 3 and 4, and, infrequently, 5 (Piotrowski et al. 1996; Miller et al. 2000), suggesting quantitative, rather than qualitative differences in dependence on ET-1 signaling between branchial arches. This appears to be reflected in the expression of dHAND in sucker mutant embryos, which is abolished in the first branchial arch, but only partly down-regulated in more posterior arches (Miller et al. 2000). Whether dHAND would be classified as an anterior arch gene remains to be established, as dHAND mutant zebrafish, like dHAND mutant mice (Srivastava et al. 1997), die before their craniofacial phenotype can be assessed (Yelon et al. 2000).
Role of Dlx6 in regulation of dHAND
We have shown that the activity of the dHAND arch 1/2 enhancer, like that of the endogenous dHAND gene, is critically dependent on EdnrA function, on the one hand, and on the integrity of putative homeodomain binding sites in the enhancer, on the other. Our data suggest that the homeodomain transcription factor Dlx6 forms the connection between these two observations. First, although branchial arch expression of several homeobox genes is down-regulated in EdnrA−/− mice (Clouthier et al. 1998, 2000), our EMSAs with SspBam208 oligonucleotides revealed a single EdnrA-dependent transcription factor binding to each of the homeodomain binding sites, which was identified as Dlx6 based on comigration and on reactivity with anti-Dll. Second, although no data are available for Dlx6 specifically, Dlx proteins can activate or repress transcription depending on the target gene involved (e.g., Benson et al. 2000 and references therein). Finally, Dlx6 expression is abolished in the ventral part of EdnrA−/− branchial arches, where dHAND is normally expressed. Together, these data strongly support the conclusion that Dlx6 is an activator of dHAND transcription in at least the first branchial arch, acting as a transducer of EdnrA-mediated signaling. The specificity of the interaction between Dlx6 and SspBam208 observed in EMSAs is underscored by the observation that, although expression of Dlx5 in the ventral part of the branchial arches is, like that of Dlx6, dependent on EdnrA function, dHAND expression in the branchial arches of Dlx5−/− embryos is not obviously affected (data not shown). A direct test of the epistatic relationship between Dlx6 and dHAND must await targeted inactivation of the Dlx6 gene.
The intracellular signaling pathways that operate in neural crest cells to transduce the signal from the activated endothelin receptor A to ET-1 target genes are unknown; however, the craniofacial phenotype caused by loss of function of the G protein α-subunits Gαq and Gα11 may hint at their possible involvement in this pathway (Offermanns et al. 1998).
Complex control of dHAND expression
Because Dlx6 expression in the branchial arches is more extensive than that of dHAND, and only the ventral part of the Dlx6 expression domain is ET-1-dependent, additional factors must act on SspBam208 in conjunction with Dlx6 to restrict expression to the proper domain. Although Dlx6 appears to be the major positively acting factor controlling dHAND branchial arch expression as identified by our EMSAs, these experiments also revealed an unexpectedly complex set of potentially negative inputs on SspBam208 that are regulated by the endothelin signaling pathway. We have not determined the binding sites for these factors in any detail, and currently have no corroborating evidence for the importance of their interaction with SspBam208 in vivo. It is clear, however, that some of these factors do not require ATTA motifs, whereas binding sites for others may in fact overlap the ATTA motifs used by Dlx6, suggesting that endothelin-dependent expression of dHAND is stringently controlled, requiring removal of repressors as well as binding of Dlx6. Further experiments are necessary to test this hypothesis.
Our findings suggest that binding of Dlx6 is necessary for activity of the dHAND branchial arch enhancer, but it is clearly not sufficient, as deletion of the SspI–Sau3AI fragment from SspBam208 abolishes enhancer function, despite the fact that all Dlx6 binding sites are retained in the remaining Sau3AI–BamHI fragment. Whether a similar statement can be made with regard to endothelin signaling is less obvious. Although it is conceivable that an equilibrium between positive and negative regulators that are differentially controlled by endothelin signaling could be sufficient to generate the proper dHAND expression domain, it is more likely that additional signaling pathways converge on SspBam208, because although both dHAND and eHAND are dependent on endothelin signaling, their expression domains overlap only in the distal part of the branchial arches (Cserjesi et al. 1995; Thomas et al. 1998; Clothier et al. 2000). This suggests that they respond differentially to additional, positional information within the branchial arches, perhaps to signals emanating from a distal signaling center. Comparative analysis of the dHAND and eHAND branchial arch enhancers should be helpful in further elucidating these upstream regulatory pathways.
Dlx and HAND genes, endothelins, and cell fate specification
It has been hypothesized that the neural crest evolved from Rohon–Beard-like sensory neurons, which emigrate from the neural tube at an early stage of development (Fritzsch and Northcutt 1993), and this appears to be supported by recent genetic evidence (Artinger et al. 1999). How these cells acquired the potential to differentiate into cartilage and bone is a question upon which no experimental data currently seem to bear in an obvious way. It is, however, intriguing that the Dll gene of the acraniate amphioxus, which does not have a neural crest, is highly up-regulated at the border of the embryonic ectoderm and neuroectoderm (Holland et al. 1996), apparently analogous to expression of Dlx5 in presumptive premigratory neural crest in mouse (Yang et al. 1998), and that besides their expression in the branchial arches, both Dlx5 and Dlx6 are expressed in differentiating osteoblasts in all developing bones (see, e.g., Acampora et al. 1999). Dlx5 has been shown to regulate genes involved in osteogenesis in vitro (e.g., Benson et al. 2000), and knockout studies have confirmed that it is involved in bone differentiation in addition to being necessary for development of the craniofacial skeleton (Acampora et al. 1999; Depew et al. 1999).
Interpretation of cranial cartilage morphogenesis in the zebrafish anterior arch mutants, including sucker, has led Kimmel and coworkers to speculate that one effect of ET-1 signaling is the specification of a ventral identity in skeletogenic neural crest, which would become dorsal by default in the absence of endothelin signaling (Kimmel et al. 1998, 2001). This would fit with the finding in chick that dorsally fated neural crest cells give rise to ventral cartilages when allowed to migrate into the ventral part of the branchial arch (Baker et al. 1997), suggesting environmental control of ventral cell fate. However, in mouse, loss of EdnrA function also affects putatively dorsal skeletal elements, although less severely than ventral structures (Clouthier et al. 1998). Whether these discrepancies are caused by species-specific differences in dorsoventral patterning or interdependence of morphogenesis of dorsal and ventral elements, or by compensating activity of other, as yet unidentified, endothelin isoforms in zebrafish, remains to be established. Although ectopic expression studies are necessary to confirm that ET-1 actually induces ventral cell fates, the results described here, and the effects of deletion of the dHAND branchial arch enhancer (H. Yanagisawa, J. Charité, D. McFadden, D. Clouthier, and E.N. Olson, in prep.) suggest that dHAND could be a key downstream selector gene for the ventral cell fate.
Forward genetics studies in mouse have uncovered essential functions for several transcription factors in development of the craniofacial skeleton, loss of function often resulting in partially overlapping defects in patterning of neural crest-derived structures. None of these essential patterning genes has yet been implicated in congenital malformations in man, possibly owing to the severity of their loss-of-function phenotypes (Wilkie and Morriss-Kay 2001). Given the early embryonic lethality of dHAND null mice, it is likely that mutations in the dHAND coding region that cause significant loss of function will not be recovered among live births. Whether mutations in the branchial arch enhancer described here may be causally involved in congenital malformations, and whether arch-1/2-specific disorders such as Treacher–Collins syndrome involve dHAND as a downstream effector, are issues that bear further investigation.
Materials and methods
DNA constructs
Reporter constructs 1–5 and 8–14 contain dHAND genomic sequences cloned upstream of the lacZ gene in plasmid pAUGLacZ, using a BssHII site most proximal to the ATG in the 5′-untranslated region of dHAND. Construct 1 has been described previously (McFadden et al. 2000). Deletion constructs were generated using convenient genomic clones (constructs 2, 14), internal restriction sites, or digestion with exonuclease III (constructs 9, 10, 12). For construct 7, a XhoI–BamHI dHAND genomic fragment was cloned upstream of the hsp68 promoter in plasmid hsp68lacZ. Vector sequences were removed prior to microinjection.
For pSG5–Dlx6, the complete murine Dlx6 cDNA was constructed by RT–PCR, sequenced (Pfeffer et al. 2001), and cloned into the EcoRI site of the pSG5 expression vector (Stratagene).
Chick dHAND genomic clones were isolated from a chick genomic library (Clontech) using the chick bHLH region as a probe.
In situ hybridization
In situ hybridization on mouse embryo sections was performed as described previously (Shelton et al. 2000), using 35S-labeled mX6EX3/4 probe. This cDNA probe was engineered to contain the entire exon 3 and part of exon 4 of mouse Dlx6, excluding the flanking homeobox and repetitive sequences. Specificity of this probe was tested by Northern analysis.
Generation and analysis of transgenic embryos and mice
F0 transgenic mouse embryos were generated as described previously (McFadden et al. 2000). Stable transgenic lines carrying construct 1 were established in the FVB strain. These lines were crossed with EdnrA mutant mice on the 129S6SvEv background (Clouthier et al. 1998), and progeny were intercrossed for a few generations, generating double heterozygous mice on a mixed FVB/129 background. Embryos were fixed and stained with Xgal as described in Charité et al. (1998), but for embryos of 11.5 d and older, the staining solution was supplemented with 0.1% NP-40, and for embryos of 13.5 d and older, 0.2% sodium deoxycholate was included as well.
Whole-cell extracts and electrophoretic mobility shift assays
Whole-cell extracts from COS-1 cells, untransfected or transfected with pSG5–Dlx6 using Fugene-6 reagent (Roche), were prepared as described by Charité et al. (1998). For whole cell branchial arch extracts, the distal parts of the first branchial arches of several E10.25–E10.5 embryos were dissected in TBS (Charité et al. 1998), pooled, and stored in liquid nitrogen until preparation of extracts. To prepare extracts from EdnrA null and matching wild-type and/or heterozygous branchial arches, arch pairs from individual embryos were stored in liquid nitrogen, and embryos were genotyped by PCR or Southern analysis or by using a construct 1 transgene crossed into the EdnrA knockout background as an indicator, scoring for expression of the β-galactosidase reporter in the second branchial arch. Arches were then taken up in a small volume of TBS, pooled according to genotype, and processed as described above.
Electrophoretic mobility shift assays were performed essentially as described by Charité et al. (1998), using 1 μL of extract per reaction and incubation for 10–15 min at room temperature. Then 1 μL of rabbit anti-Dll antibody (Panganiban et al. 1995) was added before addition of the probe, or after formation of the complex, with similar results. Oligonucleotides were (plus strand given): S1, TAAGCTTGATATTTCTCCCTCTTTT TTTTT; S2, TTTTTGAGAATTTGTCCCTGTCTTGCTTTA; S3, TCTTGCTTTATTTGTTTGTGTGAAATTTAG; S4, TGA AATTTAGTTTGAAAGGAGATCAGAGAG; S5, GATCAG AGAGAGCAGGAGGGGTTTGTAATA; S6, GTTTGTAATA AGAGAATGACCGAATGACCC; S7, CGAATGACCCAAAGGCTGATTATCTATCAT; S8, TATCTATCATTAACTGGA GACTTGGTGTAC; S9, CTTGGTGTACCCAAATTACCAGGAAGACCT; S10, AGGAAGACCTCAGTCTGCCGGACTAGTTCT; S6Sac, GTTTGAGCTCAGAGAATGACCGAATGACCC; S7Cla, CGAATGACCCAAAGGCTATCGATCTAT CAT; S8Xho, TATCTATCTCGAGCTGGAGACTTGGTGTAC; S9Hind, CTTGGTGTACCCAAAGCTTCAGGAAGACCT.
Acknowledgments
We thank John Rubenstein, Tom Sargent, Grace Panganiban, and Richard Behringer for kindly providing us with DNA constructs, antibodies, and mutant embryos; Amy Wu, Heather Lee, and John McAnally for microinjections; Jeffery Stark for histology; and Paul Overbeek for generating the construct 1 transgenic lines. J.C. was supported by a long-term EMBO fellowship, and D.G.M. is supported by a Medical Scientist Training Program grant from the NIH. G.M. is the recipient of a Career Award from Telethon-Italy (grant no. 03/cp). G.L. is supported by grants from Telethon (Italy), AIRC, CNR, and from the E.U. consortium GENOSPORA (QLK-1999–02108). M.Y. is an investigator of the Howard Hughes Medical Institute. This work was supported by grants from the National Institutes of Health and the Donald W. Reynolds Cardiovascular Clinical Research Center, Dallas, Texas to E.N.O.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL eolson@hamon.swmed.edu; FAX (214) 648-1196.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.931701.
References
- Acampora D, Merlo GR, Paleari L, Zerega B, Postiglione MP, Mantero S, Bober E, Barbieri O, Simeone A, Levi G. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development. 1999;126:3795–3809. doi: 10.1242/dev.126.17.3795. [DOI] [PubMed] [Google Scholar]
- Artinger KB, Chitnis AB, Mercola M, Driever W. Zebrafish narrowminded suggests a genetic link between formation of neural crest and primary sensory neurons. Development. 1999;126:3969–3979. doi: 10.1242/dev.126.18.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CVH, Bronner-Fraser M, Le Douarin NM, Teillet M-A. Early- and late-migrating cranial neural crest cell populations have equivalent developmental potential in vivo. Development. 1997;124:3077–3087. doi: 10.1242/dev.124.16.3077. [DOI] [PubMed] [Google Scholar]
- Benson MD, Bargeon JL, Xiao G, Thomas PE, Kim A, Cui Y, Franceschi RT. Identification of a homeodomain binding element in the bone sialoprotein gene promoter that is required for its osteoblast-selective expression. J Biol Chem. 2000;275:13907–13917. doi: 10.1074/jbc.275.18.13907. [DOI] [PubMed] [Google Scholar]
- Charité J, De Graaff W, Consten D, Reijnen MJ, Korving J, Deschamps J. Transducing positional information to the Hox genes: Critical interaction of cdx gene products with position-sensitive regulatory elements. Development. 1998;125:4349–4358. doi: 10.1242/dev.125.22.4349. [DOI] [PubMed] [Google Scholar]
- Charité J, McFadden DG, Olson EN. The bHLH transcription factor dHAND controls Sonic hedgehog expression and establishment of the zone of polarizing activity during limb development. Development. 2000;127:2461–2470. doi: 10.1242/dev.127.11.2461. [DOI] [PubMed] [Google Scholar]
- Christiansen JH, Coles EG, Wilkinson DG. Molecular control of neural crest formation, migration and differentiation. Curr Opin Cell Biol. 2000;12:719–724. doi: 10.1016/s0955-0674(00)00158-7. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Hosoda K, Richardson JA, Williams SC, Yanagisawa H, Kuwaki T, Kumada M, Hammer RE, Yanagisawa M. Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development. 1998;125:813–824. doi: 10.1242/dev.125.5.813. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Williams SC, Yanagisawa H, Wieduwilt M, Richardson JA, Yanagisawa M. Signaling pathways crucial for craniofacial development revealed by endothelin-A receptor-deficient mice. Dev Biol. 2000;217:10–24. doi: 10.1006/dbio.1999.9527. [DOI] [PubMed] [Google Scholar]
- Cserjesi P, Brown D, Lyons GE, Olson EN. Expression of the novel basic helix–loop–helix gene eHAND in neural crest derivatives and extraembryonic membranes during mouse development. Dev Biol. 1995;170:664–678. doi: 10.1006/dbio.1995.1245. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Liu JK, Long JE, Presley R, Meneses JJ, Pedersen RA, Rubenstein JLR. Dlx5 regulates regional development of the branchial arches and sensory capsules. Development. 1999;126:3831–3846. doi: 10.1242/dev.126.17.3831. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Moon RT, Raible DW. Environmental signals and cell fate specification in premigratory neural crest. BioEssays. 2000;22:708–716. doi: 10.1002/1521-1878(200008)22:8<708::AID-BIES4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Fritzsch B, Northcutt RG. Cranial and spinal nerve organization in amphioxus and lampreys: Evidence for an ancestral craniate pattern. Acta Anatomica. 1993;148:96–109. doi: 10.1159/000147529. [DOI] [PubMed] [Google Scholar]
- García-Castro M, Bronner-Fraser M. Induction and differentiation of the neural crest. Curr Opin Cell Biol. 1999;11:695–698. doi: 10.1016/s0955-0674(99)00038-1. [DOI] [PubMed] [Google Scholar]
- Holland ND, Panganiban G, Henyey EL, Holland LZ. Sequence and developmental expression of AmphiDll, an amphioxus Distal-less gene transcribed in the ectoderm, epidermis and nervous system: Insights into evolution of craniate forebrain and neural crest. Development. 1996;122:2911–2920. doi: 10.1242/dev.122.9.2911. [DOI] [PubMed] [Google Scholar]
- Howard M, Foster DN, Cserjesi P. Expression of HAND gene products may be sufficient for the differentiation of avian neural crest-derived cells into catecholaminergic neurons in culture. Dev Biol. 1999;215:62–77. doi: 10.1006/dbio.1999.9450. [DOI] [PubMed] [Google Scholar]
- Howard MJ, Stanke M, Schneider C, Wu X, Rohrer H. The transcription factor dHAND is a downstream effector of BMPs in sympathetic neuron specification. Development. 2000;127:4073–4081. doi: 10.1242/dev.127.18.4073. [DOI] [PubMed] [Google Scholar]
- Kempf H, Linares C, Corvol P, Gasc J-M. Pharmacological inactivation of the endothelin type A receptor in the early chick embryo: A model of mispatterning of the branchial arch derivatives. Development. 1998;125:4931–4941. doi: 10.1242/dev.125.24.4931. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Miller CT, Kruze G, Ullmann B, BreMiller RA, Larison KD, Snyder HC. The shaping of pharyngeal cartilages during early development of the zebrafish. Dev Biol. 1998;203:245–263. doi: 10.1006/dbio.1998.9016. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Miller CT, Moens CB. Specification and morphogenesis of the zebrafish larval head skeleton. Dev Biol. 2001;233:239–257. doi: 10.1006/dbio.2001.0201. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Kurihara H, Suzuki H, Kodama T, Maemura K, Nagai R, Oda H, Kuwaki T, Cao W-H, Kamada N, et al. Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature. 1994;368:703–710. doi: 10.1038/368703a0. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Kurihara H, Oda H, Maemura K, Nagai R, Ishikawa T, Yazaki Y. Aortic arch malformations and ventricular septal defect in mice deficient in endothelin-1. J Clin Invest. 1995;96:293–300. doi: 10.1172/JCI118033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin NM, Kalcheim C. The neural crest. 2nd ed. Cambridge, UK: Cambridge University Press; 1999. [Google Scholar]
- Maemura K, Kurihara H, Kurihara Y, Oda H, Ishikawa T, Copeland NG, Gilbert DJ, Jenkins NA, Yazaki Y. Sequence analysis, chromosomal location, and developmental expression of the mouse preproendothelin-1 gene. Genomics. 1996;31:177–184. doi: 10.1006/geno.1996.0029. [DOI] [PubMed] [Google Scholar]
- McFadden DG, Charité J, Richardson JA, Srivastava D, Firulli AB, Olson EN. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development. 2000;127:5331–5341. doi: 10.1242/dev.127.24.5331. [DOI] [PubMed] [Google Scholar]
- Miller CT, Schilling TF, Lee K-H, Parker J, Kimmel CB. sucker encodes a zebrafish endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127:3815–3828. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- Offermanns S, Zhao L-P, Gohla A, Sarosi I, Simon MI, Wilkie TM. Embryonic cardiomyocyte hypoplasia and craniofacial defects in Gαq/Gα11-mutant mice. EMBO J. 1998;17:4304–4312. doi: 10.1093/emboj/17.15.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban G, Sebring A, Nagy L, Carroll S. The development of crustacean limbs and the evolution of arthropods. Science. 1995;270:1363–1366. doi: 10.1126/science.270.5240.1363. [DOI] [PubMed] [Google Scholar]
- Pfeffer U, Ferro P, Pavia V, Trombino S, Dell'Eva R, Merlo G, Levi G. The coding region of the human DLX6 gene contains a polymorphic CAG/CCG repeat. Int J Oncol. 2001;18:1293–1297. doi: 10.3892/ijo.18.6.1293. [DOI] [PubMed] [Google Scholar]
- Piotrowski T, Schilling TF, Brand M, Jiang Y-J, Heisenberg C-P, Beuchle D, Grandel H, Van Eeden FJM, Furutani-Seiki M, Granato M, et al. Jaw and branchial arch mutants in zebrafish II: Anterior arches and cartilage differentiation. Development. 1996;123:345–356. doi: 10.1242/dev.123.1.345. [DOI] [PubMed] [Google Scholar]
- Shah NM, Groves AK, Anderson DJ. Alternative neural crest cell fates are instructively promoted by TGFβ superfamily members. Cell. 1996;85:331–343. doi: 10.1016/s0092-8674(00)81112-5. [DOI] [PubMed] [Google Scholar]
- Shelton JM, Lee M-H, Richardson JA, Patel SB. Microsomal triglyceride transfer protein expression during mouse embryonic development. Lipid Res. 2000;41:532–537. [PubMed] [Google Scholar]
- Srivastava D, Cserjesi P, Olson EN. A subclass of bHLH proteins required for cardiac morphogenesis. Science. 1995;270:1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- Stemple DL, Anderson DJ. Isolation of a stem cell for neurons and glia from the mammalian neural crest. Cell. 1992;71:973–985. doi: 10.1016/0092-8674(92)90393-q. [DOI] [PubMed] [Google Scholar]
- Thomas T, Kurihara H, Yamagishi H, Kurihara Y, Yazaki Y, Olson EN, Srivastava D. A signaling cascade involving endothelin-1, dHAND and Msx1 regulates development of neural-crest-derived branchial arch mesenchyme. Development. 1998;125:3005–3014. doi: 10.1242/dev.125.16.3005. [DOI] [PubMed] [Google Scholar]
- Trainor PA, Tam PPL. Cranial paraxial mesoderm and neural crest cells of the mouse embryo: Co-distribution in the craniofacial mesenchyme but distinct segregation in branchial arches. Development. 1995;121:2569–2582. doi: 10.1242/dev.121.8.2569. [DOI] [PubMed] [Google Scholar]
- Wilkie AOM, Morriss-Kay GM. Genetics of craniofacial development and malformation. Nat Rev Genet. 2001;2:458–468. doi: 10.1038/35076601. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Hammer RE, Richardson JA, Williams SC, Clouthier DE, Yanagisawa M. Role of endothelin-1/endothelin-A receptor-mediated signaling pathway in the aortic arch patterning in mice. J Clin Invest. 1998a;102:22–33. doi: 10.1172/JCI2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa H, Yanagisawa M, Kapur RP, Richardson JA, Williams SC, Clouthier DE, De Wit D, Emoto N, Hammer RE. Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development. 1998b;125:825–836. doi: 10.1242/dev.125.5.825. [DOI] [PubMed] [Google Scholar]
- Yang L, Zhang H, Hu G, Wang H, Abate-Shen C, Shen MM. An early phase of embryonic Dlx5 expression defines the rostral boundary of the neural plate. J Neurosci. 1998;18:8322–8330. doi: 10.1523/JNEUROSCI.18-20-08322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelon D, Ticho B, Halpern ME, Ruvinsky I, Ho RK, Silver LM, Stainier DYR. The bHLH transcription factor Hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development. 2000;127:2573–2582. doi: 10.1242/dev.127.12.2573. [DOI] [PubMed] [Google Scholar]