Abstract
Background
HLA mismatch antigens are major targets of alloreactive T cells in HLA-incompatible stem-cell transplantation, which can trigger severe graft-versus-host disease and reduce survival in transplant recipients. Our objective was to identify T-cell subsets with reduced in vitro reactivity to allogeneic HLA antigens.
Design and Methods
We sorted CD4 and CD8 T-cell subsets from peripheral blood by flow cytometry according to their expression of naive and memory markers CD45RA, CD45RO, CD62L, and CCR7. Subsets were defined by a single marker to facilitate future establishment of a clinical-grade procedure for reducing alloreactive T-cell precursors and graft-versus-host disease. T cells were stimulated in mixed lymphocyte reactions against HLA-deficient K562 cells transfected with single HLA-A/-B/-C/-DR/-DQ mismatch alleles. Alloreactivity was measured by interferon-γ spot production and cell proliferation.
Results
We observed that allogeneic HLA-reactivity was preferentially derived from subsets enriched for naïve T cells rather than memory T cells in healthy donors, irrespective of the HLA mismatch allele. This separation was most efficient if CD45RA (versus other markers) was used for sorting. The numbers of allogeneic HLA-reactive effector cells were in median 7.2-fold and 16.6-fold lower in CD45RAneg memory CD8 and CD4 T cells than in entire CD8 and CD4 T cells, respectively. In contrast, proliferation of memory T cells in response to allogeneic HLA was more variably reduced (CD8) or equivalent (CD4) when compared to that of naïve T cells. We also demonstrated in HLA-matched donor-patient pairs that leukemia-reactive CD8 cytotoxic T-lymphocytes were mainly derived from subsets enriched for naïve T cells compared to memory T cells.
Conclusions
Memory T-cell subsets of most healthy individuals showed decreased allogeneic HLA-reactivity, but lacked significant anti-leukemia responses in vitro. The clinical use of memory or naïve-depleted T cells might be beneficial for HLA-mismatched patients at high risk of graft-versus-host disease and low risk of leukemia relapse. Preferred allografts are those which contain leukemia-reactive memory T cells. Alternatively, replenishment with leukemia-reactive T cells isolated from naïve subsets is desirable.
Keywords: alloreactivity, leukemia-reactive T cells, T-cell subsets, naïve T cells, immunotherapy
Introduction
Separation of graft-versus-leukemia from graft-versus-host immune reactions remains the major challenge in allogeneic hematopoietic stem-cell transplantation (HSCT).1 Both effects are mainly mediated by T cells that are contained in donor-derived stem-cell grafts and lymphocyte products with a very broad repertoire of allospecificities.2 Therefore, refined in vitro strategies have been developed that selectively deplete graft-versus-host-reactive (i.e. alloreactive) donor T cells while sparing beneficial reactivity of T cells to infectious agents and leukemia.3 Although several of these approaches have already been demonstrated to be feasible and produce significant reductions of graft-versus-host disease (GvHD) in clinical trials,4–8 selective depletion of alloreactive T cells is still a complex technical approach and requires the in vitro culture of donor T cells for several days.
A less complex, culture-independent approach for GvHD prophylaxis would be to eliminate naïve T cells from the allograft, because they should contain most alloreactive precursors due to the enormous diversity of the naive T-cell receptor (TCR) repertoire.9 Studies in mice have established this concept by demonstrating a much higher rate of GvHD if naive T cells compared to memory T cells of unprimed animals were adoptively transferred into allogeneic recipients.10–12 In vitro experiments with human T cells confirmed that the CD62Lpos subset containing naïve and central memory T cells showed stronger alloresponses than the CD62Lneg effector memory counterpart.13 These data, however, raise important questions. First, which marker(s) of T-cell differentiation should be used clinically for depleting naïve T cells with the aim of minimizing alloreactive precursors in human allografts? Second, is the superior alloresponse of naïve precursors similar in CD4 and CD8 T-cell subsets? Lastly, will residual memory T cells still mediate reactivity to leukemia?
Naïve T cells have the phenotype CD45RApos CD45ROneg CCR7pos CD62Lpos; CCR7 and CD62L are also expressed on central memory T cells.14,15 We, therefore, decided to investigate in vitro alloreactivity of CD4 and CD8 T-cell subsets which were enriched for naïve (i.e. CD45RApos and CD45ROneg) as well as for naïve and central memory T cells (i.e. CCR7pos and CD62Lpos) by flow cytometric cell sorting. The counterpart fractions mainly containing memory T cells as well as entire CD4 and CD8 T cells were also included in the experiments. Because alloreactivity is very complex and can be directed to a very diverse panel of major and minor histocompatibility antigens mismatched between donor and recipient, we chose single non-self-HLA (allo-HLA) molecules as surrogate alloantigens. To detect pure alloreactive T-cell responses against an individual HLA mismatch allele and to minimize the interference by other T-cell specificities, HLA-deficient K562 cells were applied as standard recipient cells and were transfected with single HLA class I (-A/-B/-C) or class II (-DR/-DQ) molecules before use.
Design and Methods
Primary cells and cell lines
This study was approved by the local Ethics Committee and was performed according to the Declaration of Helsinki. Healthy peripheral blood mononuclear cell (PBMC) donors were selected based on their HLA type (Online Supplementary Table S1). PBMC were isolated from buffy coats by Ficoll separation. Primary AML blasts were separated by Ficoll centrifugation from leukapheresis products of patients with a white blood cell count greater than 105/μL.
The K562 cell clone transfected with HLA-A*02:01 cDNA has been previously described.16 Similarly produced K562 cell clones for HLA-B*35:03 and HLA-C*03:03 were provided by Dr. T. Wölfel (Mainz, Germany). Cells showed stable HLA transgene expression in G418-containing medium [median positive cells, 83 (44–98) %]. K562-HLA class II transfectant cells were generated by mRNA transfection. Briefly, 2×107 K562 cells were electroporated (400V, 5ms; Gene Pulser XcellTM, Bio-Rad, Hercules, CA, USA) with 10 μg RNA encoding the α and β chains of HLA-DR (DRA1*01:01, DRB1*07:01) and HLA-DQ (DQA1*01:02, DQB1*06:02) molecules, respectively. RNA was synthesized by in vitro transcription from pST1 cDNA vectors containing full-length HLA class II genes (provided by Dr. U. Sahin, Mainz, Germany) according to the manufacturer’s instructions (mMESSAGE mMACHINE T7 Ultra, Ambion, Austin, TX, USA). The procedure resulted in transient HLA class II expression for up to 1 week with reliably strong levels 12 h after electroporation [median positive cells, 70 (22–85) %]. Aliquots of K562-HLA class II transfectant cells were frozen and used directly after thawing and lethal irradiation (100 Gy). Epstein-Barr virus-transformed B-lymphoblastoid cell lines were generated according to standard in vitro procedures.
Cell sorting and allostimulation of CD8 and CD4 T-cell subsets in vitro
Cryopreserved PBMC were thawed and stained with fluorochrome-conjugated monoclonal antibodies for CD3 and CD8 or CD4, in combination with those for CD45RA, CD45RO, CCR7, or CD62L (Beckman Coulter, Brea, CA; BD Biosciences, San Jose, CA, USA; R&D Systems, Minneapolis, MN, USA). After washing, cells were sorted on a BD FACSAria cell sorter (BD Biosciences) into the indicated T-cell subsets (Figure 1C). Allogeneic mixed lymphocyte reactions (MLR) were performed at 37°C in AIM-V medium (GIBCO-BRL, Carlsbad, CA, USA) supplemented with 10% heat-inactivated human serum. At day 0 (d0), 5×105 sorted CD8 or CD4 T cells were stimulated with 5×104 lethally irradiated mismatched K562-HLA transfectant cells in 48-well plates. Irradiated (35 Gy) autologous PBMC (5×105/well) were added as feeder cells. MLR responder populations were re-stimulated weekly with irradiated K562-HLA cells at a responder:stimulator ratio of 10:1. Cultures contained 10 ng/mL human interleukin (hIL)-7, 1 ng/mL hIL-12, and 5 ng/mL hIL-15 (all from Miltenyi Biotec, Bergisch-Gladbach, Germany) during the first week. From d7 onwards, IL-12 was omitted, and hIL-2 was added at 100 IU/mL (ProleukinTM; Novartis, Nürnberg, Germany).
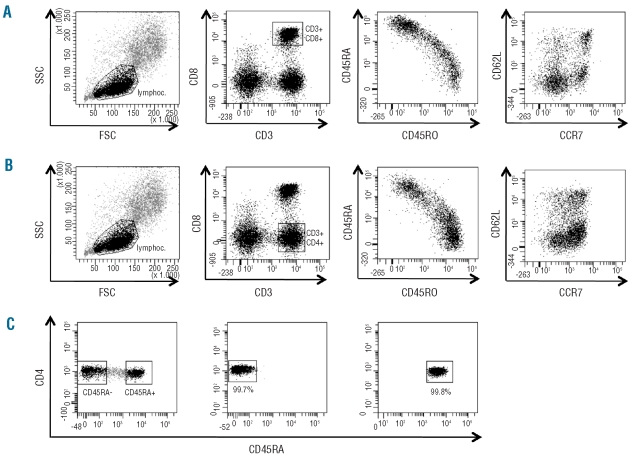
Figure 1.
Flow cytometric staining and sorting of T-cell subsets. PBMC from healthy donors were stained with monoclonal antibodies for CD3 and CD8 together with monoclonal antibodies for T-cell differentiation markers (i.e. CD45RA, CD45RO, CD62L, CCR7). (A, B) Gating of CD3 CD8 T cells (A) and CD3 CD4 T cells (B) with subsequent analysis of CD45RA and CD45RO as well as CD62L and CCR7 co-expression. Note the reciprocal expression of CD45RA and CD45RO, and the unequal distribution of CD62L and CCR7 expression, indicating that CD62L and CCR7 identify different T-cell subsets. Representative results with PBMC of donor SIB 369 are shown. (C) Gating strategy for cell sorting. After gating of CD3 CD8 T cells or CD3 CD4 T cells, the gates for a single T-cell differentiation marker were set according to strong or absent expression of this marker (>0.5 log difference in fluorescence intensity). The sorting gates and resulting fractions for CD45RA in CD4 T cells of donor 372 are shown here. For sorting gates of CD45RO, CD62L and CCR7 see Online Supplementary Figure S1.
For the generation of leukemia-reactive CD8 T-cell lines, 5×105 cells of sorted CD8 T-cell subsets from healthy donors were stimulated in allogeneic mixed lymphocyte-leukemia cultures (MLLC) with 5×105 irradiated (35 Gy) HLA class I-matched primary acute myeloid leukemia (AML) blasts. Irradiated autologous PBMC (5×105/well) of donor origin were added as feeder cells. MLLC were supplemented with the same cytokines as described for the MLR. Additionally, 10 ng/mL hIL-21 (Biomol, Hamburg, Germany) was added to improve in vitro priming of leukemia-reactive T-cell precursors.17 Cultures were re-stimulated weekly with irradiated AML blasts at a responder:stimulator ratio of 1:1.
T-cell expansion was measured by counting viable cells every week with trypan blue staining. Statistical analysis between different experimental arms was performed with SPSS15.0 software. Wilcoxon’s signed-rank test was used to calculate P values.
Flow cytometric analysis
Cells were incubated for 20 min at 4°C with fluorochrome-conjugated monoclonal antibodies (Beckman Coulter, BD Biosciences, R&D Systems). Staining was analyzed on a BD FACSCanto flow cytometer (BD Biosciences). After gating on viable lymphocytes,104 events were evaluated by BD FACSDivaTM software (BD Biosciences) and EXPO32TM software (Beckman Coulter) for re-analysis. To determine the percentage of regulatory T cells (Treg) in CD4 T cells, co-staining for CD25, CD127 (Beckman Coulter), and FOXP3 (eBioscience, San Diego, CA, USA) was performed.
Interferon-γ enzyme-linked immunosorbent spot assay
Twenty-hour interferon-γ (IFN-γ) enzyme-linked immunosorbent spot (ELISpot) assays were performed as described previously.18 T cells were seeded at 1×103 to 4×104/well and target cells at 5×104/well in AIM-V medium. To demonstrate HLA-restricted T-cell reactivity, the following murine monoclonal antibodies were added:19 W6/32, an anti-HLA class I IgG2a, PA2.1, an anti-HLA-A2 IgG1, B1.23.2, an anti-HLA-B and -C IgG2a, L243, an anti-HLA-DR IgG2a, and SPV-L3, an anti-HLA-DQ IgG2a.20 Allo-HLA reactivity of MLR cultures was determined by subtraction of spot numbers in wells with parental K562 cells from those with K562-HLA transfectant cells. The results shown are means ± standard deviation (SD) of duplicate experiments. Statistical analysis was performed with SPSS15.0 software and Wilcoxon’s signed-rank test.
51Chromium-release assay
51Chromium-release assays with an effector-to-target (E/T) incubation of 5 h were conducted as previously described.19 Data shown data are means ± SD of duplicate experiments.
Results
Measuring alloreactivity of CD8 T-cell subsets to single HLA mismatch alleles
We separated CD8 T cells from PBMC of healthy donors by flow cytometric sorting according to the strong or absent expression of CD45RA, CD45RO, CD62L, or CCR7, respectively (Figure 1). Eight different CD8 T-cell subsets with a median purity of 98.7 (77.1–100) % (n=56) were obtained and were analyzed for alloreactivity to single HLA-A, HLA-B, and HLA-C mismatch alleles by allogeneic MLR in vitro. Sorted total CD8 T cells were also included in the MLR experiments. For in vitro stimulation against allo-HLA, we used HLA-deficient K562 cells expressing single HLA molecules (K562-HLA) after transfection. MLR populations were regularly screened for allo-HLA-reactive CD8 T cells by IFN-γ ELISpot assay on d12 of cultures. We detected HLA-mismatch reactivity primarily in MLR that were initiated with CD8 subsets enriched for naïve T cells (Figure 2A). Recognition was clearly restricted to the allo-HLA mismatch allele used during primary in vitro stimulation. This was demonstrated by including parental K562 cells and K562 transfectant cells expressing third-party HLA class I alleles as control targets in all screening assays. In addition, allorecognition could be regularly blocked by monoclonal antibodies binding to the allo-HLA stimulator allele (Figure 2A).
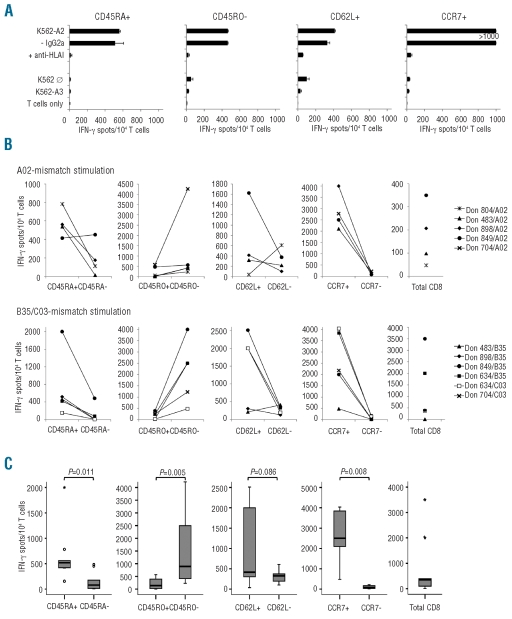
Figure 2.
HLA class I allele-specific allorecognition of CD8 T-cell subsets. MLR cultures were tested for allo-HLA reactivity in an IFN-γ ELISpot assay on d12 (i.e. 5 days after first allo-HLA restimulation on d7). (A) Reactivity to original K562-HLA mismatch stimulator cells, as well as to parental K562 cells and K562 transfectant cells carrying an irrelevant HLA allele. To demonstrate HLA-restriction of detected reactivity, monoclonal antibodies blocking T-cell receptor-HLA interactions (and IgG isotype controls) were used. Representative results with naïve-enriched CD8 T-cell subsets of donor 898 for allo-HLA-A*02:01 are shown. (B) Numbers of spot-forming cells from d12 cultures initiated with sorted subsets (CD45RA, CD45RO, CD62L, CCR7) or entire CD8 T cells from six healthy donors. Allo-HLA mismatch alleles used for in vitro stimulation were HLA-A*02:01 (upper panels), HLA-B*35:03 and HLA-C*03:03 (lower panels). (C) Box plots and P values of data presented in (B). Median (line), 25th to 75th percentile (box), minimum and maximum values (error bars) are indicated. If PBMC numbers were limited, MLR stimulations were restricted to fewer allo-HLA alleles or T-cell subsets.
Superior allogeneic HLA reactivity in subsets enriched for naïve compared to memory CD8 T cells
We next used the MLR-ELISpot test system to screen subsets enriched for naïve or memory CD8 T cells for reactivity to single HLA-A/-B/-C mismatch alleles in six different donors (for HLA types see Online Supplementary Table S1). We observed that much higher numbers of HLA mismatch-reactive CD8 T cells were derived from the CCR7pos subset than from the CCR7neg counterpart [median IFN-γ spot-forming cells (SFC) 2500 versus 74 per 104 MLR responder cells; Figure 2B,C]. This difference was detected in all donor/allo-HLA combinations tested (P=0.008; Figure 2C). Other subsets enriched for naïve CD8 T cells (i.e. CD45RApos, CD45ROneg, CD62Lpos) were also more responsive to allo-HLA stimulation than related memory fractions, with the difference being statistically significant for CD45RA (P=0.011) and CD45RO (P=0.005). The median level of allo-HLA reactivity was higher for the HLA-B/-C used than for the HLA-A mismatch alleles (2000 versus 556 SFC per 104 MLR responders for all naïve-enriched CD8 subsets). In single subsets enriched for memory CD8 T cells, however, alloreactivity could exceed that of naïve counterparts (e.g. Don 804/A02 for CD62L). Most importantly, the alloreactivity of MLR cultures derived from memory-enriched CD8 subpopulations was considerably lower than that of cultures of entire CD8 T cells. Median numbers of SFC per 104 MLR/allo-HLA I responder cells were 74 (range, 15–219) for memory CCR7neg CD8 T cells, 79 (range, 6–484) for memory CD45RAneg CD8 T cells, and 350 (range, 5–3500) for entire CD8 T cells.
As a further parameter of the alloreactive CD8 T-cell response, we measured the expansion of MLR populations during 3 weeks of in vitro stimulation with single HLA class I mismatch alleles. Again, 11 donor/allo-HLA combinations were analyzed. The strongest alloproliferation was observed for CD62Lpos and CCR7pos fractions (median d21: 12.9×107 and 9.7×107 cells), respectively (Online Supplementary Figure S2A,B). In contrast, the CD62Lneg and CCR7neg counterparts showed significantly lower cell expansion (median d21: 3.6×107 and 1.7×107 cells; P=0.015 and P=0.037). Similar differences in alloproliferation were not found for CD45RA and CD45RO subsets. We also analyzed the expression of T-cell differentiation markers in all MLR populations on d14 of culture by flow cytometry. As expected, subsets enriched for naïve CD8 T cells retained expression of early differentiation markers in a higher proportion of MLR responders compared to memory counterparts (data not shown).
Stronger leukemia reactivity in CCR7pos compared to CCR7neg CD8 T cells
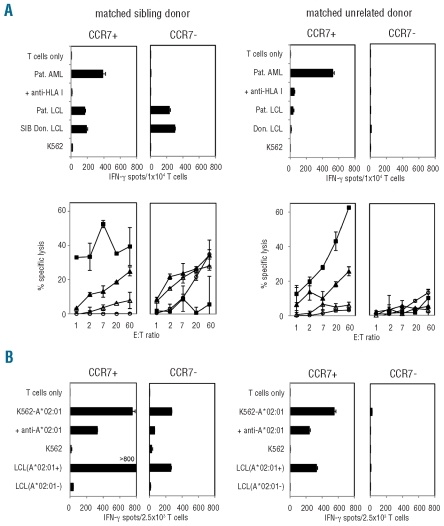
Because the CCR7pos CD8 T-cell subset of healthy individuals contained most allo-HLA-reactive effector cells (Figure 2), depletion of this compartment from entire donor lymphocytes would potentially reduce graft-versus-host-reactivity of HLA-mismatched allografts. However, elimination of donor CCR7pos CD8 T cells may also influence T-cell reactivity to leukemia cells restricted by HLA alleles matched between the donor and leukemia patient. We, therefore, investigated sorted CCR7pos and CCR7neg CD8 T cells of three healthy donors for reactivity to primary AML blasts in donor-patient pairs with complete HLA match by allogeneic MLLC. In parallel experiments, reactivity of both subsets to K562 cells expressing a single HLA class I mismatch allele was also analyzed by MLR. We observed high numbers of AML-reactive CD8 T cells in CCR7pos-derived MLLC, both in HLA-matched sibling and unrelated donors (Figure 3A). Responding CD8 T cells showed cytolytic activity against AML blasts, but did not lyse natural killer target K562. A lower level of lysis was observed against patient-derived B-lymphoblastoid cell lines. In contrast, we did not detect AML-reactive CD8 T cells in CCR7neg-derived MLLC. As expected from our previous data, allo-HLA reactivity mainly resided in CCR7pos fractions (Figure 3B). Similar MLLC results were obtained if CD45RO was used for sorting naïve and memory CD8 T cells (data not shown).
Figure 3.
Parallel analysis of CD8 T-cell subsets for reactivity to HLA-matched AML blasts and to K562-HLA mismatch cells. MLLC and MLR were initiated with sorted CCR7pos and CCR7neg CD8 T-cell subsets from healthy individuals. Stimulator cells were primary AML blasts with complete HLA class I-match (4 digits) in MLLC and K562-HLA mismatch cells in MLR. (A) Representative functional data from MLLC of a sibling donor/patient pair SIB 369/MZ369-AML (left panels) and an unrelated donor/patient pair Don 069/MZ653-AML (right panels) are shown. Cultures were analyzed on d19 (i.e. 5 days after second AML restimulation on d14) in IFN-γ ELISpot (upper panel) and 51Cr-cytotoxicity assays (lower panel). Targets of 51Cr-assays were AML blasts (■), patient-derived LCL (▴), donor-derived LCL (▵), and K562 cells (○). (B) Results from MLR of donors SIB 369 and Don 069, in which HLA-A*02:01 was used as the mismatch allele (for HLA types see Online Supplementary Table S1) are shown. Cultures were screened on d19 for allo-HLA-A*02:01-reactive CD8 T cells in IFN-γ ELISpot assays. LCL from HLA-A*02:01-positive and -negative third-party donors were included as control targets.
Separation of allogeneic HLA reactivity in subsets enriched for naïve and memory CD4 T cells
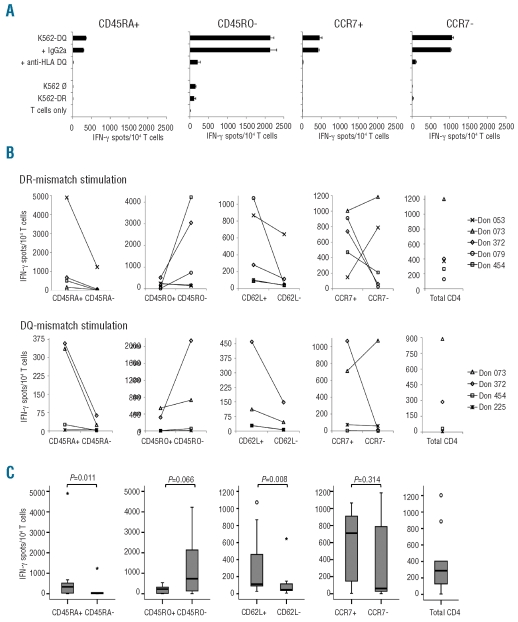
CD4 T cells of six healthy donors were sorted into subsets enriched for naïve or memory T cells using a strategy analogous to that described for CD8 T cells (Figure 1), with a median purity of 99.8 (82.8–100) % for all fractions (n=48). The subsets were stimulated weekly in MLR with K562 cells expressing single HLA-DR/-DQ mismatch alleles in nine donor/allo-HLA combinations (Online Supplementary Table S1). Again, allorecognition was measured by IFN-γ ELISpot assay in the second week of cultures (d12). We observed that the numbers of allo-HLA class II reactive T cells were usually higher in subsets enriched for naïve T cells than in those enriched for the memory counterparts (Figure 4B–C). Statistical significance was found for the CD62L (P=0.008) and CD45RA (P=0.011) subsets. The median level of mismatch reactivity was clearly higher for the used HLA-DR than for the HLA-DQ allele (601 versus 93 SFC per 104 MLR responders for all naïve CD4 subsets). In all experiments, allo-HLA class II reactivity could be blocked by monoclonal antibodies binding to the class II molecule used for primary in vitro stimulation (Figure 4A). Furthermore, significant alloresponses to parental K562 cells as well as to transfectants carrying third-party HLA alleles were not observed, again demonstrating the specificity of the MLR/K562-HLA screening approach. As for the CD8 experiments, alloreactivity of MLR cultures derived from memory-enriched CD4 T-cell subsets was substantially lower than that of total CD4 T cells. The median numbers of SFC per 104 MLR/allo-HLA II responder cells were 25 (range, 2–1237) for memory CD45RAneg CD4 T cells, 46 (range, 6–644) for memory CD62Lneg CD4 T cells, and 287 (range, 4–1204) for entire CD4 T cells. In addition to IFN-γ, CD4 MLR responder populations of individual donors secreted low levels of IL-4 and tumor necrosis factor-α, with a tendency of stronger production in MLR derived from naïve-enriched subsets (data not shown).
Figure 4.
HLA class II allele-specific allorecognition of CD4 T-cell subsets. MLR cultures were tested for allo-HLA reactivity in IFN-γ ELISpot assays on d12 (i.e. 5 days after first allo-HLA restimulation on d7). (A) Reactivity to original K562-HLA mismatch stimulator cells, as well as to parental K562 cells and K562 transfectant cells carrying an irrelevant HLA allele. To demonstrate HLA-restriction of detected reactivity, monoclonal antibodies blocking T-cell receptor-HLA interactions (and IgG isotype controls) were used. Representative results with naïve-enriched CD4 T-cell subsets of donor 372 for allo-HLA-DQB1*06:02 are shown (B) Numbers of IFN-γ spot-forming cells from d12 cultures of MLR started with sorted subsets (CD45RA, CD45RO, CD62L, CCR7) or total CD4 T cells from six healthy donors. HLA class II mismatch alleles used for allostimulation were HLA-DRB1*07:01 (upper panel) and HLA-DQB1*06:02 (lower panel). (C) Box plots and P values of data presented in (B); for explanation see legend to Figure 2C. If PBMC numbers were limited, MLR stimulations were restricted to fewer allo-HLA alleles.
The proliferation of MLR cultures from CD4 T-cell subsets toward single HLA class II mismatch alleles was overall stronger (Online Supplementary Figure S3) than that of cultures from similar allo-HLA class I/CD8 experiments. This was particularly detectable for all memory fractions. Significant differences in alloproliferation between MLR cultures derived from subsets enriched for naïve or memory CD4 T cells were not observed. Furthermore, flow cytometric analysis of MLR on d14 showed that subsets enriched for naïve CD4 T cells retained the expression of early differentiation markers in a higher proportion of MLR responders compared to subsets enriched for memory counterparts (data not shown). In order to determine the distribution of naturally occurring Treg to memory versus naïve CD4 T-cell subsets, CD45RApos and CD45RAneg fractions from five donors were co-stained for CD25high+ FOXP3pos CD127low. Of entire Treg, a median of 33.5 (19–41) % was found in CD45RApos T cells and a median of 66.5 (59–81) % in CD45RAneg T cells. The median percentage of Treg was 3.4 (2.8–3.9) % in total CD4 T cells (data not shown).
Finally, we combined the allo-HLA recognition data of CD8 and CD4 T cells (Figures 2 and 4) to identify which subset marker would allow for most efficient separation of alloreactivity in entire T cells. As shown in Online Supplementary Figure S4A, sorting strategies based on all four markers resulted in stronger allorecognition in subsets enriched for naïve compared to memory T cells (CD45RA: P=0.000; CD45RO: P=0.000; CD62L: P=0.006; CCR7: P=0.002). The memory-enriched fractions showed significantly lower allorecognition than entire T cells (e.g. for CD45RAneg versus entire T cells, P=0.016; median reduction in IFN-γ spots, 7.4-fold). In contrast, similar differences between T-cell fractions were not observed, if alloproliferation data from CD4 and CD8 T cells (Online Supplementary Figures S2 and S3) were combined (Online Supplementary Figure S4B). The median reductions in alloproliferation between total CD4/CD8 T cells and memory fractions were 0.7-fold for CD45RAneg (P=0.31), 1.1-fold for CD45ROpos (P=0.46), 1.2-fold for CD62Lneg (P=0.29), and 1.3-fold for CCR7neg (P=0.04).
Discussion
Here we introduce a rapid screening approach to detect T-cell reactivity of donors to single HLA mismatch alleles. It uses a combination of short-term MLR with a sensitive IFN-γ ELISpot readout system. The main components are HLA-deficient K562 cells that are transfected with ‘off-the-shelf’ HLA class I and II molecules, resulting in stable or transient HLA expression depending on the favored transfection method. By in vitro stimulation of T cells of healthy individuals with single HLA mismatch alleles, we observed that the number of allo-HLA-reactive IFN-γproducing T cells is usually higher in subsets enriched for naïve cells compared to memory T cells. This finding was very consistent, regardless of whether CD8 or CD4 T cells were analyzed. We hereby confirm a previous report showing stronger alloreactivity in human CD62Lpos compared to CD62Lneg T-cell subsets in vitro.13 This earlier study used a mixture of several HLA-mismatched B-lymphoblastoid cell lines as MLR stimulator cells, which has the limitation compared to the K562-HLA test system that detected alloreactivity cannot be attributed to single HLA alleles. Furthermore, if residual HLA molecules are matched between donor T cells and recipient B-lymphoblastoid cell lines, alloreactivity interferes with T-cell responses against recipient antigens presented by shared HLA. Such responses (e.g. against Epstein Barr virus) are most likely not equally distributed between donor-derived naïve and memory T cells.
It has long been debated which combination of differentiation markers spans the naïve T-cell pool most precisely.15 We used the common definition of naïve T cells by CD45 isoforms (i.e. RApos, ROneg) and also included CCR7 and CD62L comprising naïve and central memory T cells in the analysis.14 We found that depletion of central memory T cells additionally to naïve precursors (as represented by CD62Lneg or CCR7neg subsets) did not lead to significantly decreased allo-HLA responses compared to central memory containing CD45RAneg (or CD45ROpos) T cells. On the other hand, we observed overall stronger allo-HLA IFN-γ responses in naïve and central memory containing CCR7pos T-cell fractions compared to naïve-enriched CD45RApos T-cell subsets devoid of central memory cells. The latter finding pointed at least in part to significant alloreactivity residing in the central memory compartment.
Of all subsets, only data for CD45RA-sorted naïve and memory populations were significantly different (P<0.05) for both CD8 and CD4 T cells. In addition, the number of allo-HLA-reactive effector cells was reduced in median by 7.2-fold and 16.6-fold in CD45RAneg memory CD8 and CD4 T cells compared to in entire CD8 and CD4 T cells, respectively. We also observed that a higher fraction of naturally occurring Treg resided in the CD45RAneg CD4 subset than in the CD45RApos one and might have contributed to lower alloreactivity mediated by memory CD4 T-cell fractions.
The proliferative response of sorted memory T-cell subsets to allo-HLA was more variably reduced (CD8) or equivalent (CD4) than the IFN-γ response when compared to naïve T-cell subsets. This finding requires further consideration, since proliferation is a much more complex feature of alloreactivity than IFN-γ secretion. We, therefore, investigated whether the lack of correlation between d21 alloproliferation and d12 allorecognition (IFN-γ ELISpot) data might be a result of the prolonged culture period that promoted a switch from naïve to memory T cells in MLR. However, we did not find significant differences in allo-proliferation between naïve and memory T-cell subsets during the first week of MLR when using cell counting and CFSE staining as readout assays (data not shown).
Our data also suggest that depletion of naïve T cells will likely impair the generation of donor-derived CD8 cytotoxic T lymphocytes recognizing leukemia antigens in the context of shared HLA molecules. This was demonstrated in HLA-matched sibling and unrelated donor-patient pairs by primary MLLC with naïve CD8 subsets either including (i.e. CCR7pos) or devoid of (i.e. CD45ROneg) central memory T cells.14,15 We and others have reported that cytotoxic T lymphocyte-mediated reactivity to leukemia cells mainly derives from naïve CD8 T cells in healthy unprimed individuals19,21–23 and that such cytotoxic T lymphocytes target polymorphic minor histocompatibility antigens (mHag) and non-polymorphic leukemia-associated antigens (LAA).21,22,24 These findings are further supported by HLA-peptide multimer data in ex vivo PBMC showing most precursors of LAA-reactive CD8 T cells in the naïve subset of healthy individuals.25 In contrast, other groups detected HLA-peptide multimer binding leukemia-reactive CD8 T cells also in the antigen-experienced memory pool. This was demonstrated for LAA specificities in healthy donors26 as well as for mHag specificities in donors with a history of alloantigen priming in vivo (e.g. multiparous females,27 cord blood grafts28). Ideally, this would suggest memory T cells from donors previously primed against hematopoietically expressed mHag and LAA by natural immunization or vaccination should be used for transplantation.
Although the concept of ‘memory T-cell therapy’ is promising in terms of the expected lower GvHD potential, we detected single donors who had strong alloreactivity in individual memory subsets. Because memory T cells have the capability of rapid expansion upon secondary antigen challenge,29 the formation of alloreactive memory T cells may have serious consequences in terms of GvHD. Pre-sensitization would be a particular problem if pathogen-specific T cells with cross-reactivity to alloantigens are triggered by an encounter with infectious agents.30–32
We propose the use of the K562-HLA-based MLR approach for the screening of donor T-cell reactivity to individual HLA class I and II mismatch alleles in situations in which a donor with full HLA match is lacking. Such donor-patient HLA disparities, particularly at the A/B/C/DR loci, increase the risk of graft rejection and severe GvHD, and may even decrease survival after allogeneic HSCT.33–36 The approach might help to select a HLA-mismatched donor (e.g. in haploidentical HSCT) with the lowest level of reactivity against non-shared recipient HLA antigens. With the test system, we detected much stronger allorecognition of HLA-A/-B/-C/-DR versus HLA-DQ alleles, which is consistent with the clinical observation that HLA-DQ mismatches are better tolerated and do not adversely affect the outcome of allogeneic HSCT.35,36
In conclusion, allo-HLA mismatch reactivity was preferentially derived from subsets enriched for naïve compared to memory T cells in most healthy individuals. This was demonstrated by using single alleles from all HLA-A/-B/-C/-DR/-DQ loci currently included in donor-patient typing for allogeneic HSCT. We also observed that in vitro strategies for depleting naïve T cells could use several major differentiation markers expressed by naïve T cells (i.e. CD45RApos, CD45ROneg, CD62Lpos, CCR7pos) to develop a suitable sorting strategy. In our hands, the most efficient results for entire T cells were obtained with CD45RA. We also demonstrated the major limitation of the approach, which is the concurrent impairment of T-cell responses against leukemia. This problem might be addressed by using donor allografts which contain leukemia-reactive T cells in the memory compartment due to previous priming against mHag and LAA during natural immunity or vaccination. Alternatively, memory T cells could be replenished with leukemia-reactive T cells previously isolated from the naïve T-cell subset of donors by primary in vitro stimulation.19,22,23 The establishment of a Good Manufacturing Practice procedure for depleting naïve T cells (or selecting memory T cells) in vitro appears feasible. This would pave the way for clinical trials that investigate whether depletion of naïve T cells from donor-derived stem-cell grafts or lymphocyte products can indeed reduce GvHD in HSCT patients. Most suitable study populations are HLA-mismatched patients at high risk of GvHD and low risk of leukemia relapse, as well as patients with diseases other than leukemia.
Acknowledgments
we thank Magda Brikc for excellent technical assistance. We are very grateful to Dr. Ugur Sahin, Mainz, for providing cDNA vectors of HLA class II molecules, and to Dr. Thomas Wölfel, Mainz, for providing K562-HLA-B/-C transfectant cells.
Footnotes
Funding: this work was supported by grant n. KFO183-TP5 from the Deutsche Forschungsgemeinschaft to ED and WH.
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Alyea EP. Modulating graft-versus-host disease to enhance the graft-versus-leukemia effect. Best Pract Res Clin Haematol. 2008;21(2):239–50. doi: 10.1016/j.beha.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 2.Felix NJ, Allen PM. Specificity of T-cell alloreactivity. Nat Rev Immunol. 2007;7 (12):942–53. doi: 10.1038/nri2200. [DOI] [PubMed] [Google Scholar]
- 3.Rezvani K, Barrett AJ. Characterizing and optimizing immune responses to leukaemia antigens after allogeneic stem cell transplantation. Best Pract Res Clin Haematol. 2008;21(3):437–53. doi: 10.1016/j.beha.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonini C, Ferrari G, Verzeletti S, Servida P, Zappone E, Ruggieri L, et al. HSV-TK gene transfer into donor lymphocytes for control of allogeneic graft-versus-leukemia. Science. 1997;276(5319):1719–24. doi: 10.1126/science.276.5319.1719. [DOI] [PubMed] [Google Scholar]
- 5.Guinan EC, Boussiotis VA, Neuberg D, Brennan LL, Hirano N, Nadler LM, et al. Transplantation of anergic histoincompatible bone marrow allografts. N Engl J Med. 1999;340(22):1704–14. doi: 10.1056/NEJM199906033402202. [DOI] [PubMed] [Google Scholar]
- 6.Andre-Schmutz I, Le Deist F, Hacein-Bey-Abina S, Vitetta E, Schindler J, Chedeville G, et al. Immune reconstitution without graft-versus-host disease after haemopoietic stem-cell transplantation: a phase 1/2 study. Lancet. 2002;360(9327):130–7. doi: 10.1016/S0140-6736(02)09413-8. [DOI] [PubMed] [Google Scholar]
- 7.Solomon SR, Mielke S, Savani BN, Montero A, Wisch L, Childs R, et al. Selective depletion of alloreactive donor lymphocytes: a novel method to reduce the severity of graft-versus-host disease in older patients undergoing matched sibling donor stem cell transplantation. Blood. 2005;106(3):1123–9. doi: 10.1182/blood-2005-01-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amrolia PJ, Muccioli-Casadei G, Huls H, Adams S, Durett A, Gee A, et al. Adoptive immunotherapy with allodepleted donor T-cells improves immune reconstitution after haploidentical stem cell transplantation. Blood. 2006;108(6):1797–808. doi: 10.1182/blood-2006-02-001909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikolich-Zugich J, Slifka MK, Messaoudi I. The many important facets of T-cell repertoire diversity. Nat Rev Immunol. 2004;4 (2):123–32. doi: 10.1038/nri1292. [DOI] [PubMed] [Google Scholar]
- 10.Anderson BE, McNiff J, Yan J, Doyle H, Mamula M, Shlomchik MJ, et al. Memory CD4+ T cells do not induce graft-versus-host disease. J Clin Invest. 2003;112(1):101–8. doi: 10.1172/JCI17601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen BJ, Cui X, Sempowski GD, Liu C, Chao NJ. Transfer of allogeneic CD62L-memory T cells without graft-versus-host disease. Blood. 2004;103(4):1534–41. doi: 10.1182/blood-2003-08-2987. [DOI] [PubMed] [Google Scholar]
- 12.Beilhack A, Schulz S, Baker J, Beilhack GF, Wieland CB, Herman EI, et al. In vivo analyses of early events in acute graft-versus-host disease reveal sequential infiltration of T-cell subsets. Blood. 2005;106(3):1113–22. doi: 10.1182/blood-2005-02-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster AE, Marangolo M, Sartor MM, Alexander SI, Hu M, Bradstock KF, et al. Human CD62L- memory T cells are less responsive to alloantigen stimulation than CD62L+ naive T cells: potential for adoptive immunotherapy and allodepletion. Blood. 2004;104(8):2403–9. doi: 10.1182/blood-2003-12-4431. [DOI] [PubMed] [Google Scholar]
- 14.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–63. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 15.Appay V, van Lier RA, Sallusto F, Roederer M. Phenotype and function of human T lymphocyte subsets: consensus and issues. Cytometry A. 2008;73(11):975–83. doi: 10.1002/cyto.a.20643. [DOI] [PubMed] [Google Scholar]
- 16.Britten CM, Meyer RG, Kreer T, Drexler I, Wolfel T, Herr W. The use of HLA-A*0201-transfected K562 as standard antigen-presenting cells for CD8(+) T lymphocytes in IFN-gamma ELISPOT assays. J Immunol Methods. 2002;259(1–2):95–110. doi: 10.1016/s0022-1759(01)00499-9. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Bleakley M, Yee C. IL-21 influences the frequency, phenotype, and affinity of the antigen-specific CD8 T cell response. J Immunol. 2005;175(4):2261–9. doi: 10.4049/jimmunol.175.4.2261. [DOI] [PubMed] [Google Scholar]
- 18.Wehler TC, Nonn M, Brandt B, Britten CM, Grone M, Todorova M, et al. Targeting the activation-induced antigen CD137 can selectively deplete alloreactive T cells from antileukemic and antitumor donor T-cell lines. Blood. 2007;109(1):365–73. doi: 10.1182/blood-2006-04-014100. [DOI] [PubMed] [Google Scholar]
- 19.Distler E, Wolfel C, Kohler S, Nonn M, Kaus N, Schnurer E, et al. Acute myeloid leukemia (AML)-reactive cytotoxic T lymphocyte clones rapidly expanded from CD8(+) CD62L((high)+) T cells of healthy donors prevent AML engraftment in NOD/SCID IL2Rgamma(null) mice. Exp Hematol. 2008;36(4):451–63. doi: 10.1016/j.exphem.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Spits H, Borst J, Giphart M, Coligan J, Terhorst C, De Vries JE. HLA-DC antigens can serve as recognition elements for human cytotoxic T lymphocytes. Eur J Immunol. 1984;14(4):299–304. doi: 10.1002/eji.1830140404. [DOI] [PubMed] [Google Scholar]
- 21.Quintarelli C, Dotti G, De Angelis B, Hoyos V, Mims M, Luciano L, et al. Cytotoxic T lymphocytes directed to the preferentially expressed antigen of melanoma (PRAME) target chronic myeloid leukemia. Blood. 2008;112(5):1876–85. doi: 10.1182/blood-2008-04-150045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bleakley M, Otterud BE, Richardt JL, Mollerup AD, Hudecek M, Nishida T, et al. Leukemia-associated minor histocompatibility antigen discovery using T-cell clones isolated by in vitro stimulation of naive CD8+ T cells. Blood. 2010;115(23):4923–33. doi: 10.1182/blood-2009-12-260539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albrecht J, Frey M, Teschner D, Carbol A, Theobald M, Herr W, et al. IL-21-treated naive CD45RA+ CD8+ T cells represent a reliable source for producing leukemia-reactive cytotoxic T lymphocytes with high proliferative potential and early differentiation phenotype. Cancer Immunol Immunother. 2011;60(2):235–48. doi: 10.1007/s00262-010-0936-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolfel C, Lennerz V, Lindemann E, Hess G, Derigs HG, Huber C, et al. Dissection and molecular analysis of alloreactive CD8+ T cell responses in allogeneic haematopoietic stem cell transplantation. Cancer Immunol Immunother. 2008;57(6):849–57. doi: 10.1007/s00262-007-0421-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffioen M, Kessler JH, Borghi M, van Soest RA, van der Minne CE, Nouta J, et al. Detection and functional analysis of CD8+ T cells specific for PRAME: a target for T-cell therapy. Clin Cancer Res. 2006;12(10):3130–6. doi: 10.1158/1078-0432.CCR-05-2578. [DOI] [PubMed] [Google Scholar]
- 26.Rezvani K, Grube M, Brenchley JM, Sconocchia G, Fujiwara H, Price DA, et al. Functional leukemia-associated antigen-specific memory CD8+ T cells exist in healthy individuals and in patients with chronic myelogenous leukemia before and after stem cell transplantation. Blood. 2003;102(8):2892–900. doi: 10.1182/blood-2003-01-0150. [DOI] [PubMed] [Google Scholar]
- 27.Verdijk RM, Kloosterman A, Pool J, van de Keur M, Naipal AM, van Halteren AG, et al. Pregnancy induces minor histocompatibility antigen-specific cytotoxic T cells: implications for stem cell transplantation and immunotherapy. Blood. 2004;103(5):1961–4. doi: 10.1182/blood-2003-05-1625. [DOI] [PubMed] [Google Scholar]
- 28.Mommaas B, Stegehuis-Kamp JA, van Halteren AG, Kester M, Enczmann J, Wernet P, et al. Cord blood comprises antigen-experienced T cells specific for maternal minor histocompatibility antigen HA-1. Blood. 2005;105(4):1823–7. doi: 10.1182/blood-2004-07-2832. [DOI] [PubMed] [Google Scholar]
- 29.Chandok MR, Farber DL. Signaling control of memory T cell generation and function. Semin Immunol. 2004;16(5):285–93. doi: 10.1016/j.smim.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 30.Burrows SR, Khanna R, Burrows JM, Moss DJ. An alloresponse in humans is dominated by cytotoxic T lymphocytes (CTL) cross-reactive with a single Epstein-Barr virus CTL epitope: implications for graft-versus-host disease. J Exp Med. 1994;179 (4):1155–61. doi: 10.1084/jem.179.4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koelle DM, Chen HB, McClurkan CM, Petersdorf EW. Herpes simplex virus type 2-specific CD8 cytotoxic T lymphocyte cross-reactivity against prevalent HLA class I alleles. Blood. 2002;99(10):3844–7. doi: 10.1182/blood.v99.10.3844. [DOI] [PubMed] [Google Scholar]
- 32.Amir AL, D’Orsogna LJ, Roelen DL, van Loenen MM, Hagedoorn RS, de Boer R, et al. Allo-HLA reactivity of virus-specific memory T cells is common. Blood. 2010;115(15):3146–57. doi: 10.1182/blood-2009-07-234906. [DOI] [PubMed] [Google Scholar]
- 33.Anasetti C, Amos D, Beatty PG, Appelbaum FR, Bensinger W, Buckner CD, et al. Effect of HLA compatibility on engraftment of bone marrow transplants in patients with leukemia or lymphoma. N Engl J Med. 1989;320(4):197–204. doi: 10.1056/NEJM198901263200401. [DOI] [PubMed] [Google Scholar]
- 34.Sasazuki T, Juji T, Morishima Y, Kinukawa N, Kashiwabara H, Inoko H, et al. Effect of matching of class I HLA alleles on clinical outcome after transplantation of hematopoietic stem cells from an unrelated donor. Japan Marrow Donor Program. N Engl J Med. 1998;339(17):1177–85. doi: 10.1056/NEJM199810223391701. [DOI] [PubMed] [Google Scholar]
- 35.Flomenberg N, Baxter-Lowe LA, Confer D, Fernandez-Vina M, Filipovich A, Horowitz M, et al. Impact of HLA class I and class II high-resolution matching on outcomes of unrelated donor bone marrow transplantation: HLA-C mismatching is associated with a strong adverse effect on transplantation outcome. Blood. 2004;104(7):1923–30. doi: 10.1182/blood-2004-03-0803. [DOI] [PubMed] [Google Scholar]
- 36.Lee SJ, Klein J, Haagenson M, Baxter-Lowe LA, Confer DL, Eapen M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110 (13):4576–83. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]