Abstract
Ten-eleven translocation 2 (TET2) mutations have been involved in myeloid malignancies. This retrospective study aims at evaluating the frequency and impact of TET2 mutations in 247 secondary acute myeloid leukemia cases referred to as myelodysplasia-related changes (n=201) or therapy-related (n=46) leukemias. Mutation of at least one copy of the TET2 gene was detected in 49 of 247 (19.8%) patients who presented with older age, higher hemoglobin level, higher neutrophil and monocyte counts, and lower platelet count. TET2 mutations were significantly less frequent in therapy-related (8.7%) than myelodysplasia-related changes (22.3%; P=0.035) leukemias and strongly associated with normal karyotype (P<0.001). TET2 mutations did not significantly associate with NPM1, FLT3-ITD or FLT3-D835, WT1, or N- or K-RAS mutations. Complete remission was achieved in 57% of evaluable patients who had received intensive chemotherapy. In this group, TET2 mutations did not influence the complete remission rate or overall survival.
Keywords: secondary AML, TET2 mutations, characteristics, prognosis
Introduction
Mutations of the ten-eleven translocation 2 (TET2) gene, which encodes a 2-oxoglutarate/Fe2+ oxygenase that catalyses the conversion of methylcytosine to hydroxymethylcytosine, were recently identified in myeloid malignancies.1 They involve 19–26% of myelodysplastic syndromes (MDS),2,3,4 12–15% of myeloproliferative neoplasms (MPN) or MDS/MPN disorders,1,5,6,7 and 8–19% of de novo adult acute myeloid leukemias (AML).3,6,8,9 Impact of TET2 mutation on prognosis in either de novo or secondary AML remains controversial.6,8,9 In addition, the frequency and impact of TET2 mutations on initial features and response to treatment in secondary AML (sAML) have not yet been fully examined.
In this retrospective study, we analyzed the TET2 gene coding sequence in a cohort of 247 sAML recorded as myelodysplasia-related changes (MRC) AML and therapy-related (TR) AML based on the WHO 2008 classification.10 Patients with TET2 mutations (19.8%) presented with particular characteristics and had the same prognosis as patients with wild-type TET2.
Design and Methods
Patients
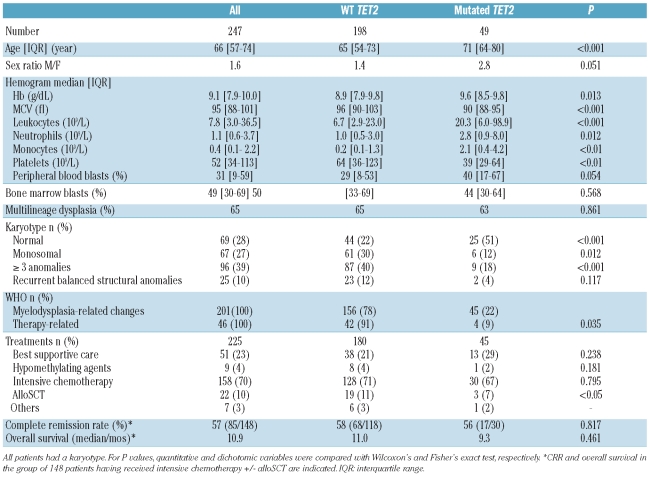
Between 2000 and 2008, bone marrow (BM) mononuclear cells were collected at diagnosis from 247 patients from 4 French centers of the Groupe Ouest-Est d’étude des Leucémies Aiguës et Autres Maladies du Sang (GOELAMS). Among these, 158 patients received intensive chemotherapy (IC) with anthracycline-cytarabine. Among 148 of 158 evaluable patients, 85 achieved complete remission (CR). Informed consent was obtained from all patients in accordance with the Declaration of Helsinki. This study was approved by the Paris Centre ethics committee (number 2346). Main patients’ characteristics are summarized in Table 1.
Table 1.
Clinical and biological characteristics of patients with secondary AML according to TET2 status.
Genotyping
Analysis of TET2 sequence variations was performed by direct sequencing of the PCR products.2 Germinal DNA or DNA after complete remission were unavailable. Frameshifts or nonsense mutations, mutations in splice site, insertions and missense mutations affecting the conserved regions only were considered. Single nucleotide polymorphisms (SNP) either previously published or recorded in the National Center for Biotechnology Information SNP database (www.ncbi.nih.gov/projects/SNP) were excluded. NPM1, FLT3-ITD or FLT3-D835, N/K-RAS ex1/2, CKIT-D816V and WT1 ex7/9 mutations were identified as recommended by Dohner et al.11
Statistical analysis
Complete remission rate was scored according to Cheson et al.12 Continuous and dichotomic variables were compared with the Wilcoxon’s and Fisher’s exact tests, respectively. Overall survival was estimated using the Kaplan-Meier method and compared with the log rank test. All tests were two-sided and P<0.05 was considered significant.
Results and Discussion
The TET2 gene was sequenced in 247 patients with sAML (MRC-AML n=201, TR-AML n=46). Diagnosis of MRC-AML was based on a past history of MDS, MPN or MPN/MDS in 95, a cytogenetics of MDS in 56, and/or multilineage dysplasia (MD) in 48 patients, respectively. Only 9 patients met the diagnosis of MRC-AML with MD as unique criteria (i.e. no antecedent of MDS, MPN or MPN/MDS and a normal karyotype). Excluding variations corresponding to SNP allowed the identification of 69 abnormalities in 49 (19.8%) patients, including 29 frameshifts inducing a stop codon, 24 nonsense mutations, 2 insertions, 2 mutations at a splice site, and 12 missense mutations in conserved domains. Most of these have been previously reported.8,9,13 As already shown, mutations spread throughout the coding sequence (Online Supplementary Figure S1). Only 5 of 69 mutations (7%) were recurrent, and 20 of 49 (40%) patients had two anomalies (Online Supplementary Table S1). The overall frequency of TET2 mutations was comparable to that reported in smaller cohorts of AML or de novo AML.8,9 However, a recent study including 783 patients with AML aged 16 to 60 years showed a lower frequency (7.6%) than that observed in our cohort in which the median age was 66 years (range 57–74).14
Clinical and biological characteristics of patients harboring a TET2 mutation are described in Table 1. Mutated patients were more frequently male than female (P=0.051) and were significantly older than other patients (71 years vs. 65 years; P<0.001) suggesting that TET2 mutations could be linked to aging. TET2 mutations were associated with significantly higher Hb level (9.6 vs. 8.9 g/dL; P=0.013), higher leukocyte (20.3 vs. 6.7×109/L; P=0.002), neutrophil (2.8 vs. 1.0×109/L; P=0.012) and monocyte (2.1 vs. 0.2×109/L; P<0.01) counts, and a lower MCV (89.7 vs. 96 fL; P<0.001) and platelet count (39 vs. 64×109/L; P=0.003). Percentages of blast cells in blood or bone marrow were the same in the two groups. Older age and high monocyte count have been previously reported in TET2 mutated MPN, or MDS/MPN.5,15 We found no difference in FAB subtype, and MD frequency was the same in the two groups. In contrast, the frequency of TET2 mutations was significantly lower in TR-AML (8%) than MRC-AML (22%; P=0.035). Patients with TR-AML had poor prognostic factors (high leukocyte count, circulating or BM blasts, abnormal karyotype, FLT3 mutations) and a 5.5-month median overall survival (OS), compared to MRC-AML (median OS 11 months, Online Supplementary Table S2).
In the whole cohort (n=247), 69 (28%) patients had a normal karyotype (NK). The percentage of patients with NK was significantly higher in the TET2 mutated group (25 of 49; 51%) than in the non-mutated group (44 of 198; 22%) (P<0.001). The association with normal karyotype has not been reported in MDS or in de novo AML. The presence of a monosomal or a complex (≥ 3 anomalies) karyotype was less frequent in the TET2 mutated group (P=0.012 and P<0.0001, respectively). Recurrent translocations were uncommon (26 of 247; 10.5%). Strikingly, only 2 TET2 mutated patients presented a known recurrent chromosome anomaly, both of them involving the 3q26 region. Thus, TET2 mutations did not associate with cytogenetic abnormalities. In NK patients, TET2 mutations also associated with older age (68 vs. 60 years; P=0.024), higher Hb level (9.9 vs. 9.0 g/dL; P=0.018) and leukocyte count (19.9 vs. 5.7×109/L; P=0.024), and a lower platelet count (44 vs. 70×109/L; P=0.048). These results suggest that the significant changes in biological parameters of TET2 mutated patients are independent of cytogenetic abnormalities.
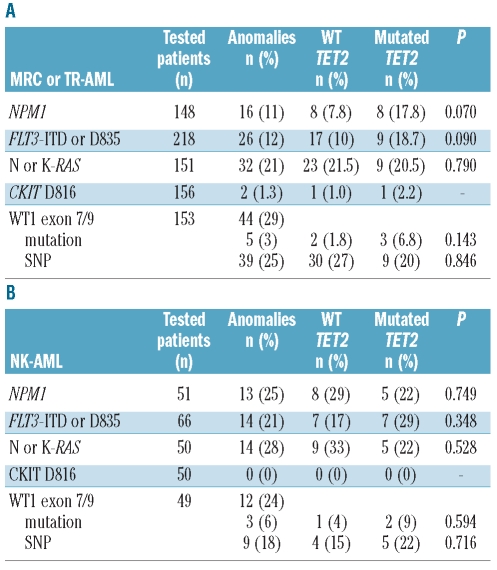
We looked at classical AML mutations in 148 cases for NPM1, 218 for FLT3-ITD and FLT3-D835, 151 for N or K-RAS, 156 in CKIT-D816V and 153 for WT1 exon 7/9 at diagnosis (Tables 2A and B). Except for N or K-RAS mutations, which were observed at the same frequency in secondary and de novo AML, other mutations were less common in sAML than expected from the analysis of de novo AML cohorts. Our data are consistent with the frequencies reported in sAML.16 In the cohort of patients with MRC or TR-AML, we found no difference in the repartition of FLT3-ITD or FLT3-D835 [17 of 170 (10%) vs. 9 of 48 (18.7%)], N or K-RAS [23 of 107 (21.5%) vs. 9 of 44 (20.5%)], or WT1 [2 of 109 (1.8%) vs. 3 of 44 (6.8%)] mutations between TET2 non-mutated and mutated groups, respectively (Table 2A). Although NPM1 associated with TET2 mutations in de novo AML,9 this link did not reach statistical significance in our series [8 of 103 (7.8%) vs. 8 of 45 (17.8%); P=0.070] or in the mixed de novo and sAML cohort reported by others.8 CKIT-D816V mutations are rare in AML. However, they appeared to be more frequent in TET2 mutated patients as reported in systemic mastocytosis.17 In NK patients, TET2 was not associated with NPM1 or FLT3-ITD or FLT3-D835 mutations (Table 2B). In TR-AML, TET2 mutations are rare (8%) compared to TP53 mutations which are detected in more than 20% of patients, as previously reported.18
Table 2.
Frequencies of classical molecular events according to TET2 status. (A) MRC or TR-AML. (B) AML with normal karyotype (NK). Mutations or SNP for WT1 exon 7/9. Fisher’s exact test for P value.
Information on treatment was available for 225 (91%) patients. Treatment was best supportive care (n=51), hypomethylating agents (n=9), IC (n=158) followed by alloSCT (n=22), others (n=7). There was no difference in the repartition of treatments between mutated and wild-type patients, except for alloSCT which was less frequent in the TET2 mutated group, probably because of older age (P<0.05; Table 1).
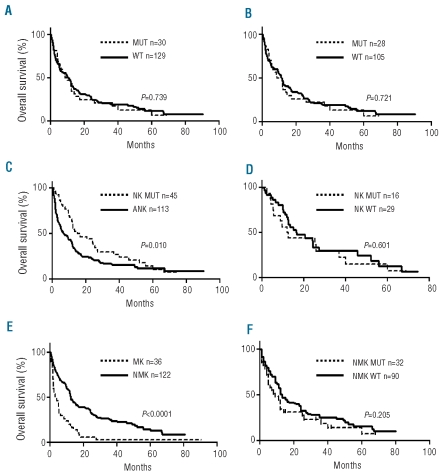
In the group of 158 patients treated with intensive chemotherapy, mutated patients had the same clinical and biological features as mutated patients in the whole cohort. The rate of complete remission was 57% (85 of 148 evaluable patients). Early death after induction was observed in 4 patients (3 wild-type and one mutated). There was no difference in complete remission rate between TET2 mutated and wild-type patients (Fisher’s exact test P=0.817). When the analysis was restricted to MRC-AML, TET2 status did not influence the complete remission rate (P=0.319). With a median follow up of 12.4 months, median overall survival from diagnosis was 11 months in wild-type (n=129) and 9.3 months in mutated patients (n=30; P=0.461). Although the median follow up was short, the size of the cohort allowed a Kaplan-Meier analysis to be performed which demonstrated that TET2 mutations had no predictive value on overall survival in the group of patients with MRC-AML or TR-AML treated with intensive chemotherapy (log rank test; P=0.739; HR 1.04 [95% CI: 0.71–1.52]) (Figure 1A). In addition, TET2 mutations had no impact on overall survival in the group of MRC-AML (Figure 1B), while their impact in TR-AML could not be analyzed.
Figure 1.
Overall survival. (A) and (B) impact of TET2 mutation in MRC-AML or TR-AML (A) and MRC-AML (B). (C) Impact of normal karyotype. (D) Impact of TET2 in AML with normal karyotype. (E) Impact of monosomal karyotype. (F) Impact of TET2 on non-monosomal karyotype AML. MUT: mutated TET2; WT: wild-type TET2; NK: normal karyotype; ANK: abnormal karyotype; MK: monosomal karyotype; NMK: non-monosomal karyotype. Log rank test for P value.
Finally, we looked at overall survival in AML treated by intensive chemotherapy depending on the presence of either a normal or a monosomal karyotype (MK) according to Breems et al.19 NK improves overall survival (Figure 1C) and TET2 had no impact on overall survival in NK AML (Figure 1D). MK was a very poor prognostic factor (Figure 1E). Although the median overall survival was three months in wild-type TET2 patients (n=46) and 11.4 months in mutated TET2 patients (n=4) with MK, the trend to better survival of TET2 mutated patients was not significant (P=0.305), possibly because of the small size of the MK subgroup (Figure 1F). These results are consistent with two reports showing that overall survival was independent of TET2 status6,9 and differ from the findings of Abdel-Wahab et al. which demonstrated an unfavorable impact.8
In conclusion, the frequency of TET2 mutations appears to be higher in sAML than in de novo AML and associated with MRC-AML rather than TR-AML. They did not associate with other mutations reported as prognostic factors in AML or modify the poor prognosis of sAML.
Acknowledgments
the authors would like to thank the investigators of the Groupe Ouest-Est d’étude des Leucémies Aiguës et des Maladies du Sang (GOELAMS) for recording patients, Dr Sophie Grabar for help in the statistical analysis, and Lamya Slama, Gaelle Herledan, and Julien Brard for technical assistance.
Footnotes
Funding: this work was supported by grants from the Institut National du Cancer (INCa 2008) and the Direction Régionale de la Recherche Clinique AP-HP (PHRC Cancer 2009).
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylacytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289–301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 3.Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41(7):838–42. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 4.Kosmider O, Gelsi-Boyer V, Cheok M, Grabar S, Della-Valle V, Picard F, et al. TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDS) Blood. 2009;114(15):3285–91. doi: 10.1182/blood-2009-04-215814. [DOI] [PubMed] [Google Scholar]
- 5.Tefferi A, Pardanani A, Lim KH, Abdel-Wahab O, Lasho TL, Patel J, et al. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia. 2009;23(5):905–11. doi: 10.1038/leu.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jankowska AM, Szpurka H, Tiu RV, Makishima H, Afable M, Huh J, et al. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009;113(25):6403–10. doi: 10.1182/blood-2009-02-205690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith AE, Mohamedali AM, Kulasekararaj A, Lim Z, Gaken J, Lea NC, et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low abundance mutant clones with early origins, but indicates no definite prognostic value. Blood. 2010;116(9):3923–32. doi: 10.1182/blood-2010-03-274704. [DOI] [PubMed] [Google Scholar]
- 8.Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114(1):144–7. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nibourel O, Kosmider O, Cheok M, Boissel N, Renneville A, Philippe N, et al. Incidence and prognostic value of TET2 alterations in de novo acute myeloid leukemia (AML) achieving complete remission. Blood. 2010;116(7):1132–5. doi: 10.1182/blood-2009-07-234484. [DOI] [PubMed] [Google Scholar]
- 10.Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–51. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 11.Döhner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):454–74. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 12.Cheson BD, Bennett JM, Kopecky KJ, Buchner T, Willman CL, Estey EH, et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J Clin Oncol. 2003;21(24):4642–9. doi: 10.1200/JCO.2003.04.036. [DOI] [PubMed] [Google Scholar]
- 13.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468(7325):839–43. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaidzik VI, Schlenk RF, Paschka P, Köhne C-H, Held G, Hadbank M, et al. TET2 mutations in acute myeloid leukemia (AML): Results on 783 patients treated within the AML HD98A study of the AML study group (AMLSG). Abstract 97, ASH meeting; 2010. [Google Scholar]
- 15.Kosmider O, Gelsi-Boyer V, Ciudad M, Racoeur C, Jooste V, Vey N, et al. TET2 gene mutation is a frequent and adverse event in chronic myelomonocytic leukemia. Haematologica. 2009;94(12):1676–81. doi: 10.3324/haematol.2009.011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dicker F, Haferlach C, Sundermann J, Wendland N, Weiss T, Kern W, et al. Mutation analysis for RUNX1, MLL-PTD, FLT3-ITD, NPM1 and NRAS in 269 patients with MDS or secondary AML. Leukemia. 2010;24(8):1528–32. doi: 10.1038/leu.2010.124. [DOI] [PubMed] [Google Scholar]
- 17.Tefferi A, Levine RL, Lim KH, Abdel-Wahab O, Lasko TL, Patel J, et al. Frequent TET2 mutations in systemic mastocytosis: clinical, KIT D816V and FIP1L1-PDGFRA correlates. Leukaemia. 2009;23(5):900–4. doi: 10.1038/leu.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christiansen DH, Andersen MK, Pedersen-Bjergaard J. Mutations with loss of heterozygosity of p53 are common in therapy-related myelodysplasia and acute myeloid leukemia after exposure to alkylating agents and significantly associated with deletion or loss of 5q, a complex karyotype, and a poor prognosis. J Clin Oncol. 2001;19 (5):1405–13. doi: 10.1200/JCO.2001.19.5.1405. [DOI] [PubMed] [Google Scholar]
- 19.Breems DA, Van Putten WL, De Greef GE, Van Zelderen-Bhola SL, Gerssen-Schoorl KB, Mellink CH, et al. Monosomal karyotype in acute myeloid leukemia: a better indicator of poor prognosis than a complex karyotype. J Clin Oncol. 2008;26(29):4791–7. doi: 10.1200/JCO.2008.16.0259. [DOI] [PubMed] [Google Scholar]