Abstract
In the yeast Saccharomyces cerevisiae, DNA double strand break (DSB) repair by nonhomologous end-joining (NHEJ) requires the DNA end-binding heterodimer Yku70p–Yku80p and the ligase Dnl4p associated with its cofactor Lif1p. NHEJ efficiency is down-regulated in MATa/MATα cells relative to MATa or MATα cells, but the mechanism of this mating type regulation is unknown. Here we report the identification of Lif2p, a S. cerevisiae protein that interacts with Lif1p in a two-hybrid system. Disruption of LIF2 abolishes the capacity of cells to repair DSBs by end-joining to the same extent than lif1 and dnl4 mutants. In MATa/MATα cells, Lif2p steady-state level is strongly repressed when other factors involved in NHEJ are unaffected. Increasing the dosage of the Lif2p protein can suppress the NHEJ defect in a/α cells. Together, these results indicate that NHEJ regulation by mating type is achieved, at least in part, by a regulation of Lif2p activity.
Keywords: DNA repair, end-joining, HO, I-SceI, a/α repression
In eucaryotes, DNA double strand breaks (DSBs) can be induced by metabolic processes, DNA damaging agents, or specific endonucleases. If unrepaired or wrongly repaired, such breaks lead to cell death or genomic rearrangements. DSBs can be repaired by at least two pathways, homologous recombination or nonhomologous end-joining (NHEJ). Homologous recombination is a mostly error-free process that involves the presence of homologous sequences. In the nonhomologous repair mechanism, DNA ends are joined with little or no base pairing at the junction and the end-joining product may be accompanied with insertions or deletions (Paques and Haber 1999; Wood et al. 2001). In addition to its major role in resistance to DNA damage, NHEJ is required for V(D)J recombination in mammals (Frank et al. 1998).
To date, genetic and biochemical studies have established that end-joining in yeast and mammals requires the same core set of proteins, the DNA end-binding proteins KU70 and KU80 that form the KU heterodimer, as well as DNA ligase IV and its associated factor XRCC4 (called Dnl4p/Lig4p and Lif1p, respectively, in S. cerevisiae) (Schar et al. 1997; Teo and Jackson 1997; Wilson et al. 1997; Herrmann et al. 1998; Wood et al. 2001). In addition, NHEJ in mammals also required DNA-PKcs (Wood et al. 2001). Lif1p interacts with the carboxy-terminal domain of Dnl4p and is required for its activity in vivo (Herrmann et al. 1998). Lif1p is detectable near DNA ends, suggesting that Lif1p binds DNA in cooperation with KU and targets Dnl4p to DSB (Teo and Jackson 2000). In S. cerevisiae, a complex formed by Rad50p, Mre11p, and Xrs2p is essential to NHEJ (Moore and Haber 1996; Boulton and Jackson 1998). In the fission yeast Schizosaccharomyces pombe and in vertebrates, the homologs of these proteins do not appear to be required for NHEJ, suggesting that the role played by this complex in the end-joining reaction is not conserved in all eucaryotes (Wilson et al. 1999; Yamaguchi-Iwai et al. 1999; Harfst et al. 2000; Manolis et al. 2001).
In S. cerevisiae, NHEJ level is influenced by mating-type status. Diploid cells, expressing simultaneously MATa and MATα, exhibit levels of NHEJ lower than haploid cells expressing either MATa or MATα (Astrom et al. 1999; Lee et al. 1999; Clikeman et al. 2001). Deletions of the SIR2, SIR3, and SIR4 genes, which are involved in transcriptional silencing, have been reported to reduce NHEJ (Tsukamoto et al. 1997). This effect is largely attributable to the derepression of the silent mating-type loci HMLα and HMRa, resulting in the coexpression of a and α information in haploid cells (Astrom et al. 1999; Lee et al. 1999). To date, the genes controlled by mating type that carry out this NHEJ regulation are unknown.
To dissect the mechanism of end-joining, we set out to isolate proteins interacting with Lif1p in a two-hybrid assay in S. cerevisiae. We identified Lif2p as a new Lif1p-interacting factor and showed that Lif2p is required for DSB repair by end-joining. Lif2p expression is specifically reduced in MATa/MATα cells and its overexpression restores higher NHEJ efficiency.
Results
Isolation of LIF2
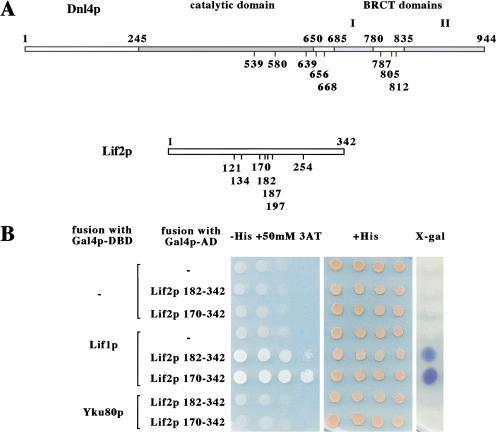
To investigate the function of the Lif1p–Dnl4p complex, we used the yeast two-hybrid system to search for yeast proteins that interact with Lif1p. Plasmid pGBT9-LIF1, encoding full-length Lif1p fused to the DNA-binding domain of Gal4p (Gal4p–DBD) was used for screening a library of random genomic fragments fused to the Gal4p-activation domain (Gal4p–AD). Eight different positive clones were isolated with plasmids encoding Gal4p–AD fused in-frame with the DNL4 gene (Fig. 1A). Clones start from amino acid 539 to 812 and extend over the carboxy-terminal end of Dnl4p, all encompassing the entire second BRCT domain. This result is in agreement with the previously described interaction between the BRCT domains of Dnl4p and Lif1p (Herrmann et al. 1998; Teo and Jackson 2000).
Figure 1.
Identification of LIF2 by a two-hybrid screen. (A) Schematic representation of Dnl4p and Lif2p with the positions of the carboxy-terminal fusions interacting with Lif1p. The boundaries of the catalytic and BRCT domains of Dnl4p are reproduced from Schar et al. (1997). (B) Specificity of the Lif1p–Lif2p interaction in the two-hybrid assay. Two Lif2p–Gal4p–AD fusions isolated in the screen were reintroduced into the Y190 two-hybrid strain in the presence of the Gal4p–DBD alone or fused to Lif1p or Yku80p. The two-hybrid interaction is revealed by growth on plates lacking histidine complemented with 50 mM 3-amino-triazole, and by X-gal staining.
In addition, seven different plasmids encoding Gal4p–AD fused in-frame with the YLR265c ORF were isolated (Fig. 1A). Hereafter, YLR265c is designated as LIF2 , encoding a 342 amino acid protein. The fusion domains of the different clones start at amino acids ranging from 121 to 254 and extend over the carboxy-terminal end of Lif2p. The two-hybrid interaction between Lif1p and Lif2p was reproduced after reintroduction of fusion protein encoding plasmids into yeast (Fig. 1B). These results suggest that the carboxy-terminal part of Lif2p specifically interacts with Lif1p. They also suggest a potential role of Lif2p in end-joining.
Lif2p is essential for NHEJ
To examine whether LIF2 could play a role in end-joining, we created null strains by gene disruption. lif2Δ mutant strains were viable and displayed no obvious growth defect at 30 and 37°C (data not shown). We analyzed NHEJ efficiency in lif2Δ mutants using two DSB-inducer systems in two distinct genetic backgrounds. The HO system was developed by Lee and colleagues in a DBY745 background (Lee et al. 1998). Under conditions of mating-type switching in yeast, HO endonuclease physiologically creates a DSB at the mating type (MAT) locus. The break is normally repaired by homologous recombination with one of the two silent mating-type loci, HML and HMR. Here, we used haploid strains lacking HML and HMR, in which the DSB can only be repaired by NHEJ. The gene encoding HO was placed under the control of a galactose regulatable promoter. Addition of galactose to the medium induces a single DSB at MAT (Moore and Haber 1996; Lee et al. 1998). The I-SceI system was developed in a W303 genetic background. A restriction site for the I-SceI endonuclease was introduced upstream of URA3 on chromosome V and the cells were transformed with a plasmid expressing I-SceI under the control of a galactose regulatable promoter. Galactose addition to the medium induces a single DSB, which should only be repaired by end-joining.
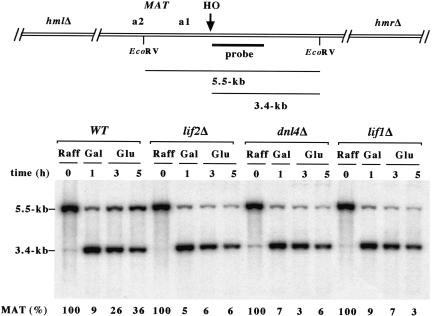
First, we examined religation efficiency of an HO cut by Southern blot analysis in wild-type, dln4Δ, lif1Δ, and lif2Δ cells. Cells were induced to express HO for 1 h. Galactose was then replaced by glucose to turn off the expression of the endonuclease. To allow for an unbiased quantification of the repair products, cells were maintained blocked in late G1 phase with α-factor during the whole experiment. In wild-type cells, the amount of intact DNA molecules dropped after HO induction and then increased after glucose repression of the endonuclease (Fig. 2). After 4 h in glucose containing medium, the DSB has been repaired by religation in about one-third of the cells. In cells lacking LIF2, as in cells lacking DNL4 and LIF1, the reformation of intact DNA molecules was not observed. In addition, cell viability at the end of the experiment has dropped to ∼5% in dnl4Δ, lif1Δ, and lif2Δ cells from ∼45% in wild-type cells (data not shown). Thus, like Lif1p and Dnl4p, Lif2p is required for the religation of a DSB by end-joining.
Figure 2.
LIF2 is required for the religation of an HO cut at MAT in the absence of a homologous sequence. Yeast strains Lev348 (WT), Lev406 (lif2Δ), Lev349 (dnl4Δ), and Lev379 (lif1Δ) grown exponentially in raffinose-containing medium were blocked in late G1 phase by the addition of α factor (Sigma, 60 ng/ml) for 4 h and exposed to galactose for an additional 1 h. Cells were resuspended in glucose-containing medium and maintained in G1 phase with α factor for an additional 4 h. Genomic DNAs were digested with EcoRV, separated by electrophoresis on a 1% agarose gel, and blotted onto a nitrocellulose membrane. The membrane was probed with a 700-bp fragment immediately adjacent to the HO site. After exposure, the membrane was rehybridized with a TRP1 fragment (data not shown). The signal from the uncut/repaired MAT band at 5.5 kb was quantified with Image-Quant using the signal at TRP1 as an internal control for each lane and normalized for each strain.
When HO and I-SceI endonucleases are expressed continuously, complementary ends cannot simply religate, as the restored site will be cut again. When the DSB occurs on an essential chromosome, this continuous cleavage leads to cell death (Fairhead and Dujon 1993; Moore and Haber 1996). Only in cells competent for NHEJ short-range mutations altering the endonuclease cleavage site can occur, giving rise to survivors (Moore and Haber 1996; see below). The frequency of survival on galactose plates can therefore be used to examine the role of LIF2 in NHEJ. In wild-type haploid cells, we found that cell viability on galactose was at ∼1 × 10−3 and ∼2 × 10−3 with HO and I-SceI, respectively (Table 1, line 1).1 In dnl4Δ, lif1Δ,and lif2Δ strains, cell viability fell by two orders of magnitude indicating that these particular religation events are abolished to the same level in all three mutants (Table 1, lines 2–4). After HO induction, cell viability was not reduced further in a lif1Δ lif2Δ double-mutant strain, indicating that LIF1 and LIF2 act in the same pathway (Table 1, lines 3–5). To ensure specificity of the lif2 disruption effect, we analyzed the rescue by a centromeric plasmid encoding Lif2p (pCEN–LIF2). This low copy number plasmid restored wild-type viability in lif2Δ cells when HO and I-SceI were expressed continuously (Table 1, line 6). These results confirm the specific requirement for Lif2p in NHEJ.
Table 1.
Survival frequency in strains with a continuously induced DSB
| Relevant genotype
|
Survival (×106)a with HO
|
Survival (×106)a with I-SceI
|
|---|---|---|
| Haploid MATa strains | ||
| 1 wild type | 970 ± 140 | 1700 ± 610 |
| 2 dn14Δ | 10 ± 7 | 14 ± 8 |
| 3 lif1Δ | 8 ± 6 | n.d. |
| 4 lif2Δ | 6 ± 3 | 7 ± 5 |
| 5 lif1Δ lif2Δ | 14 ± 9 | n.d. |
| 6 lif2Δ pCEN-LIF2 | 960 ± 150 | 2600 ± 680 |
| 7 wild-type p2μ-LIF2 | 990 ± 160 | 2600 ± 400 |
| Haploid MATa/MATα strains | ||
| 8 wild type | n.d. | 30 ± 20 |
| 9 wild type p2μ-LIF2 | n.d. | 190 ± 70 |
| 10 dnl4Δ | n.d. | 11 ± 3 |
| 11 dnl4Δ p2μ-LIF2 | n.d. | 38 ± 26 |
| Diploid MATa/MATα strains | ||
| 12 wild type | 150 ± 50 | n.d. |
| 13 wild type p2μ-LIF2 | 870 ± 140 | n.d. |
| 14 dnl4Δ/dnl4Δ | <1 | n.d. |
| 15 dnl4Δ/dnl4Δ p2μ-LIF2 | <2 | n.d. |
All haploid strains expressing HO were isogenic to Lev 348. Diploid strains were obtained by crossing Lev 348 × Lev 360 (wild type) and Lev 349 × Lev 363 (dn14Δ/dn14Δ). All strains expressing I-SceI were isogenic to Lev 338. (n.d.) Not determined.
The mean and standard deviation were calculated by averaging three samples.
Finally, we performed plasmid rejoining assays (Boulton and Jackson 1996) and compared lif2Δ mutant with isogenic dnl4Δ mutant. The yeast plasmid pRS313 was linearized by the restricition endonuclease EcoRI in a region without sequence homology to chromosomal DNA. Competent cells were then transformed in parallel with limiting amounts of cut and uncut plasmid DNA, and the number of transformants was determined after selection for a plasmid-expressed genetic marker (HIS3). The relative transformation efficiency obtained with linear versus circular plasmid DNA was used as a measure of end-joining. From three independent experiments, the relative transformation efficiency in wild-type cells was estimated at 28 ± 7%. In dnl4Δ and lif2Δ cells, this level was two orders of magnitude lower to 0.24 ± 0.04% and 0.23 ± 0.05%, respectively. This result confirms that LIF2 is required for NHEJ to the same extent as DNL4.
Lif2p expression is down-regulated in MATa/MATα cells
A systematic analysis by DNA microarray indicates that YLR265c/LIF2 mRNA steady-state level is repressed in MATa/MATα cells compared with MATa or MATα cells (Galitski et al. 1999). The MAT loci encode a1 and α2, which dimerize to form a transcriptional repressor. The a1/α2 repressor recognizes a consensus sequence (ACTACATNNANWNNTGTRC) present in the promoter region of haploid-specific genes (Goutte and Johnson 1988). In the LIF2 gene, a perfect match of this consensus is located 130 bp upstream of the putative start codon. These observations prompted us to examine directly whether Lif2p dosage is affected by mating type.
By Western blot, we compared epitope-tagged protein steady-state levels in MATa haploid cells, in MATa haploid cells harboring an integrated copy of MATα and in MATa/MATα diploid cells. As shown in Figure 3, the amount of Lif2p fell dramatically in MATa/MATα cells compared with MATa cells by at least 25-fold. Among the other proteins required for NHEJ that were tested, only Lif1p displayed a very modest repression in MATa/MATα cells. The levels of Dnl4p, Yku70p, and Yku80p were unaffected by mating type and slightly reduced in diploid cells, as expected, because only one of the two gene copies was tagged.
Figure 3.
Lif2p is down-regulated in MATa/MATα cells. Haploid MATa strains Lev412 (Lif2pmyc), Lev413 (Dnl4pmyc), Lev368 (Lif1pmyc), Lev359 (Yku70pmyc), and Lev355 (Yku80pmyc) were transformed with an integrative plasmid bearing MATα (sp427) or crossed with MATα strain Lev360. Total proteins were extracted with TCA and 30 μg were analyzed by Western blotting using monoclonal 9E10 anti-myc antibody. All tagged alleles behaved as wild type when tested for NHEJ (data not shown). (h) Haploid; (d) diploid.
The striking and specific regulation of Lif2p level by mating type correlates with the known repression of NHEJ in MATa/MATα cells. Could Lif2p overexpression bypass this regulation?
Higher dosage of LIF2 suppresses NHEJ defect in MATa/MATα cells
First,we quantified the repression of NHEJ by MAT heterozygosity in the two DSB inducer systems used in this study. On continuous expression of HO, MATa/MATα diploid strain containing a cleavable site on each homolog displayed an ∼sixfold reduction in cell viability relative to a MATa haploid strain (Table 1, line 12). On continuous expression of I-SceI, the integration of a copy of MATα in MATa haploid cells caused a ∼60-fold reduction in cell viability (Table 1, line 8). To increase the dosage of LIF2, cells were transformed with a 2μ-based plasmid containing the LIF2 sequence under the control of its own promoter (p2μ-LIF2). The plasmid was maintained at ∼20 copies per cell (data not shown). On continuous induction of HO and I-SceI, higher dosage of LIF2 increased cell viability by approximately sixfold in MATa/MATα cells (Table 1, lines 9 and 13). An improvement of survival by a higher dosage of LIF2 was not observed in MATa/MATα cells lacking DNL4 (Table 1, lines 10–11 and 14–15) and in wild-type MATa haploid cells (Table 1, line 7). Thus, even though LIF2 on the plasmid is still subject to a/α repression, its increased dosage was able to at least partially suppress the NHEJ repression by MAT heterozygosity.
Processing associated with NHEJ following I-SceI cleavage
The repair events obtained when I-SceI was expressed continuously were characterized by sequencing the region around the cleavage site in the survivors. In all analyzed cells, changes were limited to a few nucleotides in the sequence recognized by the endonuclease. As shown in Figure 4, in NHEJ-competent MATa haploid cells displaying a higher viability (wild-type, lif2Δ pCEN–LIF2 and wild-type p2μ-LIF2), almost all mutations were small deletions of 1 to 9 bases within the restriction site. The insertion of one or two bases was also observed. In NHEJ-deficient MATa haploid cells (dnl4Δ and lif2Δ), the site in most cases was either intact or affected by a single base-pair substitution. An intact site indicates that the event responsible for survival occurred elsewhere (e.g., on the endonuclease expressing plasmid) prior to plating on galactose. Base-pair substitutions at the cleavage site could also have pre-existed or be particular products of attempts at repairing the break (e.g., polymerase error during a gene conversion repair event using information from the uncleaved sister chromatid; Strathern et al. 1995).
Figure 4.
Modifications of the I-SceI site arising in survivors. The 600 base pairs surrounding the site were amplified by PCR and sequenced. For each context, 8 to 10 independent clones were analyzed. The original sequence with the 18-bp site recognized by I-SceI is represented. The arrow indicates the position of cleavage. The deletions are drawn in blue, the insertion in red, and the base substitution in yellow.
These data indicate that repair events associated with limited sequence loss at the DSB site are specific products of the NHEJ pathway. We found that in MATa/MATα wild-type cells, small deletions within the I-SceI site were not present, and in this regard these cells behaved as NHEJ-deficient mutants (Fig. 4). Higher dosage of LIF2 restored the occurrence of small deletions in MATa/MATα cells. This was not observed in MATa/MATα cells lacking DNL4, indicating that the restoration of small deletions was a product of NHEJ.
Discussion
Position of LIF2 in the NHEJ pathway
Lif2p is a previously uncharacterized protein of S. cerevisiae that we identified through its two-hybrid interaction with Lif1p, a protein associated to the NHEJ ligase Dnl4p. The LIF2 gene is essential to the precise rejoining of a DSB induced by a transient expression of the HO endonuclease in the absence of homologous template. LIF2 is also required for the imprecise end-joining events observed with continuous cleavage of the DNA by HO and I-SceI endonucleases, and for the rejoining of a plasmid linearized with an endonuclease prior to transformation into yeast cells. The end-joining defects observed in cells lacking LIF2 are undistinguishable from the ones observed in cells lacking DNL4 or LIF1. We conclude that LIF2 is required for DSB repair by NHEJ.
In budding yeast, three distinct gene groups are involved to the same degree in the NHEJ pathway as follows: DNL4 and LIF1; YKU70 and YKU80; and RAD50, MRE11, and XRS2 (Boulton and Jackson 1996, 1998; Milne et al. 1996; Tsukamoto et al. 1996; Herrmann et al. 1998). In addition, the last two groups are required for a different function at telomeres and the loss of one of their genes results in telomere shortening (Boulton and Jackson 1996, 1998). YKU70 and YKU80 are involved in telomere protection and RAD50, MRE11, and XRS2 are required for wild-type telomerase-dependent telomere elongation (Gravel et al. 1998; Nugent et al. 1998; Marcand et al. 1999; Peterson et al. 2001). Behaving as dnl4 and lif1 mutants (Teo and Jackson 1997; Herrmann et al. 1998), cells lacking LIF2 displayed normal telomere length (data not shown). These data, in addition to the complete loss of Dnl4p-dependent end-joining observed in lif2Δ cells, and the interaction of Lif1p with Dnl4p and Lif2p in a two-hybrid system, indicate that LIF2 belongs to the DNL4–LIF1 group.
Database searching failed to reveal any striking similarity between Lif2p and other known protein sequences, and our data do not address its biochemical function. We were unsuccessful at demonstrating the Lif1p–Lif2p interaction biochemically by coimmunoprecipitation of epitope-tagged proteins expressed at endogenous levels. Interestingly, double-tagged Lif2p-myc13 Dnl4p-HA4 strains were partially defective in NHEJ when single-tagged strains were not (data not shown). This negative genetic interaction could be interpreted as an evidence for a Lif2p–Dnl4p complex important to NHEJ and disrupted by the two epitope tags. In conclusion, although strongly suggested by genetic evidence, the physical interaction between Lif2p and the Lif1p–Dnl4p complex remains to be unambiguously proven.
Lif2p may fulfill a role in association with Lif1p–Dnl4p, possibly acting as a mediator between DNA ends and ligation at DSBs. In vertebrates, the crystal structure of XRCC4, the Lif1p homolog, shows a strikingly elongated dumbell-like tetramer that can bind Ligase IV and DNA simultaneously (Junop et al. 2000). In addition, the KU heterodimer is required for targeting Lif1p–Dnl4p to DNA ends in vivo (Teo and Jackson 2000). A protein-like Lif2p could be involved in the recruitment of these proteins at DSBs, or in the alteration in chromatin structure to permit DNA access. Future experiments are needed to elaborate a model of Lif1p binding to Dnl4p, Lif2p, and DNA and the predicted assembly of these components in NHEJ.
A mechanism for NHEJ regulation by mating type
Here we present evidence that Lif2p is a target of NHEJ regulation by mating type. First, Lif2p steady-state level was strongly repressed by MAT heterozygosity when other factors involved in the NHEJ pathway were unaffected. Second, Lif2p overexpression restores higher repair efficiency in MATa/MATα cells. DSB repair in these conditions was bona fide NHEJ, as it required DNL4 and the molecular events associated with repair were indistinguishable from those observed in MATa wild-type cells.
The inhibition of Lif2p activity by mating type provides a molecular mechanism for the regulation of the channeling of DNA ends into either homologous or nonhomologous repair pathways. In nature, cells expressing a or α mating type would almost always be haploid and a/α cells would be diploid. In haploid cells, NHEJ would be the only efficient pathway to repair a DSB occurring prior to DNA replication. In diploid cells, error-free recombination using the homologous chromosome would be preferred and it should be advantageous to repress the alternative pathway.
In agreement with this model, DNA repair by homologous recombination is enhanced by mating type heterozygosity (Heude and Fabre 1993; Fasullo et al. 1999; Lee et al. 1999; Clikeman et al. 2001). In diploid cells lacking YKU70, DSB-induced allelic recombination is still reduced in a/a compared with a/α cells, suggesting that homologous recombination regulation by mating type is not entirely due to the repression of the competing NHEJ pathway (Clikeman et al. 2001). Identification of other repair proteins submitted to a/α regulation should further increase our understanding of how the choice between pathways is controlled.
The efficiency of the essential but unknown function performed by Lif2p in NHEJ seems particularly sensitive to the dosage of the protein. This would explain why this protein, and not Dnl4p or Lif1p, is a regulatory target. Homologs of Lif2p in other organisms remain to be identified. In theory, they too could be the targets of different forms of regulation. For instance, in the fission yeast S. pombe, end-joining between defective telomeres was observed in cells arrested in an uncommitted G1 state, but not in cycling cells (Godhino Ferreira and Cooper 2001). It will be interesting to determine whether this regulation of NHEJ during the cell cycle involves a Lif2p homolog.
Material and methods
Strains
The yeast strains used in this study are listed in Table 2. Gene disruption and tagging used to generate these strains were done according to Longtine et al. (1998). Strain Lev348 results from the transformation of JKM139 with the linearized plasmid CY807+TRP1 (bar1∷TRP1; Aparicio and Gottschling 1994). Strain Lev360 results from the transformation of JKM179 with the linearized plasmid sp59 (Marcand et al. 1997). Strain Lev338 results from the transformation of Lev306 with the linearized plasmid sp392. Strains Lev414 and Lev415 result from the transformation of Lev338 and Lev339, respectively, with the linearized plasmid sp433.
Table 2.
Yeast strains
| Strain
|
Genotype
|
|---|---|
| Y190 | MATa gal4 gal80 his3 trp1-901 ade-101 ura3-52 leu2-3,112 URA3::GAL1-lacZ LYS2::GAL4(UAS)-HIS3 (Harper et al. 1993) |
| JKM139 | MATa hmrΔ::ADE1 hmlΔ::ADE1 ade1-100 leu2-3,112 lys5 trp1::hisG ura3-52 ade3::GAL-HO (Lee et al. 1998) |
| JKM179 | MATα hmrΔ::ADE1 hmlΔ::ADE1 ade1-100 leu2-3,112 lys5 trp1::hisG ura3-52 ade3::GAL-HO (Lee et al. 1998) |
| Lev 348 | JKM139 bar1::TRP1 |
| Lev 349 | JKM139 bar1::TRP1 dnl4Δ::KAN |
| Lev 379 | JKM139 bar1::TRP1 lif1Δ::KAN |
| Lev 406 | JKM139 bar1::TRP1 lif2Δ::KAN |
| Lev 411 | JKM139 bar1::TRP1 lif1Δ::klURA3 lif2Δ::KAN |
| Lev 360 | JKM179 adh4::URA3::TEL |
| Lev 363 | JKM179 adh4::URA3-TEL dnl4Δ::KAN |
| Lev 355 | JKM139 bar1::TRP1 YKU80-13myc::KAN |
| Lev 359 | JKM139 bar1::TRP1 YKU70-13myc::KAN |
| Lev 368 | JKM139 bar1::TRP1 LIF1-13myc::KAN |
| Lev 412 | JKM139 bar1::TRP1 LIF2-13myc::KAN |
| Lev 413 | JKM139 bar1::TRP1 DNL4-13myc::KAN |
| W303-1a | MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 can1-100 rad5-535 |
| Lev 306 | W303-1a cdc5ad |
| Lev 338 | Lev 306 GEA2::I-SceI::URA3 |
| Lev 339 | Lev 306 GEA2::I-SceI::URA3 dnl4::KAN |
| Lev 408 | Lev 306 GEA2::I-SceI::URA3 lif2Δ::KAN |
| Lev 414 | Lev 306 GEA2::I-SceI::URA3 TRP1::MATα |
| Lev 415 | Lev 306 GEA2::I-SceI::URA3 dnl4Δ::KAN TRP1::MATα |
Plasmids
The region upstream of URA3 was amplified by PCR and inserted into sp59 (Marcand et al. 1997) cut with HindIII and SalI, creating plasmid sp391 with a BglII site adjacent to the HindIII site. An I-SceI site was inserted upstream of URA3 sequences at the BglII site of plasmid sp391 using oligonucleotides Isce 1 (5′-GATCCTAGGGATAACAGGGTAATA-3′) and Isce 2 (5′-GATCTATTACCCTGTTATCCCTAG-3′), creating plasmid sp392 with a unique BglII site distal of URA3. Plasmid sp392 was linearized with SalI and BamHI prior to transformation into yeast. Plasmid p373 (pGAL–I-SceI) was created by inserting a BamHI fragment from pPEX7 (Fairhead and Dujon 1993) into p413GAL1 (CEN, HIS3).
The LIF2 gene was amplified by PCR (including 360 bp upstream of start codon and 330 bp downstream of the stop codon) and inserted into pRS315 (CEN, LEU2) and pRS425 (2μ,LEU2) creating plasmids sp421 (pCEN–LIF2) and sp431 (p2μ–LIF2), respectively.
A 4-kb fragment encompassing MATα was amplified by PCR and inserted into pRS404 (integrative, TRP1) and pRS406 (integrative, URA3), creating plasmids sp433 and sp427, respectively. Prior to transformation into yeast, sp433 and sp427 were linearized with Bsu36I and StuI, respectively.
Two-hybrid screen
The screening was done according to Flores et al. (1999). The bait construct contained the complete ORF of LIF1 cloned in pGBT9. About 7 × 106 transformed cells were selected on plates containing 50 mM 3-amino-triazole.
Cell survival
Cells were grown exponentially in raffinose containing medium, diluted in water and spread at appropriate dilutions on glucose and galactose containing plates. After 3 d at 30°C, colonies were counted. The survival rates were calculated by dividing the number of colonies on galactose by the number of colonies on glucose.
Acknowledgments
We thank James Haber and Cécile Fairhead for strains and plasmids; Michel Werner and Claire Boschiero for help with the two-hybrid screen; and Cécile Haumaitre, Bruno Fievet and Thomas Germe for their participation at the start of this work. We thank Carl Mann for continuous support, Francis Fabre for encouragement, and Serge Gangloff, Julie Smith, and Marie-Claude Marsolier for advice and critical reading of the manuscript. This work is supported by the Association pour la Recherche sur le Cancer and by Electricité de France.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL marcand@jonas.saclay.cea.fr; FAX 33-1-69-08-47-12.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.206801.
References
- Aparicio OM, Gottschling DE. Overcoming telomeric silencing: A trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes & Dev. 1994;8:1133–1146. doi: 10.1101/gad.8.10.1133. [DOI] [PubMed] [Google Scholar]
- Astrom SU, Okamura SM, Rine J. Yeast cell-type regulation of DNA repair. Nature. 1999;397:310. doi: 10.1038/16833. [DOI] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. Identification of a Saccharomyces cerevisiae Ku80 homologue: Roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clikeman J, Khalsa G, Barton S, Nickoloff J. Homologous recombinational repair of double-strand breaks in yeast is enhanced by MAT heterozygosity through yKU-dependent and -independent mechanisms. Genetics. 2001;157:579–589. doi: 10.1093/genetics/157.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhead C, Dujon B. Consequences of unique double-stranded breaks in yeast chromosomes: Death or homozygosis. Mol Gen Genet. 1993;240:170–178. doi: 10.1007/BF00277054. [DOI] [PubMed] [Google Scholar]
- Fasullo M, Bennett T, Dave P. Expression of Saccharomyces cerevisiae MATa and MATα enhances the HO endonuclease-stimulation of chromosomal rearrangements directed by his3 recombinational substrates. Mutat Res. 1999;433:33–44. doi: 10.1016/s0921-8777(98)00059-7. [DOI] [PubMed] [Google Scholar]
- Flores A, Briand JF, Gadal O, Andrau JC, Rubbi L, Van Mullem V, Boschiero C, Goussot M, Marck C, Carles C, et al. A protein-protein interaction map of yeast RNA polymerase III. Proc Natl Acad Sci. 1999;96:7815–7820. doi: 10.1073/pnas.96.14.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng HL, Davidson L, Kangaloo L, Alt FW. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- Galitski T, Saldanha AJ, Styles CA, Lander ES, Fink GR. Ploidy regulation of gene expression. Science. 1999;285:251–254. doi: 10.1126/science.285.5425.251. [DOI] [PubMed] [Google Scholar]
- Godhino Ferreira M, Cooper JP. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol Cell. 2001;7:55–63. doi: 10.1016/s1097-2765(01)00154-x. [DOI] [PubMed] [Google Scholar]
- Goutte C, Johnson AD. a1 protein alters the DNA binding specificity of α 2 repressor. Cell. 1988;52:875–882. doi: 10.1016/0092-8674(88)90429-1. [DOI] [PubMed] [Google Scholar]
- Gravel S, Larrivee M, Labrecque P, Wellinger RJ. Yeast Ku as a regulator of chromosomal DNA end structure. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- Harfst E, Cooper S, Neubauer S, Distel L, Grawunder U. Normal V(D)J recombination in cells from patients with Nijmegen breakage syndrome. Mol Immunol. 2000;37:915–929. doi: 10.1016/s0161-5890(01)00008-6. [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin- dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Herrmann G, Lindahl T, Schar P. Saccharomyces cerevisiae LIF1: A function involved in DNA double-strand break repair related to mammalian XRCC4. EMBO J. 1998;17:4188–4198. doi: 10.1093/emboj/17.14.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heude M, Fabre F. a/α-control of DNA repair in the yeast Saccharomyces cerevisiae: Genetic and physiological aspects. Genetics. 1993;133:489–498. doi: 10.1093/genetics/133.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junop MS, Modesti M, Guarne A, Ghirlando R, Gellert M, Yang W. Crystal structure of the Xrcc4 DNA repair protein and implications for end joining. EMBO J. 2000;19:5962–5970. doi: 10.1093/emboj/19.22.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Moore A, Holmes JK, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Lee SE, Paques F, Sylvan J, Haber JE. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr Biol. 1999;9:767–770. doi: 10.1016/s0960-9822(99)80339-x. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Manolis KG, Nimmo ER, Hartsuiker E, Carr AM, Jeggo PA, Allshire RC. Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J. 2001;20:210–221. doi: 10.1093/emboj/20.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Gilson E, Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- Marcand S, Brevet V, Gilson E. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 1999;18:3509–3519. doi: 10.1093/emboj/18.12.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne GT, Jin S, Shannon KB, Weaver DT. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4189–4198. doi: 10.1128/mcb.16.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Bosco G, Ross LO, Evans SK, Salinger AP, Moore JK, Haber JE, Lundblad V. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet. 2001;27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- Schar P, Herrmann G, Daly G, Lindahl T. A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes & Dev. 1997;11:1912–1924. doi: 10.1101/gad.11.15.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern JN, Shafer BK, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo SH, Jackson SP. Identification of Saccharomyces cerevisiae DNA ligase IV: Involvement in DNA double-strand break repair. EMBO J. 1997;16:4788–4795. doi: 10.1093/emboj/16.15.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Lif1p targets the DNA ligase Lig4p to sites of DNA double-strand breaks. Curr Biol. 2000;10:165–168. doi: 10.1016/s0960-9822(00)00317-1. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, Kato J, Ikeda H. Hdf1, a yeast Ku-protein homologue, is involved in illegitimate recombination, but not in homologous recombination. Nucleic Acids Res. 1996;24:2067–2072. doi: 10.1093/nar/24.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature. 1997;388:900–903. doi: 10.1038/42288. [DOI] [PubMed] [Google Scholar]
- Wilson S, Warr N, Taylor DL, Watts FZ. The role of Schizosaccharomyces pombe Rad32, the Mre11 homologue, and other DNA damage response proteins in non-homologous end joining and telomere length maintenance. Nucleic Acids Res. 1999;27:2655–2661. doi: 10.1093/nar/27.13.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TE, Grawunder U, Lieber MR. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Sonoda E, Sasaki MS, Morrison C, Haraguchi T, Hiraoka Y, Yamashita YM, Yagi T, Takata M, Price C, et al. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 1999;18:6619–6629. doi: 10.1093/emboj/18.23.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]