A definitive diagnosis of CML requires either the demonstration of the t(9;22) Philadelphia chromosome translocation (by FISH), or BCR-ABL1 fusion gene (qRT-PCR). Both techniques provide important clinical information, but are usually confined to specialist regional laboratories as they are costly, require specialist staff to produce and interpret, and take several days to complete. Since most patients with neutrophilia and/or thrombocytosis do not have CML, a rapid and simple screening test for BCR-ABL1 protein would be clinically useful. Hitherto, demonstrating the BCR-ABL1 protein has been technically difficult because of its instability,1 and the lack of a specific antibody that can reliably distinguish BCR-ABL1 protein from its normal ABL counterpart.
The BCR-ABL1 Protein kit (BD Biosciences) has been developed to detect all frequently occurring BCR-ABL1 fusion variants in human blood, including p190, p210 and p230. It uses an antibody recognizing BCR attached to a capture bead and an ABL-directed phycoerythrin (PE)–conjugated detection antibody. The presence of BCR-ABL1 protein results in a sandwich complex comprised of both the capture bead and the detection fluorophore, detectable by flow cytometry.
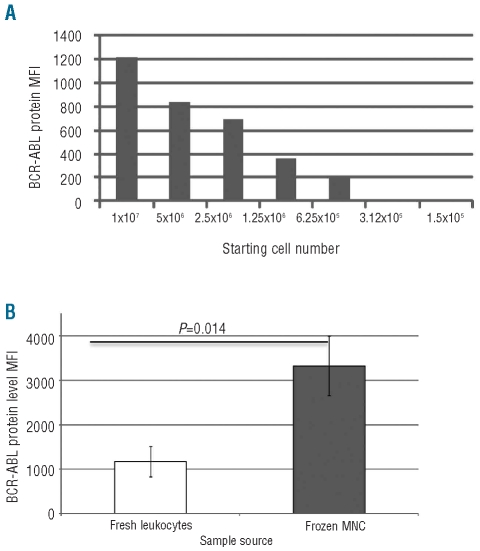
In initial optimization experiments on clinical samples, it was observed that using too few cells may result in a false negative result; thus a minimum of 1x107 cells is recommended (Figure 1A). We compared BCR-ABL1 protein detection from total leukocytes or from MNC (Figure 1B). The BCR-ABL1 MFI (mean fluorescence intensity) signal was greater in MNC than total leukocytes (P=0.014). This may be because the proteolytic activity present within mature neutrophils is reduced within the MNC cell preparation. Comparing results from fresh or frozen MNC, whereby analysis was performed immediately after thawing frozen samples, we found no significant difference in detectable BCR-ABL1 protein. However, if the thawed cells were permitted to recover overnight prior to analysis, false negative results were seen in 2 out of 5 cases tested. The difference may be due to protease activity released from neutrophil granules disrupted by cell thawing.
Figure 1.
Optimization of sample source for the detection of BCR-ABL1 protein. (A) Assessing the number of cells required to detect BCR-ABL1 protein. (B) Comparison between fresh total leukocytes and frozen MNC.
BCR-ABL1 protein is known to be particularly sensitive to degradation within primary cells,1 a feature that is not found in cell lines. It is, therefore, important to include a known BCR-ABL1 positive patient sample as a positive control.
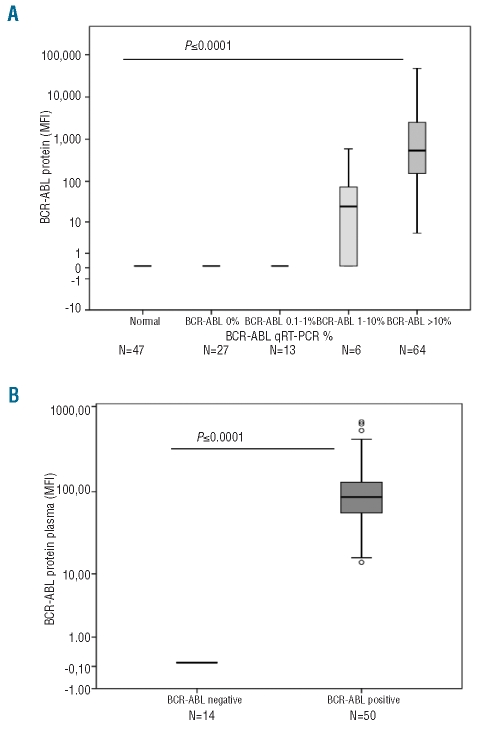
We analyzed 157 samples for both BCR-ABL1 protein and BCR-ABL1 mRNA by qRT-PCR within 24–48 h of the sample being taken. These consisted of 110 samples from patients presenting with neutrophilia and/or thrombocytosis suspected of having CML, and 47 samples from normal healthy controls the data concerning which were kept anonymous. In all samples, the BCR-ABL1 status as determined by qRT-PCR was confirmed by the BCR-ABL1 protein assay with 100% concordance. The average MFI values from normal cellular samples were 87.7±5.5 giving a cut-off value between positives and negatives set at 99 (87.7±2SD) (Figure 2A).
Figure 2.
Screening clinical samples for BCR-ABL1 protein. (A) 157 cellular samples were screened for the presence of BCR-ABL1 protein; results are stratified by the BCR-ABL qRT-PCR results for the same sample. The MFI for each sample was determined as the observed MFI, minus the MFI value of the negative control plus two standard deviations. (B) 64 plasma samples were screened for the presence of the BCR-ABL1 protein. Fourteen from patients with neutrophilia and/or thrombocytosis or healthy controls normal plasma and 50 CML patients at diagnosis were studied.
Assay specificity and sensitivity were confirmed in primary cells by assaying for BCR-ABL1 protein in patients who had commenced treatment with a TKI. In patients who were in complete cytogenetic response (CCR, defined either by standard metaphase analysis on bone marrow or as a BCR-ABL1 qRT-PCR <1%)2 the detectable BCR-ABL1 protein was either extremely low or undetectable. However, in patients at early stages of treatment but not yet in CCR (defined as a BCR-ABL1 ratio of 1–10%) BCR-ABL1 protein was readily detected (Figure 2A). In patients with newly diagnosed CML (BCR-ABL1 ratio >10%) BCR-ABL1 protein was extremely high (P=<0.0001).
The degree of decrease in BCR-ABL1 transcript level within the first three months of imatinib treatment can predict the subsequent clinical outcome.3 To assess if a change in BCR-ABL1 protein level could also be predictive of later clinical outcome, fresh MNC taken at original diagnosis were cultured in the presence of imatinib for 24 h. The change in BCR-ABL1 protein level did not correlate with achievement of CCR. Furthermore, monitoring of the BCR-ABL1 protein level once treatment had commenced provided no useful clinical information (data not shown).
Use of any screening kit in a clinical diagnostic setting must be able to reliably detect all BCR-ABL1 transcript types. BCR-ABL1 protein was detected in all transcript types tested (e1a2, e13a2, e14a2 and both e13a2 and e14a2) with no significant differences in signal intensity between BCR-ABL1 transcript types (data not shown).
The BCR-ABL1 protein assay produces a test result from lysed cellular material in 4 h. However, access to CML primary cells as positive controls are required in order to eliminate false negative reporting, and these samples may not be readily available outside specialist laboratories. To deal with this problem, we have simplified the methodology by using a plasma based assay which eliminates confounding cellular granulocytic degradation effects. It has previously been shown that leukemic cells shed their proteins, DNA and RNA into plasma.4–6 Plasma was incubated with the capture beads and detector reagent for 6 h then analyzed for the presence of BCR-ABL1 protein.
Plasma samples were analyzed from 50 newly diagnosed CML patients and from 14 patients with neutrophilia and/or thrombocytosis or healthy controls. The normal plasma sample cut-off value was determined as 98 (72.4±2SD) MFI. In all cases, and for all transcript types tested, samples that were negative by plasma were also negative by cell lysate, and the result was always in agreement with the prevailing BCR-ABL1 mRNA level as defined by qRT-PCR (Figure 2B). These data demonstrate that the simplified plasma assay is also 100% accurate at detecting BCR-ABL1 protein. The level of BCR-ABL1 protein in a plasma sample did not correlate with the sample white blood count (data not shown).
In conclusion, when screening patients presenting with high white counts for CML, it is important to have a technique which economises on time and cost, since most patients will be negative and will not require further detailed testing. Furthermore, a rapid and reliable screening test will give rise to earlier diagnosis and clinical intervention. The present data show that this assay is sufficiently reliable to provide a first-line screening test for CML diagnosis. Applying this to a plasma sample rather than a cell lysate may provide further simplification, and this may be particularly useful in developing countries whose hospitals lack definitive facilities for the detection of BCR-ABL1.
Acknowledgments
we would like to thank the University of Liverpool Biobank staff Alison Holcroft and Joanna Middleton for their help and support during this project.
Footnotes
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Patel H, Marley SB, Gordon MY. Conventional Western blotting techniques will not reliably quantify p210BCR-ABL1 levels in CML mononuclear cells. Blood. 2007;109(3):1335. doi: 10.1182/blood-2006-10-050799. [DOI] [PubMed] [Google Scholar]
- 2.Wang L, Pearson K, Pillitteri L, Ferguson JE, Clark RE. Serial monitoring of BCR-ABL by peripheral blood real-time polymerase chain reaction predicts the marrow cytogenetic response to imatinib mesylate in chronic myeloid leukaemia. Br J Haematol. 2002;118(3):771–7. doi: 10.1046/j.1365-2141.2002.03705.x. [DOI] [PubMed] [Google Scholar]
- 3.Wang L, Pearson K, Ferguson JE, Clark RE. The early molecular response to imatinib predicts cytogenetic and clinical outcome in chronic myeloid leukaemia. Br J Haematol. 2003;120(6):990–9. doi: 10.1046/j.1365-2141.2003.04200.x. [DOI] [PubMed] [Google Scholar]
- 4.Rogers A, Joe Y, Manshouri T, Dey A, Jilani I, Giles F, et al. Relative increase in leukemia-specific DNA in peripheral blood plasma from patients with acute myeloid leukemia and myelodysplasia. Blood. 2004;103(7):2799–801. doi: 10.1182/blood-2003-06-1840. [DOI] [PubMed] [Google Scholar]
- 5.Albitar M, Do K-A, Johnson MM, Giles FJ, Jilani I, O’Brien S, et al. Free circulating soluble CD52 as a tumor marker in chronic lymphocytic leukemia and its implication in therapy with anti-CD52 antibodies. Cancer. 2004;101(5):999–1008. doi: 10.1002/cncr.20477. [DOI] [PubMed] [Google Scholar]
- 6.Jilani I, Kantarjian H, Faraji H, Gorre M, Cortes J, Ottmann O, et al. An immunological method for the detection of BCR-ABL fusion protein and monitoring its activation. Leukemia Research. 2008;32(6):936–43. doi: 10.1016/j.leukres.2007.11.023. [DOI] [PubMed] [Google Scholar]