Abstract
Loss of Dmp1, an Arf transcriptional activator, leads to spontaneous tumorigenesis in mice, causing death from various forms of cancer by two years of age. Retention and expression of the wild-type Dmp1 allele in tumors arising in Dmp1+/− mice demonstrate that Dmp1 can be haplo-insufficient for tumor suppression. The mean latency of Eμ-Myc-induced B-cell lymphomas is halved on a Dmp1−/− or Dmp1+/− genetic background. Although p53 mutations or Arf deletion normally occur in ∼50% of Eμ-Myc-induced lymphomas, Dmp1 loss obviates selection for such mutations, indicating that Dmp1 is a potent genetic modifier of the Arf–p53 pathway in vivo.
Keywords: Dmp1 transcription factor, Arf, p53, Mdm2, Myc, cancer, tumor suppression
Induction of p53-dependent transcription after genotoxic stress or oncogene activation leads to expression of genes that trigger either cell cycle arrest or apoptosis, thereby guarding against tumor formation (Levine 1997). The central role of p53 in tumor suppression is underscored by the fact that about half of human cancers have p53 mutations, whereas many of the remainder exhibit particular alterations in other genes, such as Mdm2 and Arf, which disable p53 function.
The best characterized negative regulator of p53 is Mdm2, which ubiquitinates p53 and targets its proteolytic destruction (Prives 1998; Juven-Gershon and Oren 1999). Because Mdm2 is itself a p53-inducible gene, it participates in a negative feedback loop that helps to terminate the p53 response. In turn, by antagonizing functions of Mdm2, the product of the Arf tumor suppressor gene (p19Arf in mouse and p14ARF in humans) stabilizes p53 and enhances its activity (Sherr 1998; Sharpless and DePinho 1999). Arf is induced by oncogenes such as Myc, E1A, Ras, and v-Abl, and also by overexpression of E2F transcription factors whose normal role is to control the temporal expression of genes required for DNA replication. By monitoring the strength of mitogenic signals, Arf prevents hyperproliferation by diverting incipient cancer cells to alternative fates—namely, p53-dependent growth arrest, or more dramatically, apoptosis.
Arf disruption in mice predisposes to spontaneous tumor development (Kamijo et al. 1997) and enhances the rate at which mouse strains sustaining other oncogenic lesions develop tumors (Chin et al. 1997; Holland et al. 1998; Eischen et al. 1999; Jacobs et al. 1999b; Schmitt et al. 1999). Pertinent to studies described below, more than half of the Burkitt-type B-cell lymphomas arising in Eμ-Myc transgenic mice were found to sustain p53 mutations (28%) or biallelic Arf deletions (24%), whereas others lacking overt Arf or p53 mutations overexpressed Mdm2 (16%) (Eischen et al. 1999). The 25-wk mean latency for tumor formation in Eμ-Myc transgenic mice was halved on the Arf+/− background, and 80% of the resulting lymphomas lost the wild-type Arf allele. Arf-null, Eμ-Myc transgenic animals developed more aggressive lympholeukemias and died by only 8 wk of age. Together, these results emphasize the role of the Arf–Mdm2–p53 pathway in protecting against tumor emergence. However, we also observed that some Eμ-Myc lymphomas that had deleted the Arf gene or sustained p53 mutations overexpressed Mdm2, implying that the Arf–Mdm2–p53 pathway is not strictly linear. Moreover, about a quarter of the lymphomas arising in Eμ-Myc transgenic mice exhibited no identifying genetic lesion in Arf, Mdm2, or p53, suggesting that other mechanisms may deregulate the pathway.
A series of transcription factors other than E2F and Myc govern Arf expression. These include other positively acting factors, such as Dmp1 (see below) and β-catenin/Tcf (M. Oren, pers. comm.) that bind directly to the Arf promoter to induce transcription, as well as proteins such as Bmi-1 (Jacobs et al. 1999a), Tbx-2 (Jacobs et al. 2000), Twist (Maestro et al. 1999), Jun D (Weitzman et al. 2000), and p53 itself (Kamijo et al. 1998; Stott et al. 1998), which negatively regulate Arf through as yet ill-defined mechanisms. Overexpression of Twist and Tbx-2 in human rhabdomyosarcomas and breast carcinomas, respectively, suggests that active ARF repression might contribute to tumorigenesis. Epigenetic silencing of the ARF promoter has also been observed in human cancers (Esteller et al. 2001).
Dmp1 was isolated in a two-hybrid interactive screen using cyclin D2 as bait, and it encodes a 120- to 130-kD nuclear phosphoprotein with a central DNA-binding domain containing three Myb-like repeats flanked by acidic transactivation domains at both termini (Hirai and Sherr 1996; Inoue and Sherr 1998). The human and murine orthologs share 95% amino-acid sequence identity and are completely conserved throughout their Myb-like repeats (Bodner et al. 1999). Dmp1 binds to nonameric Ets consensus sequences in DNA (CCCG[G or T]ATGT) and competes with Ets-family proteins for sites that contain the GGA core. DNA binding can be antagonized by the interaction of Dmp1 with D-type, but not other cyclins. Despite the frequency of potential Dmp1-binding sites in mammalian genomes, its binding to a single site in the proximal Arf promoter induces p19Arf-dependent cell cycle arrest (Inoue et al. 1999). Conversely, Arf expression is dampened in Dmp1-null mouse embryo fibroblasts (MEFs), which resist cellular senescence, do not rapidly accumulate p53 mutations, and are susceptible to transformation by oncogenic Ras (Inoue et al. 2000). Dmp1-null mice did not spontaneously develop cancers in their first year of life but were susceptible to carcinogen- and radiation-induced tumors, implying that by regulating Arf function, Dmp1 may have tumor-suppressing activities (Inoue et al. 2000). Here, we show that Dmp1 is a bona fide tumor suppressor gene whose loss obviates the selection for Arf deletion or p53 mutation that normally occurs during Myc-induced lymphomagenesis.
Results and Discussion
Both Dmp1−/− and Dmp1+/− mice spontaneously develop tumors
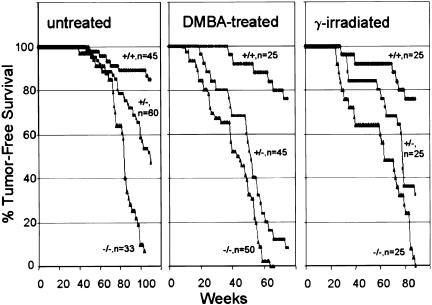
Dmp1-null mice spontaneously developed lethal tumors in their second year of life with a mean latency of 83 wk (Fig. 1). The most frequently encountered tumors were pulmonary adenomas (27% incidence) and adenocarcinomas (15%). Hepatocellular tumors (18%), B-cell lymphomas (15%), and vascular tumors, including hemangiomas (15%) and hemangiosarcomas (9%) were also relatively common. (For a detailed listing of tumor types for all cohorts, go to http://www.genesdev.org for Supplemental Materials, part A; for representative tumor histology, see Supplemental Materials, part B.) The time of appearance and variety of tumors observed in Dmp1-null animals bear no obvious relationship to those in Arf-null animals, which exhibit a different spectrum (43% sarcomas, 29% lymphomas, 17% carcinomas, 11% central nervous system tumors) and a shorter mean latency (38 wk) (Kamijo et al. 1999).
Figure 1.
Tumor-free survival in cohorts of untreated Dmp1+/+, Dmp1+/−, and Dmp1−/− mice (left) and in animals neonatally exposed to DMBA (middle) or X rays (right). The numbers of animals in each group is indicated by the corresponding survival curve.
Treating Dmp1-null mice neonatally with dimethylbenzanthracene (DMBA) or ionizing radiation accelerated tumorigenesis (Fig. 1), and many such animals developed multiple tumors of more than one histological type (Inoue et al. 2000). Lung tumors (46% adenomas and 26% adenocarcinomas) and epidermal tumors of skin (36%) predominated in DMBA-treated Dmp1-null mice. Many developed ovarian tumors mostly of granulosa cell origin (40% of females), lymphomas (14%), and melanomas (12%). Irradiated Dmp1-null mice were prone to lymphoid malignancies (32%) predominantly of T-cell origin (24%), ovarian tumors (56% of females), lung and hepatocellular tumors (each 12%).
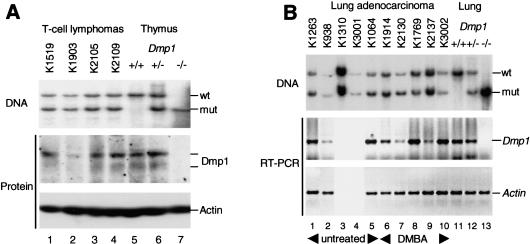
Interestingly, Dmp1+/− heterozygotes were also tumor prone (Fig. 1). A broad spectrum of tumors was seen in each of these cohorts, although the frequencies of tumor types were somewhat different from those in Dmp1-null mice (see Supplemental Materials, part A). Given the number of animals in each group, the latter differences may not be significant. However, the rates of tumor appearance in Dmp1+/− animals clearly eclipsed those of Dmp1+/+ littermate controls. These observations raised the question of whether the wild-type Dmp1 allele was lost or epigenetically suppressed, or whether Dmp1 might be haplo-insufficient for tumor suppression. The wild-type Dmp1 locus was retained in four of four T-cell lymphoblastic lymphomas from DMBA-treated Dmp1+/− mice, and Dmp1 protein was detected by immunoblotting (Fig. 2A). Lung adenocarcinoma samples from untreated and DMBA-treated Dmp1+/− mice also retained the wild-type allele and expressed Dmp1 mRNA at levels that approximated those in normal lung tissue (Fig. 2B). The relatively small differences in mean latencies of appearance of T-cell lymphomas (25 and 28 wk, respectively, for Dmp1–/− and Dmp1+/− genotypes) and lung tumors (50 and 60 wk, respectively) in DMBA-treated mice are consistent with the idea that Dmp1 is haplo-insufficient for suppression of both tumor types (see below). In two lung tumors (K938 and K2130), a reduced Southern blot signal for the wild-type Dmp1 allele correlated with a reduction in Dmp1 mRNA expression, suggesting that the normal allele was lost from some cells. Our data do not preclude such events, but argue that segregation of the wild-type allele is relatively uncommon in these tumor types.
Figure 2.
Retention and expression of the wild-type Dmp1 gene in T-cell lymphomas (A) and pulmonary adenocarcinomas (B) arising in Dmp1 heterozygotes. Tumors are designated by K numbers above the lanes. (B) (Lanes 1–5) Results with tumors obtained from untreated animals; (lanes 6–10) DMBA-treated animals. Protein was detected by direct immunoblotting of tumor cell lysates, and RNA by RT–PCR (not done for tumors K1310 and K3001 in B). Actin was used as a control for both procedures to guarantee equal loading. Results with normal tissues of the indicated genotypes appear in the right three lanes of each panel.
Because expression of the other product of the Ink4a/Arf locus (p16Ink4a) can be epigenetically silenced in lung tumors (Merlo et al. 1995; Swafford et al. 1997; Herzog et al. 1999; Patel et al. 2000), we used methylation-specific PCR assays (Herman et al. 1996) to survey the status of the Ink4a and Arf promoters (Randle et al. 2001). Importantly, lung tumors retained the Ink4a/Arf locus. Ink4a promoter methylation was detected in four of eight such samples, whereas Arf was unaffected (see Supplemental Materials, part C). Therefore, Ink4a (p16) silencing likely contributes to pulmonary carcinogenesis in the Dmp1+/− background, whereas there is no apparent selection for Arf loss.
Dmp1 loss accelerates Eμ-Myc-induced lymphomagenesis without frequent p53 mutation or Arf deletion
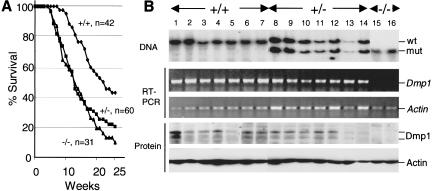
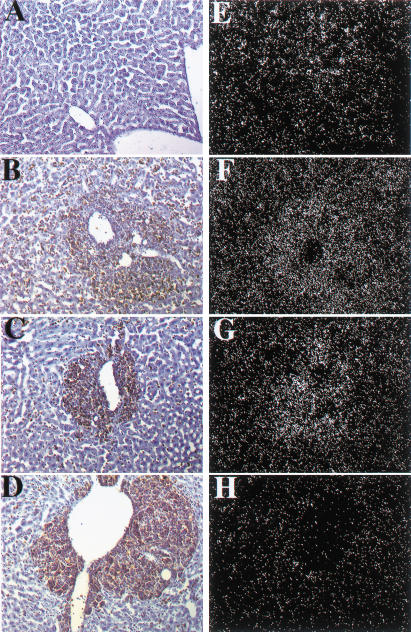
Eμ-Myc transgenic mice develop Burkitt-type B-cell tumors (Adams et al. 1985) with a mean latency of about 6 mo in our strains (Eischen et al. 1999). When crossed onto a Dmp1+/− or Dmp1−/− background, lymphomagenesis induced by the Eμ-Myc transgene was greatly accelerated (mean latency, 12 wk) with no differences between cohorts lacking one or two Dmp1 alleles (Fig. 3A). Intriguingly, the latency in the Dmp1+/− or Dmp1−/− strains mimicked that of Arf+/−, Eμ-Myc transgenic mice (Eischen et al. 1999), consistent with the idea that Dmp1 loss lowers p19Arf expression (Inoue et al. 2000). Tumors from Dmp1 heterozygotes retained and expressed the wild-type Dmp1 allele, and most contained detectable Dmp1 protein (Fig. 3B). Direct nucleotide sequencing of Dmp1 RT–PCR products from five such tumors identified no mutations in the DNA-binding domain. Gel shift assays confirmed that the residual Dmp1 protein could bind a Dmp1-specific oligonucleotide probe that does not interact with Ets family proteins (Hirai and Sherr 1996) (data not shown). In situ hybridization with an antisense (but not sense) probe confirmed that metastatic lymphomas infiltrating livers of Dmp1+/− mice (Fig. 4C,G) expressed Dmp1 mRNA, whereas background signal was seen in Dmp1-null tumors (Fig. 4D,H). These results provide strong evidence that Dmp1 is haplo-insufficient for tumor suppression.
Figure 3.
Tumor free survival (A) and Dmp1 status (B) of Eμ-Myc transgenic animals of the indicated Dmp1 genotypes. Dmp1 RNA and protein were detected by RT–PCR and immunoblotting, respectively, using actin as a control for loading.
Figure 4.
Detection of Dmp1 RNA by in situ hybridization. Liver cells from Dmp1+/+ mice (A), stained with hematoxylin (blue) express Dmp1 RNA (E). Malignant lymphoma cells in Eμ-Myc transgenic mice, visualized by antibody staining for B220 antigen (brown), surrounded central veins and invaded adjacent liver sinusoids (B–D). Regions containing metastatic B-cells in livers from both Eμ-Myc Dmp1+/+ (B) and Eμ-Myc Dmp1+/− (C) mice revealed increased hybridization signals with a Dmp1 antisense probe (F,G), respectively, relative to that in normal liver (E), consistent with continued Dmp1 expression in lymphomas from heterozygous mice. Similar results were obtained in three of three Dmp1+/− animals. Background hybridization with tissues from Eμ-Myc Dmp1−/− mice (H) was equivalent to that obtained with a control Dmp1 sense probe (data not shown).
We then evaluated the frequency of p53 mutations and Arf loss in Eμ-Myc lymphomas. The p53 gene was retained in all Eμ-Myc-induced lymphomas, regardless of their Dmp1 status (see Supplemental Materials, part D). However, both p53 and p19Arf were grossly overexpressed in 6 of 27 Dmp1+/+ tumors (22% incidence) that were suitable for analysis (see Supplemental Materials, part E). These patterns of protein expression are typical of mutant forms of p53, which neither transcriptionally activate Mdm2 to trigger p53 destruction (Haupt et al. 1997; Kubbutat et al. 1997) nor repress Arf transcription (Kamijo et al. 1998; Stott et al. 1998). Seven other Dmp1+/+ lymphomas (26%) sustained biallelic Arf deletions (see Supplemental Materials, part D). These results closely agree with those of a previous independent study in which p53 mutations and Arf deletions were observed at frequencies of 28% and 24%, respectively, in Eμ-Myc-induced lymphomas (Eischen et al. 1999).
In contrast, only 1 of 22 Eμ-Myc, Dmp1−/− lymphomas and 3 of 43 Eμ-Myc, Dmp1+/− tumors expressed mutant p53 (see Supplemental Materials, part E). The Arf gene was retained in all but one of the Eμ-Myc, Dmp1−/− tumors and in all but three of the Eμ-Myc, Dmp1+/− cases. Therefore, the combined frequencies of p53 mutation and Arf deletion in the Dmp1−/− and Dmp1+/− cohorts were 9% and 14%, respectively, versus 48% in Dmp1+/+ littermates. These results provide strong genetic evidence that loss of even a single Dmp1 allele alleviates the selection for p53 mutation and Arf loss that otherwise occurs during Eμ-Myc-induced lymphomagenesis.
Dmp1 loss did not significantly affect the patterns of Mdm2 protein expression in tumors that maintained Arf and p53 function. In those Dmp1+/+, Dmp1+/−, and Dmp1−/− Eμ-Myc-induced lymphomas that lacked evidence of p53 mutation or Arf loss, full-length Mdm2 isoforms were detected in 15%, 14%, and 20% of the respective cases (data not shown), similar to the frequency (16%) observed earlier (Eischen et al. 1999). Therefore, although Mdm2 overexpression may potentially down-regulate p53 in a subset of tumors, these effects appear to be independent of Dmp1 status.
Conclusions and implications
It is now clear that virtually all Dmp1-null mice develop cancers of many histological types in their second year of life. Despite the long latency before detection, many such tumors (e.g., pulmonary adenocarcinomas, hepatomas, lymphomas, sarcomas) were highly malignant. Dmp1 heterozygotes are also highly tumor-prone, whether exposed to carcinogens or irradiated, or left untreated. Notably, in virtually all cases where we could examine relatively pure tumor samples for Dmp1 DNA, RNA, and protein expression, we failed to obtain evidence that the wild-type Dmp1 allele was lost, silenced, or mutated. This was particularly evident in Eμ-Myc lymphomas arising in Dmp1+/− mice in which the wild-type Dmp1 allele was retained and shown, by several independent criteria, to be expressed. Although these data do not exclude the possibility that the wild-type Dmp1 allele can be lost or its expression silenced by other mechanisms, we can fairly conclude that Dmp1 is haplo-insufficient for tumor suppression. Therefore, in principle, Dmp1 loss may contribute to certain human monosomy-7 disorders (Bodner et al. 1999).
The idea that endogenous Dmp1 can physiologically control Arf first emanated from experiments with Dmp1-null MEFs, which do not rapidly up-regulate Arf upon continuous passage and mimic properties of immortalized Arf-null cells (Inoue et al. 2000). A powerful genetic argument that Dmp1 modulates Arf gene expression in living animals now stems from studies with Eμ-Myc-induced lymphomas. In cultured fibroblasts and in B-lymphocytes in culture and in vivo, Myc acts as a potent Arf inducer (Zindy et al. 1998; Eischen et al. 1999; Jacobs et al. 1999a; Schmitt et al. 1999). Selection for Arf loss attenuates Myc-induced apoptosis, facilitating immortalization of primary MEFs in culture as well as the appearance of Burkitt-type tumors in Eμ-Myc transgenic mice. About 25% of the emerging lymphomas sustain Arf deletions, whereas another 25% exhibit p53 mutations. However, in Eμ-Myc-induced Dmp1−/− and Dmp1+/− tumors, the aggregate frequencies of p53 mutation plus biallelic Arf deletion were reduced to 9% and 14%, respectively. Therefore, Dmp1 loss greatly limits disruption of the latter genes by Myc. Given the susceptibility of Dmp1+/− animals to develop tumors, one prediction is that Arf might also be found to be haplo-insufficient for tumor formation under some circumstances. Recent data indicating that p16Ink4a mutation on one chromosome (with Arf retention and expression) and monoallelic Ink4a/Arf deletion on the other can collaborate in melanomagenesis support this hypothesis (Krimpenfort et al. 2001).
These results raise several other interesting questions. First, the Arf–Mdm2–p53 signaling pathway is disrupted in the vast majority of human cancers, raising the possibility that p53 loss of function, whether partial or complete, is part of the life history of most, if not all, tumor cells. If p53 is indeed dysfunctional at some level in all cancer cells, we might well imagine two broad classes of tumors: (1) those that are disrupted (by mutation, amplification, gene silencing, etc.) in the Arf–Mdm2–p53 axis per se, and (2) those that have sustained mutations in transcription factors (or other modifiers) that modulate the activity of the Arf–Mdm2–p53 pathway. The latter class might include genes such as Dmp1, as well as the Arf-negative regulators, Twist (Maestro et al. 1999) and Tbx-2 (Jacobs et al. 2000), that are amplified and overexpressed in particular tumor types. Second, whereas human cancers lacking functional p53 are generally resistant to therapy by ionizing radiation and genotoxic drugs, those that conserve wild-type ARF, Hdm2, and p53 genes should retain the ability to respond (Schmitt et al. 1999). This can be tested using the Eμ-Myc mouse lymphoma model (Schmitt et al. 2000) and might ultimately be of prognostic significance in human cancers.
Materials and methods
Mice, genotyping, and tumor histology
Cohorts of Dmp1-null, heterozygous and wild-type mice (129SvJ × C57BL/6 background) were observed for tumor formation for 24 mo. Genotyping was performed by Southern blotting using EcoRV digestion and a 1.1 kb Eco47III genomic probe (Inoue et al. 2000). Neonatal treatment of mice with γ-irradiation or DMBA was performed as described (Kamijo et al. 1997). Dmp1-null mice were mated with Eμ-Myc transgenic mice (C57BL/6 background, kindly provided by Dr. Alan Harris [Walter & Eliza Hall Institute, Melbourne, Australia] and Dr. Charles Sidman [University of Cincinnati, OH]). F1 Dmp1+/−, Eμ-Myc transgenic mice were crossed with Dmp1+/− nontransgenic mice to create Dmp1+/+, Dmp1+/−, and Dmp1−/− Eμ-Myc transgenic animals. All mice were observed daily and sacrificed when moribund. Tumors from euthanized mice were fixed in formalin, and paraffin-embedded tissues were analyzed by light microscopy and immunohistochemistry as described previously (Inoue et al. 2000).
RNA expression
Total RNA, extracted from dissected tissues, was used as a template for cDNA synthesis, followed by PCR amplification and detection of products using murine Dmp1-specific primers. The sequence of murine Dmp1 primers were sense 5′-CTGTAGCTGAAAGAGTGGGTA-3′; antisense 5′-TGTATTATCTTCCAAGCGGGC-3′. Sequences for β-actin primers were sense 5′-GTGGGCCGCCCTAGGCACCAG-3′; antisense 5′-CTCTTTGATGTCACGCACGATTTC-3′. PCR was performed for 30 cycles for Dmp1 and 21 cycles for β-actin. Under these conditions, both Dmp1 and β-actin cDNAs were amplified in quantitative ranges.
Immunoblotting
Protein lysates were prepared from freshly isolated T-cell lymphomas, lung adenocarcinomas, or B-cell tumors from Eμ-Myc transgenic mice (Kamijo et al. 1997). Briefly, cells were sonicated 2 × 7 sec after addition of ice-cold buffer (50 mM Tris-HCl at pH 8.0, 120 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA, 1 mM PMSF, 0.4 U/mL aprotinin, 1 mM NaF, and 0.1 mM Na orthovanadate). Undissolved material was sedimented in a microcentrifuge (4°C, 15 min, 14,000 rpm). Proteins (100 μg/lane) were electrophoretically separated on polyacrylamide gels containing sodium dodecyl sulfate (SDS) and transferred onto nitrocellulose (MSI, Westboro, MA). Sites of protein binding were visualized by optimized direct immunoblotting methods (Zindy et al. 1998) with affinity-purified rabbit polyclonal antibodies to the mouse p19Arf (Quelle et al. 1995) and Dmp1 (Hirai and Sherr 1996) carboxyl termini, or with commercial antibodies to p53 (Ab-7, Calbiochem), Mdm2 (C-18, Santa Cruz Biotech), or actin (C-11, Santa Cruz Biotech).
In situ detection of Dmp1 mRNA in Eμ-Myc lymphoma cells metastatic to liver
Lymphoma-bearing mice were injected intraperitoneally with ketamine and rompun and perfused intracardially with a fixative containing 4% paraformaldehyde in 0.1 M sodium phosphate buffer (pH 7.6). Isolated livers were transferred into 25% sucrose in 0.1 M sodium phosphate buffer (pH 7.6), at 4°C for an additional 24 h. Serial sections of 12 μm-thickness cut with a cryostat were mounted on Fisher Super-frost-plus slides and stored at −20°C. To create a Dmp1-specific probe for in situ hybridization, an AvrII–Bsu36I fragment (288 bp) that had been deleted from the genome of Dmp1-null mice was cloned into the AvrII–EcoRV site of a pBluescript vector. In situ hybridization was performed as described (Zindy et al. 1997). Malignant B cells infiltrating liver were detected using a rat monoclonal antibody (1:200 dilution, 15 min) to mouse B cell CD45R/B220 (01121D, Pharmingen), biotinylated rabbit antibody (1:200 dilution, 10 min) to rat IgG (BA-4001, Vector Laboratories), and streptavidin congugated to horse radish peroxidase (K1016, DAKO) followed by a 5-min incubation with chromagen substrate (3,3′ diaminobenzidine tetrahydrochloride; K3466, DAKO). Slides were counterstained with hematoxylin, dehydrated, and coverslipped.
Acknowledgments
We thank Martine F. Roussel, Christine M. Eischen, and John D. Cleveland for helpful suggestions and insightful criticisms; Alan Harris and Charles Sidman for Eμ-Myc transgenic mice; Justine Cunningham for help with photography of in situ hybridization; and Esther van de Kamp, Cam Hornsby, Rose Mathew, and Dorothy Bush for excellent technical assistance. This work was supported in part by Cancer Center Core Grant CA21765 and by the American Lebanese Associated Charities of St. Jude Children's Research Hospital. C.J.S. is an Investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sherr@stjude.org; FAX (901) 495-2381.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.929901.
References
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Bodner SM, Naeve CW, Rakestraw KM, Jones BG, Valentine VA, Valentine MB, Luthardt FW, Willman CL, Raimondi SC, Downing JR, et al. Cloning and chromosomal localization of the gene encoding human cyclin D-binding Myb-like protein (hDMP1) Gene. 1999;229:223–238. doi: 10.1016/s0378-1119(98)00591-5. [DOI] [PubMed] [Google Scholar]
- Chin L, Pomerantz J, Polsky D, Jacobson M, Cohen C, Cardon-Cardo C, Horner JW, II, DePinho RA. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes & Dev. 1997;11:2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF–Mdm2–p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes & Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog CR, Noh S, Lantry LE, Guan KL, You M. Cdkn2a encodes functional variation of p16INK4a but not p19ARF, which confers selection in mouse lung tumorigenesis. Mol Carcinog. 1999;25:92–98. doi: 10.1002/(sici)1098-2744(199906)25:2<92::aid-mc3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Hirai H, Sherr CJ. Interaction of D-type cyclins with a novel myb-like transcription factor, DMP1. Mol Cell Biol. 1996;16:6457–6467. doi: 10.1128/mcb.16.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland EC, Hively WP, DePinho RA, Varmus HE. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes & Dev. 1998;12:3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Sherr CJ. Gene expression and cell cycle arrest mediated by transcription factor DMP1 is antagonized by D-type cyclins through a cyclin-dependent–kinase-independent mechanism. Mol Cell Biol. 1998;18:1590–1600. doi: 10.1128/mcb.18.3.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Roussel MF, Sherr CJ. Induction of ARF tumor suppressor gene expression and cell cycle arrest by transcription factor DMP1. Proc Natl Acad Sci. 1999;96:3993–3998. doi: 10.1073/pnas.96.7.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Wen R, Rehg JE, Adachi M, Cleveland JL, Roussel MF, Sherr CJ. Disruption of the ARF transcriptional activator DMP1 facilitates cell immortalization, Ras transformation, and tumorigenesis. Genes & Dev. 2000;14:1797–1809. [PMC free article] [PubMed] [Google Scholar]
- Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999a;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Jacobs JJL, Scheijen B, Vonchen J-W, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc induced apoptosis via INK4a/ARF. Genes & Dev. 1999b;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JJL, Keblusek P, Robanus-Maandag E, Kristel P, Lingbeek M, Nederlof PM, van Welsem T, van de Vijver MJ, Koh EY, Daley GQ, et al. Senescence bypass screen identified TBX2, which represses Cdkn2a (p19ARF) and is amplified in a subset of human breast cancers. Nat Genet. 2000;26:291–299. doi: 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Oren M. Mdm2: The ups and downs. Mol Med. 1999;5:71–83. [PMC free article] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo T, Bodner S, van de Kamp E, Randle DH, Sherr CJ. Tumor spectrum in ARF-deficient mice. Cancer Res. 1999;59:2217–2222. [PubMed] [Google Scholar]
- Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Loss of p16INK4aconfers susceptibility to metastatic melanoma in mice. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Maestro R, Dei Tos A, Hamamori Y, Krasnokutsky S, Sartorelli V, Kedes L, Doglioni C, Beach D, Hannon GJ. Twist is a potential oncogene that inhibits apoptosis. Genes & Dev. 1999;13:2207–2217. doi: 10.1101/gad.13.17.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumor suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- Patel AC, Anna CH, Foley JF, Stockton PS, Tyson FL, Barrett JC, Devereux TR. Hypermethylation of the p16 (Ink4a) promoter in B6C3F1 mouse primary lung adenocarcinomas and mouse lung cell lines. Carcinogenesis. 2000;21:1691–1700. doi: 10.1093/carcin/21.9.1691. [DOI] [PubMed] [Google Scholar]
- Prives C. Signaling to p53: Breaking the MDM2–p53 circuit. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Randle DH, Zindy F, Sherr CJ, Roussel MF. Differential effects of p19Arf and p16Ink4a loss on senescence of murine bone marrow-derived pre-B cells and macrophages. Proc Natl Acad Sci. 2001;98:9654–9659. doi: 10.1073/pnas.171217498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt CA, McCurrach ME, De Stanchina E, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes & Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt CA, Rosenthal CT, Lowe SW. Genetic analysis of chemoresistance in primary murine lymphomas. Nat Med. 2000;6:1029–1035. doi: 10.1038/79542. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9:22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Tumor surveillance via the ARF–p53 pathway. Genes & Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden KH, et al. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swafford DS, Middleton SK, Palmisano WA, Nikula KJ, Tesfaigzi J, Baylin SB, Herman JG, Belinsky SA. Frequent aberrant methylation of p16INK4a in primary rat lung tumors. Mol Cell Biol. 1997;17:1366–1374. doi: 10.1128/mcb.17.3.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzman JB, Fiette L, Matsuo K, Yaniv M. JunD protects cells from p53-dependent senescence and apoptosis. Mol Cell. 2000;6:1109–1119. doi: 10.1016/s1097-2765(00)00109-x. [DOI] [PubMed] [Google Scholar]
- Zindy F, Soares H, Herzog K-H, Morgan J, Sherr CJ, Roussel MF. Expression of INK4 inhibitors in cyclin D-dependent kinases during mouse brain development. Cell Growth Diff. 1997;8:1139–1150. [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes & Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]