Abstract
MicroRNAs (miRNAs) are a novel class of small noncoding RNAs that regulate gene expression by targeting mRNAs for either cleavage or translational repression. They have been shown to play important roles in a broad range of biological processes including development, cellular differentiation, proliferation and apoptosis. Conventional detection methods, such as northern blot, real-time PCR or microarray, have been used to assess miRNA expression. However, these techniques require the fixation or lysis of cells, and thus cannot be used to study the dynamic function of miRNAs in living cells. Recent remarkable advances in molecular imaging techniques have provided the capability of noninvasive repeated quantitative imaging of tumour or stem cells in living animals. The current brief discussion focuses on the reporter and fluorescent beacon imaging approaches to visualize miRNA expression in living subjects.
Keywords: MicroRNA, Molecular imaging, Reporter gene
Introduction
Mature microRNAs (miRNAs) are single-stranded, small noncoding RNAs of approximately 21 nucleotides that act as post-transcriptional regulators of gene expression in animals and plants. The biogenesis of miRNAs is initiated from primary miRNAs (pri-miRNAs) of length 1–3 kb, which are mostly transcribed from intragenic or intergenic regions by RNA polymerase II [1]. The primary transcripts undergo further processing in the nucleus, resulting in a hairpin intermediate of about 70–100 nucleotides, called pre-miRNAs [2]. The pre-miRNAs are then transported out of the nucleus to the cytoplasm and processed by another ribonuclease, Dicer, into a mature double-stranded miRNA [3]. After strand separation, the guide strand or mature single-stranded miRNA is incorporated into RNA-induced silencing complex (RISC), whereas the passenger strand (mRNA) is commonly degraded [4]. miRNAs function by complete or partial complementary base pairing with 3′ untranslated regions (UTRs) of target mRNAs, and thereby elicit mRNA degradation or translational inhibition [5]. The human genome may encode more than 1,000 miRNAs, which are estimated to regulate about 60% of mammalian genes [6–8]. A growing body of evidence suggests that miRNAs play vital regulatory roles in a vast range of biological processes, including early development, cellular differentiation, proliferation and apoptosis [9–12]. Recent studies have uncovered both tumour suppressive and oncogenic potential of a number of miRNAs [13, 14], underscoring their importance in human cancer. Consequently, intensive efforts have been made to develop miRNA-based therapy based on the rationale that restoring normal miRNA programs in the cancer cell may rewire the cell connectivity map and reverse cancer phenotypes [15].
As the miRNA field continues to evolve, efficient and reliable detection of miRNAs is an essential step towards the understanding of their roles in diverse regulatory pathways, which will certainly affect the development of miRNA-based therapies. Conventional methods including northern blot, real-time PCR and microarray have been widely used to measure the expression level of endogenous miRNAs [16–18]. Northern blot and its variants have often been used to validate miRNA expression since the discovery of miRNA [19, 20]. Other multiplexed single-miRNA approaches, such as a modified invader assay [21] and quantitative RT-PCR of pre-miRNAs [22] or mature miRNAs [23], are more sensitive and require low amounts of starting material. Later, oligonucleotide microarray-based detection platforms were developed and are the most-used approach to miRNA high-throughput profiling [24, 25]. Recently, Taqman Low Density Arrays [26, 27] and next-generation sequencing (i.e. deep sequencing) [28, 29] have emerged as the most effective high-throughput technologies for miRNA profiling.
However, these techniques are time-consuming and laborious, and cannot be performed repeatedly on the same subjects. Most importantly, they require the fixation or lysis of cells and thus cannot be used to study the dynamic function of miRNAs in living cells. Therefore, a noninvasive monitoring approach capable of repeated image acquisition is needed to evaluate miRNA expression patterns in vivo and to further translate the developments in miRNA detection into clinical practice. Here, we review recently developed miRNA imaging strategies that assist our understanding of the biogenesis and biological function of miRNAs in vivo and for monitoring miRNAs in human diseases.
Fluorescent protein-based miRNA imaging
Since green fluorescent protein (GFP) from the jellyfish Aequorea victoria was discovered in 1962, it has been used extensively to visualize subcellular compartments or to track specific molecules within cells. To date, many different fluorescent proteins with emission spectra peaks in the visible and near-infrared regions have been applied to in vivo imaging [30]. Moreover, the unique emission wavelengths of these fluorescent proteins make multicolour imaging possible without fluorescent wavelength overlap.
A typical miRNA responsive fluorescent protein reporter system fuses the DNA fragment encoding the fluorescent protein with three or four miRNA targeting sites, which are usually complementary against the particular miRNA [31]. Multiple copies of a perfectly complementary target are used to optimize the repression of the transgene in the presence of the target miRNA [32]. Using this strategy, Brown et al. [33, 34] constructed a miRNA-regulated lentiviral vector by inserting four tandem copies of a 23-bp sequence (mirT) with perfect complementarity into several miRNAs of interest into the 3′-UTR of a GFP expression cassette driven by the ubiquitously expressed phosphoglycerate kinase promoter. The suppression of GFP expression was confirmed by the presence of miRNAs using flow cytometry and fluorescence microscopy [33, 34].
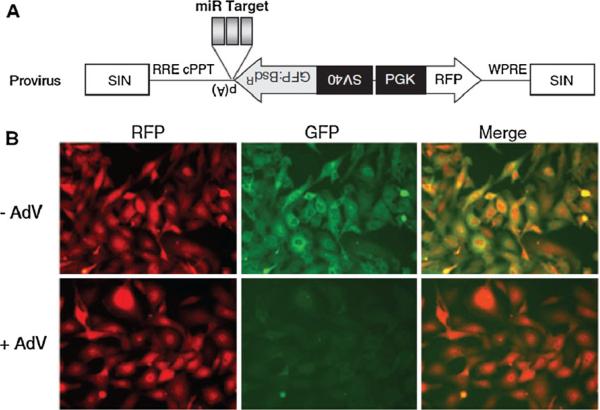
Recently, Kato et al. [35] described a two-colour monitoring system to investigate the dynamic activity of specific miRNA in living cells. In this imaging system, three copies of complementary sequences against miR-133 were inserted in tandem between the GFP and CMV promoter of a retroviral vector, which encodes two different fluorescent proteins. Since miRNA functions through sequence-specific interactions with the target mRNA, the expression of miR-133 can reduce the level of GFP whose mRNA bears the complementary sequences against miR-133, but does not affect the level of red fluorescent protein (RFP). Thus, the RFP cassette was used as an internal control for normalization. Using these two fluorescent proteins as reporters, the activity of muscle-specific miRNA, miR-133, was successfully evaluated during myogenesis in C2C12 mouse myoblast cells. The same group also developed another lentiviral vector, in which complementary sequences against specific miRNAs were cloned between GFP and the poly(A) signal of a lentiviral vector encoding two differently coloured fluorescent proteins under the bidirectional control of two distinct promoters (Fig. 1) [36]. Using this two-colour lentiviral system, adenoviral infection and consequent miRNA expression was successfully visualized by the reduction in GFP expression. In this system, RFP expression provides an endogenous reference to reduce the risk of fluctuating expression of the transgenes due to random integration of the lentiviral vector into the cellular genome.
Fig. 1.
a Scheme of the two-colour lentiviral vector system. The three tandem repeats with perfect complementarity to miRNAs are inserted between the downstream of GFP and the polyadenylation signal. b Two-colour imaging of adenovirus-derived miRNAs in living cells. HT1080 cells were transduced with the two-colour lentiviral vector. Strong expression of both GFP and RFP indicates that the lentiviral vector was efficiently transduced. At 5 days after miRNA expressing adenovirus infection, the fluorescence of GFP in cells was reduced, while the fluorescence of RFP was retained (adapted from reference [36] with permission)
However, in vivo small-animal imaging using these fluorescent proteins as reporters will suffer from autofluorescence background and limited tissue penetration of light, and consequently low signal-to-background contrast [37]. Eliminating these disadvantages has been the target of much research involving the development of new fluorescent proteins with emissions in the near-infrared range (700–900 nm), which have lower tissue absorption coefficients [30]. Using these new near-infrared proteins as reporters may be an alternative method for in vivo animal imaging.
Luciferase reporter-based miRNA imaging
The luciferase optical reporter genes, which include firefly luciferase (Fluc), Renilla luciferase (Rluc) and Gaussia luciferase (Gluc), have been widely used for the bioluminescence imaging (BLI) of living small animals. Fluc oxidizes its substrate beetle D-luciferin (benzothiazole) and emit light at about 562 nm [38], whereas Rluc and Gluc use coelenterazine as substrate and emit light at about 480 nm. Due to the better tissue penetration of photons with longer wavelengths, Fluc is better than Rluc and Gluc for in vivo imaging. Besides, the biodistribution difference from the substrates could also influence the imaging results. Gluc emits light at an intensity 1,000-fold greater than that emitted by native Fluc and Rluc. Moreover, Gluc is stable at elevated temperatures and is the smallest luciferase known [39], making it easier to use for constructing clones and cell transfection. Although lacking microscopic ability, BLI is superior to fluorescence imaging for in vivo imaging since the emitted light from BLI is virtually background-free [40] and can be used to detect very low-level signals.
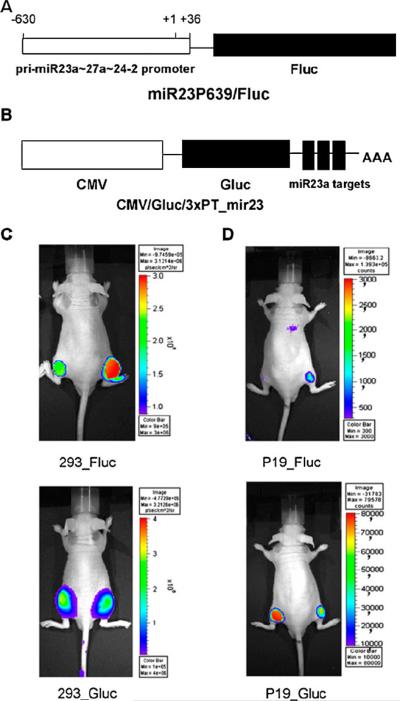
The available imaging strategies based on luciferase reporter genes have been used to investigate not only the functional action of miRNAs but also the production patterns of miRNAs including the pri-miRNAs and the mature forms of miRNAs. Previous studies have demonstrated that pri-miRNAs are transcribed from the genome by RNA polymerase II, and the 5′-upstream region of genomic miRNA controls the long pri-miRNA transcripts [1]. To acquire images of the transcription activity of primiRNAs that were transcribed from the genome, Lee et al. [41] fused the 5′-upstream promoter region of pri-miRNA-23a upstream to the cassette of pGL3-basic reporter gene vector containing a promoterless Fluc gene (Fig. 2a). This miRNA-specific reporter vector was then transfected into cultured cells and implanted into nude mice. The results from BLI in living animals showed differences in endogenous expression levels of pri-miRNA-23a transcripts in 293 and P19 cells (Fig. 2c, d upper). By applying this reporter system, the same group also monitored the highly expressed pattern of a brain-specific primary miR-9 (pri-miR-9) during neurogenesis [42].
Fig. 2.
Reporter constructs to monitor pri-miRNAs and mature miRNA activities. a miR23P639/Fluc. A segment ranging from 603 to 36 bp upstream from the transcription initiation site of miR23a;27a;24-2 gene clusters was inserted into pGL3_basic (Promega) containing a promoterless Fluc gene. b CMV/Gluc/3×PT_mir23. CMV/Gluc carries three copies of the target sequence perfectly complementary to miR-23a (3×PT_mir23) in its 39 UTR. c, d In vivo visualization of expression of pri-miR-23a transcripts and activity of mature forms of miR-23a in nude mice. 293 cells (c), and undifferentiated P19 cells (d) were transfected with miR23P639/Fluc and CMV/Gluc/3×PT_mir23 and were grafted into the right side of the mice. The same cells transfected with pGL3_basic and CMV/Gluc were grafted into the left side of the mice as controls
To investigate the expression of mature miR-221 in papillary thyroid carcinoma, Kim et al. [43] developed a Gluc imaging system in which three perfectly matched complementary sequences of miR-221 were fused immediately after the stop codon of the Gluc reporter vector. Then the transcript of this vector would be expected to be bound by endogenous miR-221, and thus the transcript of Gluc would be degraded or the translation of Gluc would be suppressed, resulting in a decrease in the bioluminescent signal. The in vivo imaging results revealed that Gluc activities were regulated according to miR-221 levels.
Previous studies have shown that several genes produce a large amount of pri-miRNAs but a low level of mature miRNAs during biogenesis, which is related to some unknown regulatory mechanisms [44]. The expression pattern of both pri-miRNAs and mature miRNAs were simultaneously assessed using a dual luciferase reporter system [41], in which the Fluc gene was under the control of miR-23a promoter to monitor the primary transcript activity of miR-23a (Fig. 2a), whereas three copies of the miR-23a complementary sequence were fused with the Gluc gene to monitor the targeting activity of mature miR-23a (Fig. 2b). After transfecting this dual luciferase reporter system into P19 cells, the Fluc signal for pri-miR-23a was increased while the Gluc signal for mature miR-23a was reduced. Conversely, the turnover from pri-miR-23a to mature miR-23a was much slower in 293 cells (Fig. 2c, d). This discrepancy in the different levels of pri-miR-23a and mature miR-23a in 293 cells is possibly due to some unknown regulatory factors that control Drosha or Dicer processing steps or block the release of pri-miR-23a from the nucleus into the cytoplasm.
Unlike the reporter systems for imaging mature miRNAs, the reporter systems for imaging miRNA targets are based on the fact that the target sequence of a particular miRNA is known and the level of the miRNA activity is measured. By doing so, the 3′-UTR of the bona fide target gene of miRNA is fused immediately after the stop codon of the luciferase reporter vector. In this way, partial interaction between miRNA and the 3′-UTR region in the reporter gene constructs results in a reduction in the bioluminescent signal. Using this reporter system, Kim et al. [45] cloned the 3′-UTR of homeobox B5 (HoxB5), which has been verified as one of the miR-221 target genes, into the Gluc reporter gene. The in vivo bioluminescent signal from the Gluc reporter gene is significantly decreased by the presence of endogenous mature miR-221 during the development of papillary thyroid carcinoma. The same group also applied the same reporter system to monitor miR-124a-mediated repression of the target gene chromosome 14 open reading frame 24 (c14orf24) during neurogenesis [46].
Fluorescence beacon-based miRNA imaging
Clinical translation of imaging based on reporter genes is limited since genetic modification of cells without substantially affecting their characteristics is a challenge. The sensitivity of this technique is determined by the reporter probe's pharmacokinetics and by the reporter gene expression level in the transfected cells. Besides, the reporters for imaging mature miRNAs or miRNA targets described above represent signal-off systems, as miRNAs bind and destabilize their target mRNAs. In this case, it is difficult to determine whether the observed signal-off data result from miRNA expression or cell death in vivo. In addition, signal-off systems are usually not sensitive due to the high background. Recently, several signal-on imaging strategies have been developed to image endogenous mRNA and miRNAs [47, 48]. Signal-on systems are advantageous for in vivo imaging because of their relatively low background.
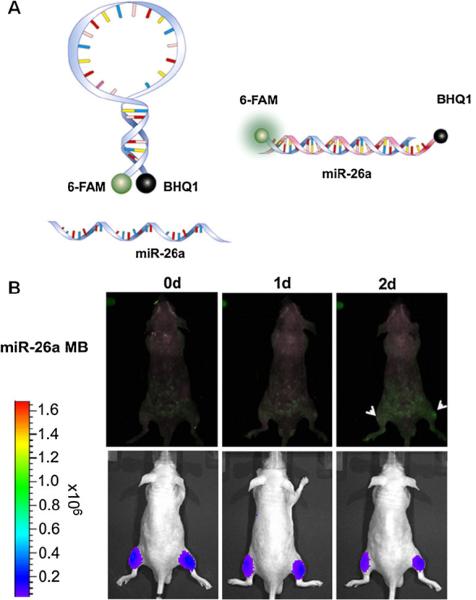
Kang et al. [49] developed a molecular beacon (MB) and successfully imaged miR-206 and miR-26a biogenesis during myogenesis both in vitro and in vivo. The MB is composed of a single-stranded and stem-loop DNA oligonucleotide that is complementary to the target miRNAs. A reporter fluorophore is at one end of and a quencher is at the other end of the MB. In the absence of target miRNAs, the fluorophore and the quencher molecules are in close proximity, which results in fluorescence energy absorption by the quencher molecules. By contrast, by combining the complementary sequences of the MB, the presence of miRNAs when mature miRNA-26a or miRNA-206 is expressed and binds to the loop of each MB, will result in a conformational change that results in the separation of the fluorophore and quencher and the recovery of the fluorescence signal (Fig. 3). This system also allows potential multiplex imaging of miRNAs since several dye–quencher pairs can be applied to a single cell simultaneously. Indeed, with cotransfection into a single cell, the miRNA-26a MB containing 6-FAM (absorbance/emission wavelengths 494/525 nm) and BHQ1 and the miRNA-206 MB containing Texas Red-X (absorbance/emission wavelengths 583/603 nm) and BHQ2, miR-26a and miR-206 expression in a single C2C12 cell were simultaneously monitored during myogenesis in vitro and in vivo.
Fig. 3.
a A schematic illustration of MB imaging miRNAs. miRNA-26a-linked MB was designed to have 6-FAM (absorbance/emission wavelengths 494/525 nm) as fluorophore and BHQ1 as a quencher. b In vivo fluorescence imaging of miR-26a (green) during differentiation of C2C12 cells. C2C12 cells were transfected with the miR-26a MB and the firefly luciferase gene simultaneously. After differentiation induction, C2C12 cells were injected into the right thigh of a nude mouse. As a control, C2C12 cells without differentiation were injected into the left thigh of a nude mouse. Fluorescence images of the miR-26a MB (excitation 490 nm, emission 520 nm) were obtained. Firefly luciferase images were acquired after injection of luciferin
The same group also reported a “smart” magnetic fluorescent nanoparticle to monitor intracellular miR-124a during neuronal differentiation. The nanoparticle was composed of cobalt ferrite in the central core and the fluorescent organic dye, rhodamine B isothiocyanate (RITC, excitation/emission 555/578 nm), coated with a silica shell. Then a partially double-stranded oligonucleotide was conjugated with the carboxyl-terminal nanoparticles via amine–carboxyl interaction. The double-stranded oligonucleotide contained a miR124a binding sequence and a quenching oligonucleotide with BHQ1 on the 3′ end. With the dual magnetic fluorescent nanoparticle imaging probe, the authors showed enhanced fluorescence signals in mice with P19 cells after induction of neuronal differentiation. In addition, magnetic resonance (MR) images allowed in vivo cellular tracking with the MF-miR124a MB [50].
Conclusion and perspectives
In view of the importance of miRNAs in diverse regulatory pathways, various noninvasive miRNA imaging techniques, based on different reporter genes, have been developed to monitor miRNA biogenesis and functions in vivo. The miRNA imaging system harbouring two different fluorescent proteins offers a useful approach to multicolour and real-time tracking of the dynamic activity of miRNAs of interest in living cells. However, when used for in vivo animal imaging, such reporter-based systems have the limitation of high background noise and low sensitivity compared to the luciferase-based reporter imaging system.
The luciferase-based imaging system, in which the promoter region of miRNAs or the miRNA target sequence are fused with the luciferase reporter genes, provides noninvasive molecular imaging information regarding the production pattern and function of miRNAs. There are, however, two major concerns about both the reported bioluminescent and fluorescent reporter gene imaging systems. One concern is that they are signal-off systems in which the presence of functional miRNA will decrease the reporter signal, as a result of miRNAs binding and degrading their target mRNAs. To overcome the limitations of the signal-off problem in the reporter gene imaging systems, we have suggested a construct containing an inhibitory element [51]. The other concern is that the optical reporter genes remain a distant goal for clinical application due to the attenuation of optical signal in deep tissue and organs. Future research work may involve the development of more clinically relevant radionuclide or MR reporters for miRNA imaging [52].
The newly developed MBs and nanoparticles are promising for direct in vivo miRNA imaging. However, the in vivo imaging using miRNA MBs discussed above is still preliminary since the MBs were incubated with cells in vitro first and a phantom was then developed with these MB-containing cells in living animals [49, 50], which is not considered as real in vivo imaging. In addition, the relatively short emission wavelength of the dye molecules used is not appropriate for in vivo imaging, either. The combination of near-infrared fluorescent dyes and the corresponding paired quencher could be used an alternative. Another shortcoming of this system is that without the use of a vectorization agent, naked oligonucleotides will not enter the cells of interest. By using a vectorization agent such as nanoparticles, the pharmacokinetics of the nano delivery system will deeply affect the whole-body detection of the miRNAs. Thus, imaging probes that are more biocompatible and deliverable need to be developed for real-time in vivo imaging of miRNAs.
No single imaging modality is sufficient to provide all details as each modality has its own limitations. To harness the strengths of different imaging methods, multimodality imaging has become attractive for both preclinical and clinical studies. Multimodality imaging enables the combination of anatomical, functional and molecular information by combining images from different modalities taken at the same point. This strategy combines the strengths of different modalities and yields a hybrid imaging platform with characteristics superior to those of any of its constituents alone [53, 54]. Therefore, study of the expression pattern of miRNAs by the application of multimodality imaging will be a promising future direction. However, there are still some concerns about clinical transfer and biocompatibility when multimodality imaging is used; for example, reporter gene systems involve genetic modification of the studied object, and MB nanoparticles are often toxic and do not distribute homogeneously in the body.
In conclusion, molecular imaging techniques are a powerful tool for noninvasive monitoring of the in vivo dynamics of miRNAs, including their localization patterns, production profiles and functional actions. Such techniques will advance our understanding of the regulatory networks of miRNAs in both malignant diseases and other pathological pathways, and eventually improve our ability to use miRNAs as diagnostic and therapeutic agents.
Acknowledgments
This work was supported in part by the Intramural Research Program of the National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health, the National Basic Research and Development Program of China (973) under grant nos. 2006CB705700 and 2011CB707702, the National Natural Science Foundation of China under grant nos. 81090272, 81090274, 81000632, 30900334 and 30970845, and the Fundamental Research Funds for the Central Universities under grant no. JY10000910002.
Footnotes
Conflicts of interest None.
References
- 1.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 3.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–6. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–70. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 7.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 10.Dostie J, Mourelatos Z, Yang M, Sharma A, Dreyfuss G. Numerous microRNPs in neuronal cells containing novel micro-RNAs. RNA. 2003;9:180–6. doi: 10.1261/rna.2141503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 12.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–5. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 13.Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winter J, Diederichs S. MicroRNA biogenesis and cancer. Methods Mol Biol. 2011;676:3–22. doi: 10.1007/978-1-60761-863-8_1. [DOI] [PubMed] [Google Scholar]
- 15.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 17.Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93:1600–8. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 19.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 20.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allawi HT, Dahlberg JE, Olson S, Lund E, Olson M, Ma WP, et al. Quantitation of microRNAs using a modified Invader assay. RNA. 2004;10:1153–61. doi: 10.1261/rna.5250604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmittgen TD, Jiang J, Liu Q, Yang L. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res. 2004;32:e43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA. 2005;11:1737–44. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 25.Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat Methods. 2004;1:155–61. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- 26.Rosero S, Bravo-Egana V, Jiang Z, Khuri S, Tsinoremas N, Klein D, et al. MicroRNA signature of the human developing pancreas. BMC Genomics. 2010;11:509. doi: 10.1186/1471-2164-11-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cervigne NK, Reis PP, Machado J, Sadikovic B, Bradley G, Galloni NN, et al. Identification of a microRNA signature associated with progression of leukoplakia to oral carcinoma. Hum Mol Genet. 2009;18:4818–29. doi: 10.1093/hmg/ddp446. [DOI] [PubMed] [Google Scholar]
- 28.Buermans HP, Ariyurek Y, van Ommen G, den Dunnen JT, 't Hoen PA. New methods for next generation sequencing based microRNA expression profiling. BMC Genomics. 2010;11:716. doi: 10.1186/1471-2164-11-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fehniger TA, Wylie T, Germino E, Leong JW, Magrini VJ, Koul S, et al. Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Res. 2010;20:1590–604. doi: 10.1101/gr.107995.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wessels JT, Yamauchi K, Hoffman RM, Wouters FS. Advances in cellular, subcellular, and nanoscale imaging in vitro and in vivo. Cytometry A. 2010;77:667–76. doi: 10.1002/cyto.a.20931. [DOI] [PubMed] [Google Scholar]
- 31.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–8. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 32.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 33.Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457–67. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 34.Brown BD, Venneri MA, Zingale A, Sergi L, Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med. 2006;12:585–91. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- 35.Kato Y, Miyaki S, Yokoyama S, Omori S, Inoue A, Horiuchi M, et al. Real-time functional imaging for monitoring miR-133 during myogenic differentiation. Int J Biochem Cell Biol. 2009;41:2225–31. doi: 10.1016/j.biocel.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kato Y, Sawata SY, Inoue A. A lentiviral vector encoding two fluorescent proteins enables imaging of adenoviral infection via adenovirus-encoded miRNAs in single living cells. J Biochem. 2010;147:63–71. doi: 10.1093/jb/mvp144. [DOI] [PubMed] [Google Scholar]
- 37.Troy T, Jekic-McMullen D, Sambucetti L, Rice B. Quantitative comparison of the sensitivity of detection of fluorescent and bioluminescent reporters in animal models. Mol Imaging. 2004;3:9–23. doi: 10.1162/15353500200403196. [DOI] [PubMed] [Google Scholar]
- 38.Gould SJ, Subramani S. Firefly luciferase as a tool in molecular and cell biology. Anal Biochem. 1988;175:5–13. doi: 10.1016/0003-2697(88)90353-3. [DOI] [PubMed] [Google Scholar]
- 39.Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11:435–43. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 40.Negrin RS, Contag CH. In vivo imaging using bioluminescence: a tool for probing graft-versus-host disease. Nat Rev Immunol. 2006;6:484–90. doi: 10.1038/nri1879. [DOI] [PubMed] [Google Scholar]
- 41.Lee JY, Kim S, Hwang do W, Jeong JM, Chung JK, Lee MC, et al. Development of a dual-luciferase reporter system for in vivo visualization of microRNA biogenesis and posttranscriptional regulation. J Nucl Med. 2008;49:285–94. doi: 10.2967/jnumed.107.042507. [DOI] [PubMed] [Google Scholar]
- 42.Ko MH, Kim S, Hwang do W, Ko HY, Kim YH, Lee DS. Bioimaging of the unbalanced expression of microRNA9 and microRNA9* during the neuronal differentiation of P19 cells. FEBS J. 2008;275:2605–16. doi: 10.1111/j.1742-4658.2008.06408.x. [DOI] [PubMed] [Google Scholar]
- 43.Kim HJ, Chung JK, Hwang do W, Lee DS, Kim S. In vivo imaging of miR-221 biogenesis in papillary thyroid carcinoma. Mol Imaging Biol. 2009;11:71–8. doi: 10.1007/s11307-008-0188-6. [DOI] [PubMed] [Google Scholar]
- 44.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–7. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim HJ, Kim YH, Lee DS, Chung JK, Kim S. In vivo imaging of functional targeting of miR-221 in papillary thyroid carcinoma. J Nucl Med. 2008;49:1686–93. doi: 10.2967/jnumed.108.052894. [DOI] [PubMed] [Google Scholar]
- 46.Ko HY, Lee DS, Kim S. Noninvasive imaging of microRNA124a-mediated repression of the chromosome 14 ORF 24 gene during neurogenesis. FEBS J. 2009;276:4854–65. doi: 10.1111/j.1742-4658.2009.07185.x. [DOI] [PubMed] [Google Scholar]
- 47.Nitin N, Santangelo PJ, Kim G, Nie S, Bao G. Peptide-linked molecular beacons for efficient delivery and rapid mRNA detection in living cells. Nucleic Acids Res. 2004;32:e58. doi: 10.1093/nar/gnh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santangelo P, Nitin N, Bao G. Nanostructured probes for RNA detection in living cells. Ann Biomed Eng. 2006;34:39–50. doi: 10.1007/s10439-005-9003-6. [DOI] [PubMed] [Google Scholar]
- 49.Kang WJ, Cho YL, Chae JR, Lee JD, Choi KJ, Kim S. Molecular beacon-based bioimaging of multiple microRNAs during myogenesis. Biomaterials. 2011;32:1915–22. doi: 10.1016/j.biomaterials.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 50.Hwang do W, Song IC, Lee DS, Kim S. Smart magnetic fluorescent nanoparticle imaging probes to monitor microRNAs. Small. 2010;6:81–8. doi: 10.1002/smll.200901262. [DOI] [PubMed] [Google Scholar]
- 51.Niu G, Chen X. Noninvasive visualization of microRNA by bioluminescence imaging. Mol Imaging Biol. 2009;11:61–3. doi: 10.1007/s11307-008-0190-z. [DOI] [PubMed] [Google Scholar]
- 52.Niu G, Gaut AW, Ponto LL, Hichwa RD, Madsen MT, Graham MM, et al. Multimodality noninvasive imaging of gene transfer using the human sodium iodide symporter. J Nucl Med. 2004;45:445–9. [PubMed] [Google Scholar]
- 53.Pichler BJ, Judenhofer MS, Pfannenberg C. Multimodal imaging approaches: PET/CT and PET/MRI. In: Semmler W, Schwaiger M, editors. Handbook of experimental pharmacology. Vol 185/1. Molecular Imaging I. Springer; Berlin: 2008. pp. 109–132. [DOI] [PubMed] [Google Scholar]
- 54.Iagaru A, Mittra E, Yaghoubi SS, Dick DW, Quon A, Goris ML, et al. Novel strategy for a cocktail 18F-fluoride and 18F-FDG PET/CT scan for evaluation of malignancy: results of the pilot-phase study. J Nucl Med. 2009;50:501–5. doi: 10.2967/jnumed.108.058339. [DOI] [PubMed] [Google Scholar]