Abstract
Methylation patterns of the mammalian genome are thought to be crucial for development. The precise mechanisms designating specific genomic loci for methylation are not known. Targeted deletion of Lsh results in perinatal lethality with a rather normal development. We report here, however, that Lsh−/− mice show substantial loss of methylation throughout the genome. The hypomethylated loci comprise repetitive elements and single copy genes. This suggests that global genomic methylation is not absolutely required for normal embryogenesis. Based on the similarity of Lsh to other SNF2 chromatin remodeling proteins, it suggests that alteration of chromatin affects global methylation patterns in mice.
Keywords: Lsh, methylation, SNF2, SWI/SNF complex, chromatin
Methylation of cytosine residues in the genome is thought to be crucial for normal development in mammals as well as plants, frogs, and fish. DNA methylation is involved in the regulation of a diverse range of biological processes such as genomic imprinting and X-chromosome inactivation (for reviews, see Yeivin and Razin 1993; Razin and Shemer 1995; Robertson and Jones 2000). Establishment of genomic methylation patterns in mammals is a highly orchestrated process, with almost complete erasure during early embryogenesis and a resetting of the pattern subsequent to implantation. In general, genes are relatively unmethylated in expressing tissues and methylated in nonexpressing tissues leading to the hypothesis that methylation controls tissue specific gene expression. Alternatively, genomic methylation may primarily protect against the expression of parasitic sequences (Walsh and Bestor 1999). Deletion of DNA methyltransferase in Xenopus leads to abnormal development of the embryo (Stancheva and Meehan 2000; Stancheva et al. 2001). Reports on mice that are deficient in DNA methyltransferases support also a role for methylation in development as these mice die early during embryogenesis (Li et al. 1992; Lei et al. 1996; Okano et al. 1999).
Lsh (lymphoid specific helicase) belongs to the family of SNF2/helicases. SNF2-like proteins are frequently involved in chromatin remodeling (Jarvis et al. 1996). Lsh shows a preferential lymphoid expression pattern in adult mice and has been shown to be important for normal lymphoid development (Geiman et al. 1998; Geiman and Muegge 2000). However, a low expression of Lsh has been found in multiple embryonal tissues suggesting a broader role for Lsh in development (Geiman et al. 2001). SNF2-containing protein complexes can alter nucleosomal structure in vitro (Peterson and Workman 2000) and recombinant SNF2 homologs on their own can cause nucleosomal sliding along DNA (Hamiche et al. 1999; Langst et al. 1999). This in vitro activity of SNF2 family members is held responsible for altering chromatin accessibility in vivo. A recent study identified the gene Ddm1 (decrease in DNA methylation), another SNF2 family member, as a modulator of genomic methylation in Arabidopsis thaliana (Jeddeloh et al. 1999). The methylation system in plants appears to be highly conserved although homologs for the chromomethylases have not been identified in mammals, possibly because mammals primarily maintain CG methylation instead of methylation at CNG sites (Lindroth et al. 2001). This latter Ddm1 study suggests a relationship between regulation of chromatin structure and genomic methylation in plants. Because Ddm1 shares about 50% identity with Lsh over the region containing the helicase domains (Jeddeloh et al. 1999), we studied the effect of Lsh on genomic methylation patterns in mice.
Results and Discussion
To determine whether Lsh has an effect on DNA methylation we examined genomic DNA derived from Lsh−/− mice (Geiman and Muegge 2000; Geiman et al. 2001) for their methylation status at multiple repetitive sequences that are highly methylated. The minor satellite sequences (50,000–100,000 copies) are located around centromeres. Analysis of genomic DNA from fibroblasts, brain, liver, intestine, heart and lung, and the whole body (Fig. 1A,B) as well as thymus (K. Dennis, T.M. Geiman, and K. Muegge, unpubl.) showed increased digestibility with HpaII, a methylation-sensitive enzyme. The pattern of the HpaII digest was comparable with the MspI digest (a methylation-insensitive isoschizomer) suggesting a substantial loss of methyl groups at the minor satellite sequence in the absence of Lsh. The defect in methylation was detectable in newborn tissue (Fig. 1B) as well as fetal tissue (Fig. 1A) as early as day 13.5 of gestation (the earliest time point we have examined).
Figure 1.
Hypomethylation of minor satellite sequences in Lsh−/− mice. (A) Southern analysis of genomic DNA derived at day 13.5 of gestation. DNA was digested with HpaII or MspI, blotted, and probed for minor satellite sequences using MR150. (B) Southern analysis of genomic DNA derived from newborn mice within 24 h after birth. Whole body comprises every tissue with the exception of the examined internal organs. DNA was digested with HpaII or MspI, blotted, and probed for minor satellite sequences using MR150.
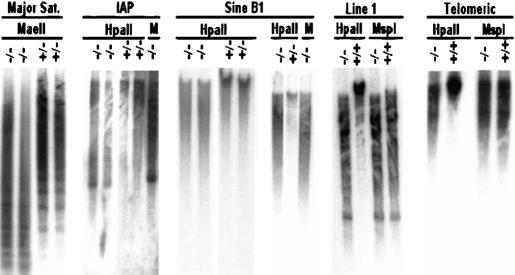
To analyze whether hypomethylation is a widespread phenomenon in the genome, other methylated sequences with high copy number were examined. The major satellite sequence, with 700,000 copies in centromeric regions, also showed substantial hypomethylation in DNA isolated from fetal brain tissue of Lsh-deleted animals (Fig. 2) as well as from MEF cultures or body tissue of newborn mice (K. Dennis and K. Muegge, unpubl.). Sequences of the intracisternal A-particle retrovirus (IAP) (1000–2000 copies) are heavily methylated in littermate controls but not in Lsh-deleted animals. Line 1, another retroviral element (50,000–100,000 copies), Sine B1, the murine homolog of human Alu repeats, as well as examination of telomeric sequences showed substantial hypomethylation in the absence of Lsh (Fig. 2). Moreover, comparison of HpaII and MspI digests of DNA from Lsh-deficient tissue appear almost indistinguishable suggesting substantial loss of methylation at the examined sites.
Figure 2.
Hypomethylation of repetitive sequences in Lsh−/− mice. Southern analysis for repetitive sequences. Genomic DNA derived from Lsh−/− or littermate controls was digested with MaeII and probed for major satellite sequences, or digested with HpaII and MspI (M) and probed for IAP, Sine B1, Line 1, or telomeric sequences.
To determine whether Lsh deficiency selectively leads to hypomethylation of repetitive elements or also effects single copy sequences a number of specific genomic loci were examined. The genes for β-Globin and Pgk-2 (phosphoglycerate kinase) are highly methylated single copy genes with a tissue-specific expression pattern. Pgk-1, another highly methylated gene, is located on the X-chromosome. The methylation sensitive sites examined were located either 5′ of the gene (Pgk-1), within the exon (Pgk-2), or 3′ of the gene (Globin) and did not contain any known repetitive sequences (Singer-Sam et al. 1990; Kafri et al. 1992; Tada et al. 1997). All three examined loci revealed a substantial loss of methylation in newborn tissue as well as embryonic tissue from Lsh−/− mice (Fig. 3A–C). In addition, the upstream region of the H19 gene was examined comprising the imprinting control region with all four CTCF (CCCTC-binding factor) binding sites (Bell and Felsenfeld 1999). This region showed substantial hypomethylation in Lsh−/− mice (Fig. 3D). In contrast the Igf2r gene showed no difference in methylation between Lsh−/− tissue and wild-type tissue (K. Dennis and K. Muegge, unpubl.). These results demonstrate that loss of the Lsh gene effects methylation of repetitive elements and single-copy sequences including the imprinted region of the H19 gene.
Figure 3.
Hypomethylation of single copy sequences in Lsh−/− mice. (A) Southern analysis of the β-Globin gene. Genomic DNA was derived from newborn mice or embryos at day 13.5 gestation. DNA was digested with BamHI with or without the methylation sensitive restriction enzyme HhaI, blotted, and probed for β-Globin. (B) Southern analysis of the Pgk-2 gene. (C) Southern analysis of the Pgk-1 gene. (D) Southern analysis of the H19 upstream imprinted region.
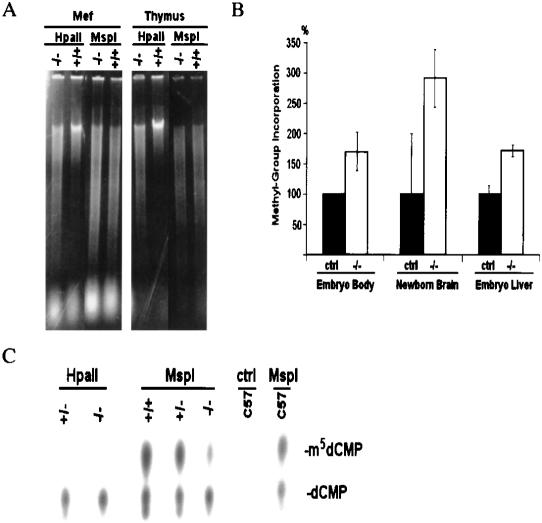
A global defect in methylation was visualized using ethidium bromide stain. Genomic DNA derived from thymus and MEF (Fig. 4A) as well as brain (K. Dennis and K. Muegge, unpubl.) of Lsh-deficient mice was highly digestible with the methylation-sensitive enzyme HpaII. To quantify the extent of hypomethylation, genomic DNA from Lsh−/− mice was tested in vitro for its ability to accept methyl groups and compared with littermates using the SssI methyltransferase. Genomic DNA derived from embryonic liver and body as well as newborn brain of Lsh-deleted mice accepted two to three times more methylation than controls (Fig. 4B). Assuming an average methylation level of 60%–70% in control samples, this difference would suggest a genome-wide methylation level of only 13%–32% in genomic DNA from Lsh-deleted mice. In addition, genomic DNA from newborn brain was digested with MspI, radiolabeled at 5′ ends, and degraded to single deoxynucleoside monophosphates. Cytosine as well as methyl-cytosine was separated by thin-layer chromatography and the resulting spots were quantified by PhosporImager. The control digest using HpaII did not show a spot for methylated cytosine indicating that the assay is highly specific. Whereas wild-type controls showed that about 57% of their genome was methylated, the Lsh-deleted mice show only 34% methylation at CCGG sites (Fig. 4C). Thus, loss of the Lsh gene resulted in a substantial loss of methylation throughout the genome.
Figure 4.
Global hypomethylation in Lsh−/− mice. (A) Genomic DNA from embryonal fibroblasts (MEF) or adult thymus from radiation chimeras (Geiman and Muegge 2000) was digested with the methylation-sensitive enzyme HpaII and the nonsensitive enzyme MspI, subjected to agarose gel electrophoresis, and visualized by ethidium bromide stain. (B) Methyl acceptance assay. Equal amounts of genomic DNA derived from day 13.5 embryonic body, embryonic liver, or from newborn brain were methylated in vitro by SssI CG methylase using radiolabeled S-adenosyl-methionine as donor. This approach allows determination of the amount of unmethylated CG sites in the genome and serves as an indirect measurement of genomic methylation levels. The amount of incorporated radiolabeled methyl groups on cytosines per microgram of DNA was measured in Lsh deleted samples and control littermates as described previously (Antoun et al. 2000). (C) Direct measurement of methyl-cytosine in genomic DNA. Equal amounts of genomic DNA derived from brain samples of Lsh−/− mice and littermate controls were digested with MspI, radiolabeled at the 5′-ends, and digested with nuclease P1 to generate 5′-deoxymononucleotides. Cytosine and methyl-cytosine were separated by thin layer chromatography (Cedar et al. 1979) and quantified using PhosphorImager analysis. The ratio of methyl-cytosine to total cytosine indicates the level of methylation at all CCGG sites. HpaII digests should not generate methyl-cytosine spots and serve as controls, indicating the specificity of the assay.
We hypothesize that Lsh protein may participate in the process of de novo or maintenance DNA methylation rather than simply altering the level of DNA methyltransferase mRNA and protein for several reasons: (1) mRNA levels for DNA methyltransferases were comparable between Lsh-deficient samples and control samples in embryonic bodies (Fig. 5A), embryonic liver, or MEF cultures (T. Fan and K. Muegge, unpubl.). (2) The level of Dnmt1 protein (whose targeted deletion results in a similar degree of genomic hypomethylation) is indistinguishable between Lsh−/− tissue and controls (Fig. 5B). Similarly, the amounts of Dnmt3a and Dnmt3b proteins were equally expressed in Lsh-deficient tissue and control sample (Fig. 5B). (3) Methyltransferase activity as measured from nuclear extracts from Lsh-deficient tissue was not significantly altered in comparison with control tissue (Fig. 5C). This also has been reported for mutants of the Ddm1 gene in A. thaliana (Jeddeloh et al. 1999) sharing a high similarity with Lsh. (4) The biological phenotype of Lsh−/− mice is very distinct from that reported for either Dnmt1- or Dnmt3a/b-deficient embryos (Li et al. 1992; Okano et al. 1999; Geiman et al. 2001).
Figure 5.
Expression of DNA methyltransferases and measurement of Mtase activity in Lsh−/− mice. (A) RT–PCR analysis. Total RNA of embryonic body (2 wild type, 1 heterozygote, and 3 knockout) derived from day 17.5 gestation was reverse transcribed and subjected to real-time PCR analysis for measurement of Dnmt1, Dnmt3a, or Dnmt3b or Gapdh transcripts as control. (CT) Cycle threshold, cycle number at which each PCR reaction reaches a predetermined fluorescence threshold, set within the linear range of all reactions. (B) Western analysis. Cellular extracts derived from fetal brain tissue were analyzed using specific antiserum against murine Dnmt1, Dnmt3a (Imgenex), Dnmt3b (Affinity Bioreagents), β-Actin (Sigma), or PCNA (Santa Cruz) as control. A similar result was obtained using lysates derived from embryonic body of day 17.5 gestation. (C) Mtase activity. Cellular extracts were prepared from indicated tissues and examined in vitro for their ability to transfer radio-labeled methyl-groups onto synthetic template poly[d(I–C)]·poly[d(I–C)] (Li et al. 1992). Embryonic bodies are from day 13.5 gestation.
A global effect on methylation of the genome has thus far only been reported for embryos deficient in DNA methyltransferase. Dnmt1 N is a targeted Dnmt1 allele that leads to a 95% reduction of the Dnmt1 protein. Mutant embryos develop to day 10.5 and show 30% reduction of genome methylation levels (Li et al. 1992). The S and C alleles of Dnmt1 are more severe and lead to an arrest in embryonic development around day 8.5 with an almost undetectable genomic methylation level. In contrast, conditional mutants that lack Dnmt1 from day 12 of gestation in neuroblasts die shortly after birth from respiratory stress without any obvious defects in the brain structure (Fan et al. 2001). The methyltransferases Dnmt3a and Dnmt3b are thought to be involved in de novo methylation. Dnmt3a−/− mice look normal at birth and die runted by 4 wk of age with no apparent change in global methylation levels (Okano et al. 1999). Dnmt3b−/− could not be recovered at birth and showed developmental abnormalities from day 9.5 on (Okano et al. 1999). Loss of Dnmt3b does not affect global methylation patterns but instead leads to hypomethylation of certain classes of sequences such as minor satellite sequences. Double mutants deficient in Dnmt3a and Dnmt3b show abnormalities from day 8.5 on and die before day 11.5. Double deficient mutants show substantial losses of methylation in retroviral sequences, and major and minor satellite repeats (Okano et al. 1999), although less severely than reported here for Lsh−/− mice (Figs. 1 and 2, cf. HpaII digests with MspI digests; Li et al. 1992; Okano et al. 1999). The defects in genomic methylation, as reported previously, lead to early arrest during embryogenesis. This is in contrast with the phenotype reported for Lsh-deficient mice (Geiman and Muegge 2000; Geiman et al. 2001): Lsh−/− mice can develop until term and die shortly after birth. Lsh−/− mice show reduced lymphoid numbers, renal lesions with signs of necrosis, and a 20% weight reduction at birth. However, otherwise they appear grossly normal by morphologic and histologic criteria despite their global methylation defect. Thus, normal methylation patterns of the genome are not absolutely required for murine development until birth. To date it remains unclear which loci in the genome require methylation for survival of the organism.
Based on its presumed chromatin remodeling activity we speculate that Lsh may regulate chromatin accessibilty for DNA methyltransferases. Because Lsh protein expression correlates with S-phase of the cell cycle (Geiman and Muegge 2000) Lsh may facilitate access of DNA methyltransferases to hemimethylated sites after replication occurs and thus co-operate to maintain methylation patterns. Alternatively, the presence of Lsh may protect against demethylase activities (Weiss et al. 1996; Ramchandani et al. 1999). Overall, Lsh−/− mice provide a valuable model in mammals for examining the relationship between chromatin remodeling and methylation.
Recent studies contributed much to our understanding of how DNA methylation can regulate chromatin accessibility (Nan et al. 1998; Ng et al. 1999; Wade et al. 1999). The deficiency in genomic methylation in Lsh−/− mice suggests that the reverse is also true, that chromatin structure can have an impact on genomic methylation levels. In further support, it has recently been observed that ATRX, another protein with SNF2 similarity, can alter methylation patterns (Gibbons et al. 2000); however, in contrast to Lsh the effect is not global, but at distinct genomic sites (ribosomal 18s locus or an Y-specific satellite) and furthermore not only hypomethylation but also hypermethylation has been reported.
Aberrant methylation patterns have been suspected to promote tumorigenesis (Jones and Gonzalgo 1997; Baylin and Herman 2000) to be involved in the process of aging (Issa 2000) or to be causative for inherited diseases such as the ICF syndrome (Okano et al. 1999; Xu et al. 1999). The observed defect in global methylation in Lsh−/− mice will be a helpful tool to increase our understanding of the mechanism and consequences of specific genomic methylation patterns in disease.
Materials and methods
Southern analysis
Genomic DNA was prepared from the indicated tissues, digested, separated by electrophoresis on 1% agarose gels, and transferred by blotting on Nytran plus membranes (Schleicher& Schuell). Membranes were hybridized overnight at 42°C in hybridization buffer (Amersham) with 32P-labeled probes and washed twice in 2×SSC/0.1%SDS at 65°C or 42°C for 30 min and twice in 0.2×SSC/0.1%SDS at 65°C or 42°C for 30 min. The following oligonucleotide probes were used for detection of repetitive sequences: Major satellite (60-mer GenBank accession no. X06899, base pair 1–60), Line1 (60-mer accession no. D84391, base pair 5670–5729), Sine B1 (27-mer accession no. AC002121, base pair 13464–13490), telomeric probe [42-mer, 5′-(TTAGGG) × 7-3′]. The minor satellite probe was a 66-mer oligonucleotide 5′-GACTGAAAAACACATTCGTTG GAAACGGGATTTGTAGAACAGTGTATATCAATGAGTTACAATG AG-3′ as described elsewhere (Tada et al. 1997). The IAP probe was 299 bp long and derived by PCR reaction as described below. The detection of the methylation sensitive sites and maps for the β-globin, Pgk-2, and Pgk-1 genes have been described elsewhere (Singer-Sam et al. 1990; Kafri et al. 1992; Tada et al. 1997). Examination of the 5′ region of the H19 gene (including a map of the region) has been described (Tada et al. 1997). The 3.8-kb SacI fragment used as a probe was amplified by PCR reaction as described below. The probe comprises the imprinting control region with all four CTCF-binding sites and examines eight distinct HhaI sites.
RT–PCR analysis
For detection of mRNA for DNA methyltransferases 2 μg of total RNA was reversed transcribed. Real-time PCR was perfomed in the ABI PRISM 7900 Sequence Detection System (Applied Biosystems) using SYBR Green I as a double-strand DNA-specific binding dye and continuous fluorescence monitoring. Each reaction contained 100 ng of cDNA template and primers at a concentration of 400 nM in a final volume of 25 μL in SYBR Green PCR Master Mix (Applied Biosystems) containing AmpliTaq Gold. PCR was initiated with one cycle 50°C for 2 min and one cycle 94°C for 10 min followed by 40 cycles: 94°C for 30 sec, 60°C for 30 sec, 72°C for 45 sec. Cycle threshold, the cycle number at which each PCR reaction reaches a predetermined fluorescence threshold, was set within the linear range of all reactions. Melting curve analysis of amplification products was performed at the end of each PCR. As an additional control, PCR products were separated by electrophoresis and detected by hybridization with an internal oligonucleotide probe. The following oligonucleotides were used as primers: dnmt1 (GenBank accession no. X14805: sense base pair 310–330, antisense base pair 681–701), dnmt3a (accession no. AF068625: sense base pair 363–382, antisense base pair 759–779), dnmt3b (accession no. AF068628: sense base pair 305–328, antisense base pair 818–838).
Quantitative measurement of global methylation
Measurement of methylated cytosine at CCGG sites was determined as described previously (Cedar et al. 1979; Li et al. 1992). Purified genomic DNA was digested with HpaII or MspI, treated with alkaline phosphatase and radiolabeled with [α-32P] at the 5′-termini using polynucleotide kinase. Phenol–chloroform-purified DNA was digested with nuclease P1 and the 5′-deoxymononucleotides were separated by thin-layer chromatography on cellulose plates. The radioactivity over methyl-cytosine and cytosine was scanned by PhosphorImager analysis. The ratio was formed between methyl-cytosine versus total cytosine indicating the percentage of methylation at CCGG sites in the genome. HpaII was used as control digest indicating the specificity of the assay; as HpaII does not recognize methylated CCGG sites and only cleaves unmethylated sites the HpaII digest should only generate cytosine and not methyl-cytosine spots. The methyl acceptor assay was performed as described previously (Antoun et al. 2000). Purified genomic DNA (200 ng) was incubated for 4 h at 37°C with 4 units of M.SssI CpG methylase, 3 μCi [methyl-3H]S-adenosyl L-methionine, and 1.5 μM nonradioactive AdoMet. After termination the mixture was spotted on Whatman glass filters washed with 5% trichloroacetic acid, followed by 70% ethanol and the incorporated radioactivity was quantified by liquid scintillation counting.
Mtase activity
Measurement of DNA methyltransferase activity was performed as described previously (Li et al. 1992). Protein extract (20 μg) was added to 5 μCi of [methyl-3H]S-adenosyl L-methionine and 4 μg of poly[d(I–C)]–poly[d(I–C)]. After incubation for 2 h at 37°C, followed by phenol–chloroform extraction the nucleic acids in the aqueous phase were denatured, neutralized, and precipitated and the radioactivity that was incorporated into DNA was quantified by scintillation counting.
Acknowledgments
We thank Drs. Scott Durum, Howard Young, Joost Oppenheim, and Timothy Bestor for their suggestions on the manuscript and helpful discussions. We thank Dr. Narayan Bhat and James Cherry for their help on using real-time PCR analysis. We thank Dr. Timothy Bestor for the generous gift of the murine Dnmt1 antiserum. We thank Dr. Chun Wun for his generous gift of the murine Dnmt3a antiserum. We are grateful to the technical assistance of Rodney Wiles and Terry Stull. This project has been funded in whole or part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract No. N01-C0–56000.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL muegge@mail.ncifcrf.gov; FAX (301) 846-7077.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.929101.
References
- Antoun G, Baylin SB, Ali-Osman F. DNA methyltransferase levels and altered CpG methylation in the total genome and in the GSTP1 gene in human glioma cells transfected with sense and antisense DNA methyltransferase cDNA. J Cell Biochem. 2000;77:372–381. [PubMed] [Google Scholar]
- Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: Epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Cedar H, Solage A, Glaser G, Razin A. Direct detection of methylated cytosine in DNA by use of the restriction enzyme MspI. Nucleic Acids Res. 1979;6:2125–2132. doi: 10.1093/nar/6.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Beard C, Chen RZ, Csankovszki G, Sun Y, Siniaia M, Biniszkiewicz D, Bates B, Lee PP, Kuhn R, et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci. 2001;21:788–797. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiman TM, Muegge K. Lsh, an SNF2/helicase family member, is required for proliferation of mature T lymphocytes. Proc Natl Acad Sci. 2000;97:4772–4777. doi: 10.1073/pnas.97.9.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiman TM, Durum SK, Muegge K. Characterization of gene expression, genomic structure, and chromosomallocalization of Hells (Lsh) Genomics. 1998;54:477–483. doi: 10.1006/geno.1998.5557. [DOI] [PubMed] [Google Scholar]
- Geiman TM, Tessarollo L, Anver MR, Kopp JB, Ward JM, Muegge K. Lsh, a SNF2 family member, is required for normal murine development. Biochim Biophys Acta. 2001;1526:211–220. doi: 10.1016/s0304-4165(01)00129-5. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, McDowell TL, Raman S, O'Rourke DM, Garrick D, Ayyub H, Higgs DR. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in thepattern of DNA methylation. Nat Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- Issa JP. CpG-island methylation in aging and cancer. Curr Top Microbiol Immunol. 2000;249:101–118. doi: 10.1007/978-3-642-59696-4_7. [DOI] [PubMed] [Google Scholar]
- Jarvis CD, Geiman T, Vila-Storm MP, Osipovich O, Akella U, Candeias S, Nathan I, Durum SK, Muegge K. A novel putative helicase produced in early murine lymphocytes. Gene. 1996;169:203–207. doi: 10.1016/0378-1119(95)00843-8. [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Stokes TL, Richards EJ. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- Jones PA, Gonzalgo ML. Altered DNA methylation and genome instability: A new pathway to cancer? Proc Natl Acad Sci. 1997;94:2103–2105. doi: 10.1073/pnas.94.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes & Dev. 1992;6:705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- Langst G, Bonte EJ, Corona DF, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacementof the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylasecomplex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Ramchandani S, Bhattacharya SK, Cervoni N, Szyf M. DNA methylation is a reversible biological signal. Proc Natl Acad Sci. 1999;96:6107–6112. doi: 10.1073/pnas.96.11.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A, Shemer R. DNA methylation in early development. Hum Mol Genet. 1995;4:1751–1755. doi: 10.1093/hmg/4.suppl_1.1751. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21:461–467. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- Singer-Sam J, Grant M, LeBon JM, Okuyama K, Chapman V, Monk M, Riggs AD. Use of a HpaII-polymerase chain reaction assay to study DNA methylation in the Pgk-1 CpG island of mouse embryos at the time of X-chromosome inactivation. Mol Cell Biol. 1990;10:4987–4989. doi: 10.1128/mcb.10.9.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancheva I, Meehan RR. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes & Dev. 2000;14:313–327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Stancheva I, Hensey C, Meehan RR. Loss of the maintenance methyltransferase, xDnmt1, induces apoptosis in Xenopus embryos. EMBO J. 2001;20:1963–1973. doi: 10.1093/emboj/20.8.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M, Tada T, Lefebvre L, Barton SC, Surani MA. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus inhybrid cells. EMBO J. 1997;16:6510–6520. doi: 10.1093/emboj/16.21.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histonedeacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- Walsh CP, Bestor TH. Cytosine methylation and mammalian development. Genes & Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A, Keshet I, Razin A, Cedar H. DNA demethylation in vitro: Involvement of RNA. Cell. 1996;86:709–718. doi: 10.1016/s0092-8674(00)80146-4. [DOI] [PubMed] [Google Scholar]
- Xu GL, Bestor TH, Bourc'his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Pequignot E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- Yeivin A, Razin A. Gene methylation patterns and expression. EXS. 1993;64:523–568. doi: 10.1007/978-3-0348-9118-9_24. [DOI] [PubMed] [Google Scholar]