Abstract
Background and Aims
Outer membrane proteins of Helicobacter pylori mediate important pathogen–host interactions such as colonization, adhesion and the inflammatory response. hopQ genotypes have been suggested to be associated with increased risk of peptic ulcer. The aim of this study was to test the relation of hopQ genotype to H. pylori-related disease and histological changes in Asian and Western countries.
Methods
hopQ genotype, cagA status and vacA genotype of H. pylori isolated from patients from Asian and Western countries were determined and the results were compared with the clinical presentation and gastric histology.
Results
Most Asian strains possessed virulent genotypes (hopQ type I, vacA s1-m1 and cagA-positive). In Western countries, hopQ type I genotype was significantly linked with vacA s1 and m1 genotypes and cagA-positive status. Inflammatory cell infiltration and atrophy scores were significantly higher in patients with hopQ type I strains than those with type II in Western patients. However, the hopQ type I genotype was not associated with an increased risk for peptic ulcer or gastric cancer, and had no additive effects to vacA genotypes or cagA-positive status.
Conclusion
The expression of multiple putative virulence factors in Asian strains likely explains the relatively high incidence of clinical outcomes including gastric cancer compared with other parts of the world. Although hopQ genotype did not improve the predictive value above other genotyping for development of H. pylori-related gastroduodenal diseases, the hopQ genotype might be able to add a useful virulence marker for gastroduodenal diseases.
Keywords: gastric cancer, Helicobacter pylori, hopQ, virulence factor
Introduction
Although histological gastritis is universal among Helicobacter pylori-infected individuals, only approximately 20% develop a clinically significant outcome such as peptic ulcer, gastric cancer and primary B-cell gastric lymphoma.1–4 Bacterial factors that predict an increased risk for a clinical outcome development have been identified (i.e. the presence of the cag pathogenicity island) and there is an active search for new ones.
There is considerable variation among H. pylori strains isolated from individuals from different geographic regions4–7 and the host–H. pylori strain interactions likely differ in relation to regulation of the resulting immune and inflammatory responses of gastric mucosa. Approximately 4% of the H. pylori genome encodes a diverse repertoire of outer membrane proteins (OMP) that have been grouped into five major families. The Helicobacter outer membrane protein (Hop) family is the largest and includes adhesins such as BabA (HopS),8 SabA (HopP),9 OipA (HopH),10 AlpAB (HopB and HopC)11 and HopQ.12 Adherence of H. pylori to the gastric mucosa plays important roles in the initial colonization and long-term persistence on the gastric mucosa as well as in the intensity of the resulting inflammatory response.
Sequence analysis of hopQ from unrelated H. pylori strains has revealed two genotypes with high levels of genetic diversity (75–80% nucleotide identity) which have been classified as type I and type II.12 In the US population with peptic ulcer, most of patients with cagA-positive and vacA s1 genotype strains also expressed the hopQ type I genotype.12 This study examined the diversity of hopQ and its correlation to gastroduodenal diseases and gastric histology among different countries and different clinical outcomes as well as with other putative virulence factors vacA and cagA.
Methods
Patients and H. pylori isolates
We examined strains and tissues from H. pylori-infected patients from Korea, Japan, Thailand, the USA and Colombia presenting with gastric cancer, gastric ulcer, duodenal ulcer or gastritis alone (Table 1). Gastritis was defined as histological gastritis with no peptic ulcers or gastric cancer. Among 557 strains examined, 358 strains had been previously examined for vacA s- and m-region genotypes and cagA status to examine the geographic distribution of H. pylori strains in the world.13 Informed consent was obtained from all patients under protocols approved by the hospitals’ ethics committees.
Table 1.
Demographic characteristics of patients and genotypes of hopQ, vacA and cagA
| Asian
|
Western
|
Total | P-value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Korea | Japan | Thailand | Total | USA | Colombia | Total | ||||
| N | 127 | 120 | 38 | 285 | 173 | 99 | 272 | 557 | ||
| Sex | Male/female | 97/30 | 75/45 | 22/16 | 194/91 | 145/28 | 56/43 | 201/71 | 395/162 | <0.01 |
| Age | (mean ± SD) | 47.5 ± 1.2 | 56.9 ± 1.3 | 52.1 ± 1.5 | 52.1 ± 0.8 | 49.2 ± 1.1 | 55.9 ± 1.5 | 51.0 ± 0.9 | 54.5 ± 0.6 | <0.01 |
| Disease | Gastritis | 27.5% | 22.5% | 44.7% | 27.7% | 53.8% | 33.3% | 46.3% | 36.8% | <0.01 |
| GU | 21.3% | 27.5% | 15.8% | 23.2% | 17.3% | 0% | 11.0% | 17.2% | ||
| DU | 19.7% | 23.3% | 26.3% | 22.1% | 23.7% | 26.3% | 24.6% | 23.3% | ||
| GC | 31.5% | 26.7% | 13.2% | 27.0% | 5.2% | 40.4% | 18.0% | 22.6% | ||
| hopQ | Type I | 83.5% | 95.0% | 86.8% | 88.8% | 63.0% | 42.4% | 55.5% | 72.5% | <0.01 |
| Type II | 1.6% | 0% | 0% | 0.7% | 27.2% | 37.4% | 30.9% | 15.4% | ||
| Mix | 2.4% | 0% | 2.6% | 1.4% | 7.5% | 14.1% | 9.9% | 5.6% | ||
| Non-detection | 12.6% | 5.0% | 10.6% | 9.1% | 2.3% | 6.1% | 3.7% | 6.5% | ||
| vacA | s1m1 | 87.4% | 97.5% | 20.5% | 88.1% | 66.6% | 65.7% | 63.6% | 76.1% | <0.01 |
| s1m2 | 12.6% | 2.5% | 39.5% | 11.9% | 10.5% | 24.2% | 16.9% | 14.4% | ||
| s2m2 | 0% | 0% | 0% | 0% | 22.9% | 10.1% | 19.5% | 9.5% | ||
| cagA | Present | 97.6% | 100% | 100% | 98.9% | 85.0% | 79.8% | 83.1% | 91.2% | <0.01 |
| Absent | 2.4% | 0% | 0% | 1.1% | 15.0% | 20.2% | 16.9% | 8.8% | ||
DU, duodenal ulcer; GC, gastric cancer; GU, gastric ulcer; mix, mix type of hopQ type I and type II; SD, standard deviation.
Preparation of H. pylori genomic DNA
Helicobacter pylori strains were grown at 37°C on brain heart infusion (BHI) (BD Diagnostic Systems, Sparks, MD, USA) plates containing 7% horse blood (Cocalico Biological, Reamstown, PA, USA). The organisms were identified as H. pylori by Gram staining, colony morphology, and positive oxidase, catalase and urease reactions. Genomic DNA was extracted with the QIAamp tissue kit (QIAGEN, Santa Clarita, CA, USA) or InstaGene Matrix (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer’s instructions. The hopQ genotypings (type I and type II) were determined by polymerase chain reaction (PCR) methods reported by Cao et al.12 Genotypings of vacA s- and m-regions and cagA status were determined by PCR as described previously.7,14,15
Histology
Gastric biopsy specimens were taken from the antrum (pyloric gland area) and the corpus (fundic gland area), and were stained with hematoxylin–eosin and modified Giemsa. The biopsy specimens were fixed in 10% buffered formalin, embedded in paraffin and cut into sequential 4-μm sections. The H. pylori density, activity of gastritis (neutrophil infiltration) and atrophy were graded from 0 (absent/normal) to 5 (maximal) as previously described.16 The score is presented as the mean scores of biopsy samples from the antral and corpus areas.
It is well known that atrophic gastritis has high risk of developing into gastric ulcer and gastric cancer, and antral dominant gastritis is associated with development of duodenal ulcer; therefore, the gastritis group contained a mixture of patients, some of whom would be destined to develop severe clinical outcomes. Recently, the Operative Link on Gastritis Assessment (OLGA) staging system was advocated, in which gastritis staging combined with H. pylori infection provided clinically relevant information on the overall status of the gastric mucosa with implications for prognosis, therapy and management.17,18 We therefore analyzed the risk of clinical outcomes by comparing with not only all gastritis patients, but also OLGA stage I and II subjects as a control group.
Data analysis
Statistical differences in demographic characteristics, frequencies of hopQ genotypes, vacA genotypes and cagA status among the different geographic groups were determined by one-way ANOVA or the χ2-test. The effects of the hopQ genotypes, vacA genotypes and cagA status on the risk for developing gastric cancer and peptic ulcer in patients were expressed as odds ratios (OR) with 95% confidence intervals (CI) with reference to gastritis alone or mild gastritis alone subjects adjusted by age and sex. P < 0.05 was accepted as statistically significant. Calculations were carried out using StatView ver. 5.0 statistical software (SAS Institute, Cary, NC, USA).
Results
We examined 557 H. pylori isolates (127 strains isolated in Korea, 120 in Japan, 38 in Thailand, 173 in the USA and 99 in Colombia). The demographic characteristics including proportion of sex, mean age, clinical outcomes, mean age and frequencies of different virulence factors genotypes (hopQ, vacA s- and m-region and cagA status) in each country are shown in Table 1. There were significant differences of sex ratio, age and clinical outcomes among different proportions (P < 0.01).
Prevalence of hopQ, vacA genotyping and cagA status
hopQ type I was present in 72.5% (404 strains) and hopQ type II was found in 15.4% (86 isolates) (Table 1). The remaining 67 cases (12.1%) were either mixed type I and II genotypes or hopQ not detected by PCR and they were excluded from analyses which examined the relation between hopQ genotype and clinical outcome. There were no significant differences in the patterns of hopQ, vacA s-region genotypes and cagA status among Western countries (the USA and Colombia) or among Asian countries (Korea, Japan and Thailand) and data from different Asian or Western countries were combined into Asian and Western strains. The patterns of vacA m-region genotypes significantly differed in Asian (P < 0.01) but not in Western countries (Table 1). hopQ type II strains were extremely rare in Asian countries (2/255; 0.7%) compared to Western countries (84/235; 35.7%) (P < 0.01); no hopQ type II strains were observed in strains from either Japan or Thailand.
Relation between hopQ genotype and other virulence factors
The hopQ type I genotype was significantly associated with vacA s1, m1 genotypes and cagA-positive status in the Western population (P < 0.01; ϕ value, >0.6) (Table 2). In fact, all but two (149/151: 98.7%) hopQ type I strains were cagA-positive; however, 50% (42/84) of hopQ type II strains were also cagA-positive (Table 2). In contrast, the ϕ value of the association between cagA status and vacA s-region genotypes was extremely high (0.897) in the Western population, suggesting that the relationship between cagA status and vacA s-region genotypes was much stronger compared with that between hopQ type I genotype and cagA-positive status, vacA s1 and m1 genotypes. One the other hand, in Asian countries, the majority of H. pylori strains showed hopQ type I genotype accompanied with cagA-positive and the vacA s1 and m1 genotype (Table 2).
Table 2.
Relationship between hopQ genotype and status of other virulence factors of Helicobacter pylori
| Virulence factor | hopQ type I | hopQ type II | P-value | ϕ value | Contingency value | ||
|---|---|---|---|---|---|---|---|
| Asian population | cagA | Positive | 251 | 2 | 0.90 | −0.008 | 0.008 |
| Negative | 2 | 0 | |||||
| vacA s-region | s1 | 273 | 2 | 1.00 | – | – | |
| s2 | 0 | 0 | |||||
| vacA m-region | m1 | 223 | 2 | 0.60 | −0.032 | 0.032 | |
| m2 | 30 | 0 | |||||
| Western population | cagA | Positive | 149 | 42 | < 0.01 | 0.613 | 0.513 |
| Negative | 2 | 42 | |||||
| vacA s-region | s1 | 148 | 37 | < 0.01 | 0.731 | 0.528 | |
| s2 | 3 | 47 | |||||
| vacA m-region | m1 | 129 | 21 | < 0.01 | 0.609 | 0.489 | |
| m2 | 22 | 63 |
Relation between hopQ, vacA genotypes or cagA status and clinical outcomes
To evaluate the predictive value of hopQ genotyping in relation to clinical outcomes we performed age- and sex-adjusted univariate logistic regression analysis. In Asian countries, no significant differences were found between hopQ genotypes, cagA status and vacA genotypes and gastrointestinal diseases which was as expected due to the almost universal prevalence of hopQ type I genotype (data not shown).
In Western countries, the prevalence of type hopQ I genotype was 57.5% (65/113) for gastritis patients, 70.4% (38/54) for duodenal ulcer patients, 75% (21/28) for gastric ulcer patients and 67.5% (27/40) for gastric cancer patients (Table 3). The carriage of hopQ type I was increased in those with peptic ulcers and gastric cancer (OR adjusted for age and sex, 1.81–2.28) but the risk was not statistically sufficient (P = 0.11–0.15) (Table 4). In contrast, the vacA s1m1 genotypes and cagA-positive status were significantly increased in gastric cancer (adjusted OR, 5.00 [P < 0.01]; and 3.26 [P = 0.04]) and cagA-positive status was also significantly increased in duodenal ulcer (adjusted OR, 2.91; P = 0.02) (Table 4).
Table 3.
Prevalence of different virulence factors in gastroduodenal diseases in Asian and Western populations
| Asian
|
Western
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gastritis (n = 66) | GU (n = 64) | DU (n = 56) | GC (n = 69) | Gastritis (n = 113) | GU (n = 28) | DU (n = 54) | GC (n = 40) | ||
| hopQ | Type I | 66 (100%) | 64 (100%) | 56 (100%) | 67 (97.1%) | 65 (57.5%) | 21 (75.0%) | 38 (70.4%) | 27 (67.5%) |
| Type II | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 2 (29%) | 48 (42.5%) | 7 (25.0%) | 16 (29.6%) | 13 (32.5%) | |
| vacA | s1m1 | 58 (87.9%) | 57 (89.1%) | 44 (78.6%) | 66 (95.7%) | 64 (56.6%) | 18 (64.2%) | 33 (61.1) | 34 (85.0%) |
| s1m2 | 8 (12.1%) | 7 (10.9%) | 12 (21.4%). | 3 (4.3%) | 20 (17.7%) | 5 (17.9%) | 11 (20.4%) | 0 (0.0%) | |
| s2m2 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 29 (25.7%) | 5 (17.9%) | 10 (18.5%) | 6 (15.0%) | |
| cagA | Present | 66 (100%) | 64 (100%) | 54 (96.4%) | 69 (98.7%) | 85 (75.2%) | 25 (89.3%) | 47 (87.0%) | 34 (85.0%) |
| Absent | 0 (0.0%) | 0 (0.0%) | 2 (3.5%) | 1 (1.3%) | 28 (24.8%) | 3 (10.7%) | 7 (13.0%) | 6 (15.0%) | |
| Combination | |||||||||
| hopQ + vacA | Type I-s1m1 | 58 (87.9%) | 57 (89.1%) | 44 (78.6%) | 64 (92.8%) | 57 (50.4%) | 18 (64.3%) | 27 (50.0%) | 27 (67.5%) |
| Type II-s2m2 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 27 (23.9%) | 5 (17.9%) | 9 (16.7%) | 6 (15.0%) | |
| Others | 8 (12.1%) | 7 (10.9%) | 12 (21.4%) | 5 (7.2%) | 29 (25.7%) | 5 (17.9%) | 18 (33.3%) | 7 (17.5%) | |
| hopQ + cagA | Type 1-positive | 66 (100%) | 64 (100%) | 54 (96.4%) | 67 (97.1%) | 64 (56.6%) | 21 (75.0%) | 37 (68.5%) | 27 (67.5%) |
| Type II-negative | 0 (0.0%) | 0 (0.0%) | 2 (3.8%) | 0 (0.0%) | 21 (18.6%) | 4 (14.3%) | 10 (18.5%) | 7 (17.5%) | |
| Others | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 2 (2.9%) | 28 (24.8%) | 3 (10.7%) | 7 (13.0%) | 6 (15.0%) | |
| hopQ + vacA; cagA | Type I-s1m1-positive | 58 (87.9%) | 57 (89.1%) | 43 (76.8%) | 64 (92.8%) | 57 (50.4%) | 18 (64.3%) | 27 (50.0%) | 27 (67.5%) |
| Type II-s2m2-negative | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 26 (23.0%) | 3 (10.7%) | 6 (11.1%) | 6 (15.0%) | |
| Others | 8 (12.1%) | 7 (10.9%) | 13 (23.2%) | 5 (7.2%) | 30 (26.6%) | 7 (25.0%) | 19 (38.9%) | 7 (17.5%) | |
DU, duodenal ulcer; GC, gastric cancer; GU, gastric ulcer.
Table 4.
Sex- and age-adjusted risks for peptic ulcer and gastric cancer in relation to hopQ type I genotype in Western population
| Duodenal ulcer (n = 54)
|
Gastric ulcer (n = 28)
|
Gastric cancer (n = 40)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | OR | 95%CI | P-value | n | OR | 95%CI | P-value | n | OR | 95%CI | P-value | |
| vs gastritis patients (n = 132) | ||||||||||||
| hopQ type I (vs type II) | 38 | 1.81 | 0.89–3.68 | 0.10 | 21 | 2.28 | 0.84–6.16 | 0.10 | 27 | 1.87 | 0.79–4.41 | 0.15 |
| vacA s1m1 (vs s2m2) | 40 | 1.90 | 0.82–4.41 | 0.13 | 19 | 1.78 | 0.56–5.70 | 0.33 | 42 | 5.00 | 1.62–15.45 | < 0.01 |
| cagA positive (vs. negative) | 60 | 2.91 | 1.16–7.29 | 0.02 | 27 | 2.91 | 0.76–11.16 | 0.12 | 43 | 3.26 | 1.09–9.77 | 0.04 |
| vs OLGA stage I and II (n = 92) | ||||||||||||
| hopQ type I (vs type II) | 38 | 2.74 | 1.26–5.94 | 0.01 | 21 | 3.61 | 1.26–10.36 | 0.02 | 27 | 2.95 | 1.15–7.55 | 0.02 |
| vacA s1m1 (vs s2m2) | 40 | 2.99 | 1.17–7.67 | 0.02 | 19 | 2.71 | 0.77–9.50 | 0.12 | 42 | 8.38 | 2.37–29.60 | < 0.01 |
| cagA positive (vs. negative) | 60 | 4.68 | 1.72–12.70 | < 0.01 | 27 | 4.39 | 1.07–17.91 | 0.04 | 43 | 5.51 | 1.63–18.58 | < 0.01 |
95%CI, 95% confidence interval; OLGA, Operative Link on Gastritis Assessment; OR, odds ratio.
When compared with gastritis patients of OLGA stage I and II who had lower risk of gastroduodenal disease development, the risk of peptic ulcer and gastric cancer developments in patients with hopQ type I, vacA s1m1 and cagA-positive strains significantly increased (Table 4).
Combined effects of hopQ genotype and other virulent factors to clinical outcomes
We evaluated the additive effects by combination analysis of hopQ genotype and other virulence factors as a marker to predict clinical outcome (Tables 3 and 5). In the Western population, the gastric cancer was significantly associated with combination of hopQ type I genotype with vacA s1m1 genotypes compared with hopQ type II genotype with vacA s2 genotype or vacA s2 genotype plus cagA-positive groups (adjusted OR, 3.41; P = 0.05) (Table 5). However, there was no additive and synergic effects with relation to peptic ulcer or gastric cancer found when the analysis was done for each virulence factor genotype (hopQ alone, vacA s and m-region alone or cagA status alone) and for the genotype consisting of a combination of genes (Tables 4 and 5).
Table 5.
Sex- and age-adjusted risks for peptic ulcer and gastric cancer in relation to combination of hopQ type I genotype with vacA and/or cagA status in Western population
| Duodenal ulcer (n = 54)
|
Gastric ulcer (n = 28)
|
Gastric cancer (n = 40)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | OR | 95%CI | P-value | n | OR | 95%CI | P-value | n | OR | 95%CI | P-value | |
| vs gastritis patients (n = 132) | ||||||||||||
| hopQ type I + vacA s1m1 | 27 | 1.46 | 0.59–3.64 | 0.42 | 18 | 1.71 | 0.52–5.63 | 0.38 | 27 | 3.41 | 1.03–11.30 | 0.05 |
| hopQ type I + cagA (present) | 37 | 1.45 | 0.61–3.44 | 0.40 | 21 | 2.63 | 0.67–10.39 | 0.17 | 27 | 2.30 | 0.73–7.29 | 0.16 |
| hopQ type I + vacA s1m1 + cagA (present) | 27 | 2.24 | 0.80–6.34 | 0.48 | 18 | 2.98 | 0.73–12.18 | 0.13 | 27 | 3.62 | 1.06–12.31 | 0.04 |
| vs OLGA stage I and II (n = 92) | ||||||||||||
| hopQ type I + vacA s1m1 | 27 | 2.63 | 0.96–7.20 | 0.06 | 18 | 3.27 | 0.91–11.76 | 0.07 | 27 | 6.97 | 1.84–26.40 | < 0.01 |
| hopQ type I + cagA (present) | 37 | 2.35 | 0.92–6.03 | 0.08 | 21 | 4.50 | 1.07–18.94 | 0.04 | 27 | 4.18 | 1.18–14.81 | 0.03 |
| hopQ type I + vacA s1m1 + cagA (present) | 27 | 4.36 | 1.36–13.98 | 0.01 | 18 | 5.93 | 1.32–26.60 | 0.02 | 27 | 7.49 | 1.90–29.58 | < 0.01 |
95%CI, 95% confidence interval; OLGA, Operative Link on Gastritis Assessment; OR, odds ratio.
When compared with gastritis patients of OLGA stage I and II, the risk of peptic ulcer and gastric cancer developments in patients with combined genotype of hopQ type I, vacA s1m1 and cagA-positive strains significantly increased (Table 5).
Histopathological findings in relation to hopQ genotype
Because the gastritis group contained a mixture of patients, some of whom would be destined to develop clinical outcomes, we performed histopathological analyses in patients with gastritis alone. In Asian countries, there was no significant association between histology and H. pylori factors due to most strains possessing hopQ type I and vacA s1m1 genotypes and were cagA-positive (data not shown). In the Western population, the hopQ type I, vacA s1m1 genotypes or cagA-positive status was significantly associated with increased scores of gastric mucosal atrophy and inflammatory cell infiltration in both the antrum and the corpus compared with patients with hopQ type II, vacA s2 and m2 genotypes or cagA-negative status, respectively (Fig. 1 and Table 6). In contrast, there was no difference in the density of H. pylori among hopQ, vacA genotypes or cagA status (data not shown). The pathological effects by combination analysis of hopQ genotype and other virulence factors had no additive or synergic effect for enhanced inflammation or progression of atrophy compared with analysis of one virulence factor genotype (hopQ alone, vacA s and m-region alone or cagA status alone) (Table 6).
Figure 1.
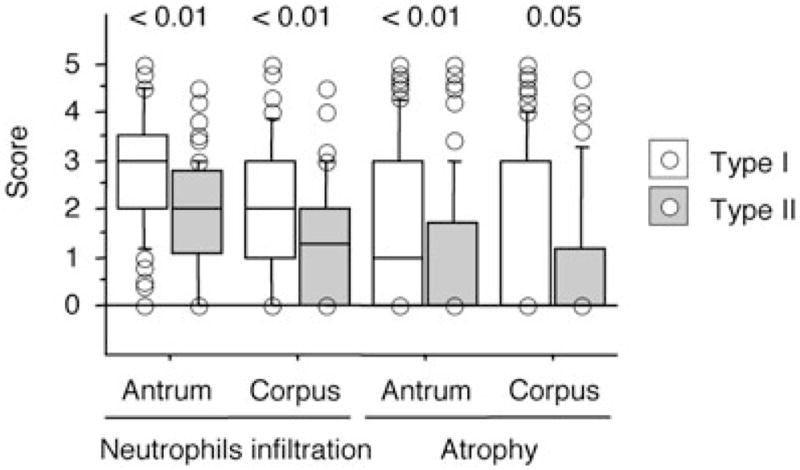
The score of gastric mucosal atrophy and inflammation activity (neutrophil infiltration) in gastric antrum and corpus among different hopQ genotype groups in gastritis patients. *P < 0.01.
Table 6.
Pathological evaluation ofHelicobacter pylori virulence factor genotype status
| Antrum
|
Corpus
|
|||
|---|---|---|---|---|
| High virulent factor | Low virulent factor | High virulent factor | Low virulent factor | |
| Neutrophil infiltration | NA | NA | NA | NA |
| hopQ type I/type II | 2.78 ± 0.10* | 1.91 ± 0.12 | 2.02 ± 0.10* | 1.38 ± 0.12 |
| vacA s1m1/s2m2 | 2.70 ± 0.09* | 1.74 ± 0.15 | 2.02 ± 0.09* | 1.13 ± 0.16 |
| cagA positive/negative | 2.62 ± 0.08* | 1.70 ± 0.16 | 1.89 ± 0.08* | 1.17 ± 0.17 |
| hopQ type1-vacA s1m1/type II-s2m2 | 2.79 ± 0.11* | 1.73 ± 0.17 | 2.11 ± 0.11* | 1.22 ± 0.18 |
| hopQ type1-vacA m1/type II-m2 | 2.76 ± 0.10* | 1.65 ± 0.17 | 2.03 ± 0.10* | 1.16 ± 0.17 |
| hopQ type1-vacA s1m1-cagA (+)/type II-s2m2-(−) | 2.79 ± 0.11* | 1.67 ± 0.17 | 2.11 ± 0.11* | 1.23 ± 0.18 |
| Atrophy | NA | NA | NA | NA |
| hopQ type I/type II | 1.62 ± 0.13* | 0.94 ± 0.16 | 1.26 ± 0.14* | 0.79 ± 0.15 |
| vacA s1m1/s2m2 | 1.73 ± 0.13* | 0.80 ± 0.16 | 1.40 ± 0.13* | 0.71 ± 0.19 |
| cagA positive/negative | 1.55 ± 0.11* | 0.66 ± 0.16 | 1.22 ± 0.11* | 0.62 ± 0.18 |
| hopQ type1-vacA s1m1/type II-s2m2 | 1.68 ± 0.15* | 0.64 ± 0.16 | 1.39 ± 0.15* | 0.57 ± 0.17 |
| hopQ type1-vacA m1/type II-m2 | 1.61 ± 0.14* | 0.58 ± 0.15 | 1.25 ± 0.14* | 0.57 ± 0.16 |
| hopQ type1-vacA s1m1-cagA (+)/type II-s2m2-(−) | 1.68 ± 0.15* | 0.55 ± 0.16 | 1.39 ± 0.15* | 0.55 ± 0.17 |
P < 0.05.
Discussion
hopQ belongs to the largest OMP family in H. pylori (Hop family) and some of them are known as adhesins such as BabA,8 SabA,9 OipA10 and AlpAB.11 However, the details of the physiological roles of hopQ are still unclear. In phylogenetic analysis, two highly divergent families of hopQ alleles were identified, and the hopQ type I alleles from Western and Asian strains were similar, and that from type II allele markedly differed.19 Analyses of synonymous and non-synonymous nucleotide substitutions suggested that there is a positive selection for HopQ amino acid diversity.19 Moreover, hopQ type II genotype was identified commonly in Western H. pylori strains, but rarely in East Asian strains as well as vacA s2 genotype and cagA-negative strain. Cao et al.12 previously reported that the carriage of type I genotype was significantly increased in subjects with peptic ulcers in the US population, suggesting that hopQ might play a biological role in the gastric mucosa. In this study, we demonstrated that neither peptic ulcer nor gastric cancer were significantly related to the hopQ type I genotype in either Western or Asian populations. However, the carriage of hopQ type I genotype was significantly associated with enhanced inflammatory cell infiltration in the gastric mucosa and gastric mucosal atrophy compared with that of hopQ type II genotype in Western populations. However, when we evaluated the effects of the hopQ genotype in combination analysis with virulence factor genotypes of cagA and vacA, the risk of peptic ulcer and gastric cancer development, as well as the severity of gastric inflammation or atrophy, we found no additive or synergic effects.
The hopQ genotypes in nearly all East Asian strains was classified as type I such that the equilibrium between hopQ type I and type II in East Asian strains of H. pylori is markedly skewed toward type I compared to Western strains. In most Asian strains, multiple genetic virulence markers, such as cagA, vacA and oipA, are present in a very high proportion of cases20–24 and our results are consistent with the notion that the hopQ type I genotype is linked to their presence. The expression of multiple putative virulence factors (e.g. hopQ type I, vacA s1 and m1 genotypes and cagA-positive) in Asian strains is associated with a high proportion of the infected population with high levels of gastric mucosal inflammation of gastric mucosa, and likely explains the relatively high incidence of clinical outcomes including gastric cancer compared with other parts of the world.
Because there was no additive effect of combined hopQ and vacA genotypes and cagA status in terms of outcome or severity of gastritis, the biological roles and interaction of hopQ and other virulence factors remain unclear. The fact that hopQ genotype I and cagA-positive status and vacA genotype were significantly correlated, precluded detection of a specific role for hopQ genotype I or II. Currently, many OMP factors whose status is closely related to cagA/vacA status, such as oipA and babA, are well-known virulence factors and their biological roles are gradually being confirmed. Identification of possible important biological roles will probably be best identified using animal models where the effects of specific combinations of putative virulence factors can be tested directly.
Acknowledgments
This material is based upon work supported in part by the Office of Research and Development Medical Research Service Department of Veterans Affairs, by Public Health Service grant DK56338 which funds the Texas Gulf Coast Digestive Diseases Center. The project described was also supported by grant no. DK 62813 from the National Institutes of Health (NIH). These contents are solely the responsibility of the authors and do not necessarily represent the official views of the Veterans Affairs (VA) and NIH. The authors thank Professor Tatsuo Yamamoto (Niigata University School of Medicine) for suitable advice.
References
- 1.Uemura N, Okamoto S, Yamamoto S, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784–9. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 2.Hopkins RJ, Girardi LS, Turney EA. Relationship between Helicobacter pylori eradication and reduced duodenal and gastric ulcer recurrence: a review. Gastroenterology. 1996;110:1244–52. doi: 10.1053/gast.1996.v110.pm8613015. [DOI] [PubMed] [Google Scholar]
- 3.Wotherspoon AC, Doglioni C, de Boni M, Spencer J, Isaacson PG. Antibiotic treatment for low-grade gastric MALT lymphoma. Lancet. 1994;343:1503. [PubMed] [Google Scholar]
- 4.Suzuki H, Hibi T, Marshall BJ. Helicobacter pylori: present status and future prospects in Japan. J Gastroenterol. 2007;42:1–15. doi: 10.1007/s00535-006-1990-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falush D, Wirth T, Linz B, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–5. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 6.Achtman M, Azuma T, Berg DE, et al. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32:459–70. doi: 10.1046/j.1365-2958.1999.01382.x. [DOI] [PubMed] [Google Scholar]
- 7.Kersulyte D, Mukhopadhyay AK, Velapatino B, et al. Differences in genotypes of Helicobacter pylori from different human populations. J Bacteriol. 2000;182:3210–8. doi: 10.1128/jb.182.11.3210-3218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ilver D, Arnqvist A, Ogren J, et al. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–7. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 9.Mahdavi J, Sonden B, Hurtig M, et al. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297:573–8. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaoka Y, Kwon DH, Graham DY. A M(r) 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc Natl Acad Sci USA. 2000;97:7533–8. doi: 10.1073/pnas.130079797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odenbreit S, Till M, Hofreuter D, Faller G, Haas R. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol Microbiol. 1999;31:1537–48. doi: 10.1046/j.1365-2958.1999.01300.x. [DOI] [PubMed] [Google Scholar]
- 12.Cao P, Cover TL. Two different families of hopQ alleles in Helicobacter pylori. J Clin Microbiol. 2002;40:4504–11. doi: 10.1128/JCM.40.12.4504-4511.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaoka Y, Orito E, Mizokami M, et al. Helicobacter pylori in North and South America before Columbus. FEBS Lett. 2002;517:180–4. doi: 10.1016/s0014-5793(02)02617-0. [DOI] [PubMed] [Google Scholar]
- 14.Atherton JC, Cao P, Peek RM, et al. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–7. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 15.Yamaoka Y, Kodama T, Kashima K, Graham DY, Sepulveda AR. Variants of the 3′ region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J Clin Microbiol. 1998;36:2258–63. doi: 10.1128/jcm.36.8.2258-2263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.el-Zimaity HM, Graham DY, al-Assi MT, et al. Interobserver variation in the histopathological assessment of Helicobacter pylori gastritis. Hum Pathol. 1996;27:35–41. doi: 10.1016/s0046-8177(96)90135-5. [DOI] [PubMed] [Google Scholar]
- 17.Rugge M, Meggio A, Pennelli G, et al. Gastritis staging in clinical practice: the OLGA staging system. Gut. 2007;56:631–6. doi: 10.1136/gut.2006.106666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rugge M, Correa P, Di Mario F, et al. OLGA staging for gastritis: a tutorial. Dig Liver Dis. 2008;40:650–8. doi: 10.1016/j.dld.2008.02.030. [DOI] [PubMed] [Google Scholar]
- 19.Cao P, Lee KJ, Blaser MJ, Cover TL. Analysis of hopQ alleles in East Asian and Western strains of Helicobacter pylori. FEMS Microbiol Lett. 2005;251:37–43. doi: 10.1016/j.femsle.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 20.Ito Y, Azuma T, Ito S, et al. Analysis and typing of the vacA gene from cagA-positive strains of Helicobacter pylori isolated in Japan. J Clin Microbiol. 1997;35:1710–4. doi: 10.1128/jcm.35.7.1710-1714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maeda S, Ogura K, Yoshida H, et al. Major virulence factors, VacA and CagA, are commonly positive in Helicobacter pylori isolates in Japan. Gut. 1998;42:338–43. doi: 10.1136/gut.42.3.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan ZJ, van der Hulst RW, Feller M, et al. Equally high prevalences of infection with cagA-positive Helicobacter pylori in Chinese patients with peptic ulcer disease and those with chronic gastritis-associated dyspepsia. J Clin Microbiol. 1997;35:1344–7. doi: 10.1128/jcm.35.6.1344-1347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Doorn LJ, Figueiredo C, Sanna R, et al. Expanding allelic diversity of Helicobacter pylori vac. A J Clin Microbiol. 1998;36:2597–603. doi: 10.1128/jcm.36.9.2597-2603.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Doorn LJ, Figueiredo C, Sanna R, Blaser MJ, Quint WG. Distinct variants of Helicobacter pylori cagA are associated with vacA subtypes. J Clin Microbiol. 1999;37:2306–11. doi: 10.1128/jcm.37.7.2306-2311.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]


