Abstract
The transcription factor TFIIB plays a central role in preinitiation complex assembly, providing a bridge between promoter-bound TFIID and RNA Polymerase II. TFIIB possesses sequence-specific DNA-binding ability and interacts with the TFIIB-recognition element (BRE), present in many promoters. Here we show that the BRE suppresses the basal level of transcription elicited by a core promoter, which increases the amplitude of transcriptional stimulation in the presence of an activator protein. Further, we find that an activator can disrupt the TFIIB–BRE interaction within a promoter-bound complex. Our results reveal a novel function for activators in the modulation of core promoter recognition by TFIIB.
Keywords: Transcription, RNA polymerase II, TFIIB, promoter, transactivation
Transcriptional activation in eukaryotes requires the displacement of nucleosomes in the promoter region of a gene and also the stimulation of the assembly of the preinitiation complex (PIC; for reviews, see Hampsey 1998; Roberts 2000). Several of the general transcription factors (GTFs) have been proposed as targets of transcriptional activators, including TFIID, TFIIB, and TFIIH. Current models propose that recruitment of GTFs by activators increases the rate/extent of PIC assembly by overcoming a rate-limiting step (Ptashne and Gann 1997). Activators have also been shown to stimulate open complex formation and facilitate promoter escape.
The GTF TFIIB has been proposed by several groups to be a target of transcriptional activators (Lin and Green 1991; Colgan et al. 1993; Kim and Roeder 1994; Wu and Hampsey 1999). Many activation domains can interact directly with TFIIB. Significantly, this interaction has been shown to be required for transcriptional activation both in vitro and in vivo (Roberts et al. 1993; Wu and Hampsey 1999; Hawkes et al. 2000). In addition to mediating TFIIB recruitment, activators can induce a conformational change in TFIIB that has the potential to drive PIC assembly forward (Roberts and Green 1994).
TFIIB can engage in sequence-specific contact with the core promoter (Lagrange et al. 1998; Qureshi and Jackson 1998). This TFIIB-recognition element (BRE) is located immediately upstream of the TATA element in a subset of promoters. A helix–turn–helix (HTH) motif at the C terminus of TFIIB mediates contact with the BRE (Lagrange et al. 1998; Tsai and Sigler 2000). The TFIIB–BRE interaction facilitates the assembly of a TFIIB–TBP–TATA complex and stimulates transcription (Lagrange et al. 1998; Qureshi and Jackson 1998). In addition, the TFIIB–BRE interaction helps to orient the PIC on the promoter (Bell et al. 1999; Littlefield et al. 1999; Tsai and Sigler 2000).
In this study we show that, both in a crude nuclear extract and in living cells, the BRE acts as a negative element. Furthermore, we show that an activation domain abates the TFIIB–BRE interaction in a DNA-bound complex. Our results suggest that activator-mediated disruption of the TFIIB–BRE interaction occurs during transcriptional activation.
Results
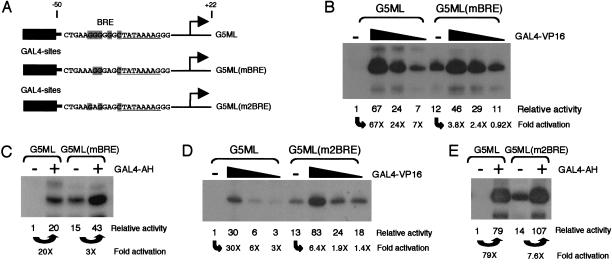
We constructed an AdML (ML) promoter derivative in which the BRE was mutated at two critical nucleotides and called it ML(mBRE). Methylation interference analysis confirmed that the ML(mBRE) construct, which harbors mutations at −34 and −38, contained a defective BRE (data not shown). ML and ML(mBRE) were linked to five GAL4 DNA-binding sites (Fig. 1A), and their transcriptional activities were compared in a crude HeLa cell nuclear extract in both the absence and presence of the activator GAL4–VP16. Surprisingly, the G5ML(mBRE) promoter derivative elicited a significantly greater basal level of transcription than the wild-type promoter (Fig. 1B). The level of transcription attained in the presence of the activator GAL4–VP16, however, was comparable to both the wild-type and mutant promoters. We observed the same effect with the activator GAL4-AH (Fig. 1C). In addition, we tested a previously characterized BRE mutation (designated m2BRE), which contains substitutions at bases −34 and −37 (Fig. 1A; Lagrange et al. 1998). Again, the level of basal transcription was significantly elevated compared with the wild-type ML promoter, but the level of transcription elicited by the activators was similar (Fig. 1D,E). Therefore, the BRE acts as a negative element when tested in a basal transcription assay using a crude HeLa nuclear extract. Furthermore, the presence of a BRE significantly increases the amplitude of transcriptional activation elicited by the acidic activators GAL4–VP16 and GAL4–AH.
Figure 1.
The BRE is a negative element that enhances the amplitude of transcriptional activation. (A) Diagram of the Adenovirus major late core promoter (ML) fragments (−50 to +22) with only the TATA (underlined) and BRE (shaded) shown. ML(mBRE) contains substitutions at positions −34 and −38 and ML(m2BRE) at positions −34 and −37. The BRE consensus sequence is; G/C-G/C-G/A-C-G-C-C-3′. (B) An in vitro transcription assay comparing the promoters from part A, in the absence or presence of GAL4–VP16 (0.2 ng, 2 ng, and 20 ng). Quantitation is shown below as activity relative to basal on the ML promoter and also fold activation relative to the basal of each respective promoter. (C) As in part B, except that GAL4-AH (0.5 μg) was used as the activator. (D) As in part B, except that the ML(m2BRE) construct was used. (E) As in part C, except that ML(m2BRE) was compared with ML.
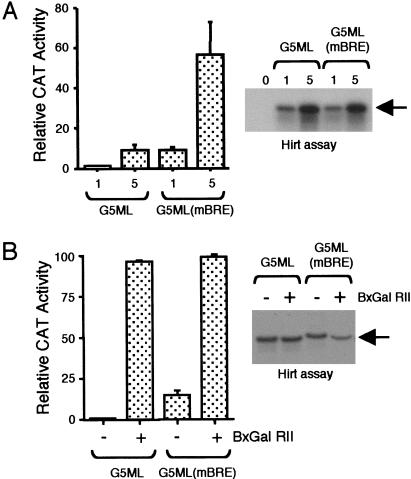
We next sought to confirm our observations in a living cell. Embryonic kidney 293 cells were transfected with either 1 μg or 5 μg each of the ML and ML(mBRE) core promoters linked to the CAT gene. The cells were harvested 48 h later and CAT activity was measured (Fig. 2A). The results show that, similar to the effect we observed in a crude nuclear extract, the BRE suppresses transcription of the ML core promoter. To confirm that equivalent amounts of each of the constructs was transfected, we performed a Hirt assay to directly assess the amount of DNA that was taken up by the cells. The transfected plasmid was recovered from washed cells and detected by primer extension (Fig. 2A, right panel). The results confirm that equivalent amounts of plasmid were transfected into the cells. We next performed a transient transfection assay to assess the effect of the activator BxGal RII (GAL4 DNA-binding domain linked to the acidic activation region II of yeast GAL4) on transcription of the ML and ML(mBRE) derivatives (Fig. 2B). This activator was chosen because it belongs to the same acidic class of activation domain as VP16, but is significantly less potent in vivo, thus allowing a more accurate comparison between basal and activated transcription. As before, the ML(mBRE) elicited a significantly greater level of basal transcription than the wild-type ML promoter. In the presence of the acidic activator BxGal RII, however, the level of transcription was similar. Taking these results together with the results of Figure 1, we conclude that, in both a crude nuclear extract and living cells, the BRE acts as a negative element, and that this, in turn, increases the amplitude of transcriptional activation.
Figure 2.
The BRE acts as a negative element in transient transfection. (A) Cells were transfected with either 1 μg or 5 μg of the G5ML or G5ML(mBRE) promoter derivatives, and CAT activity was measured. The graph is presented as activity relative to 1 μg of ML and represents the average of three independent transfection assays. The Hirt assay at right shows an autoradiograph to detect (by primer extension) plasmid recovered from the transfected cells. (0) Cells that were mock-transfected. (B) As in part A, except that only 1 μg of each promoter derivative was transfected alone or along with 1 μg of an expression vector that produces the activator BxGal RII (indicated by +). The results are the mean average of three independent experiments.
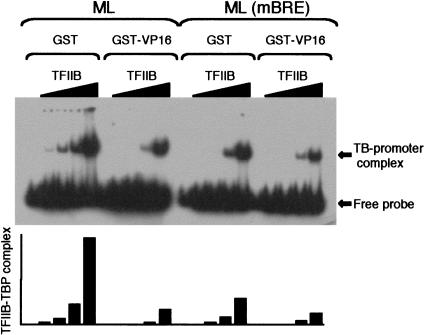
Although the BRE suppresses the basal level of transcription observed at the AdML promoter, in the presence of an activator protein the innate level of transcription is not affected by the BRE. We therefore next tested whether the TFIIB–BRE interaction was a direct target of a transcriptional activator. It has been shown previously by electrophoretic mobility shift assay that the TFIIB–BRE interaction stabilizes a TFIIB–TBP–promoter complex (Lagrange et al. 1998). We therefore reasoned that if an activator disrupts the TFIIB–BRE interaction, the presence of an activation domain as a competitor in solution should destabilize the TFIIB–TBP–promoter complex. TBP and increasing amounts of TFIIB were incubated with the ML core promoter in the presence of GST or GST–VP16. Complexes were resolved by native electrophoresis and are shown in Figure 3 (left). A bar graph is shown below the autoradiogram. The presence of the VP16 activation domain caused a marked decrease in the formation of a TFIIB–TBP–promoter complex, suggesting that an activator can, indeed, destabilize a TFIIB–TBP complex assembled at the wild-type ML promoter. If the effect of VP16 on TFIIB–TBP complex formation on the ML promoter is owing to disruption of the TFIIB–BRE interaction, then VP16 should not affect TFIIB–TBP complexes formed on the ML(mBRE) derivative. Consistent with previous results, mutation of the BRE results in less efficient TFIIB–TBP–promoter complex formation (Fig. 3, right; Lagrange et al. 1998). Moreover, GST-VP16 exhibited a reduced ability to destabilize a TFIIB–TBP–ML(mBRE) complex; compared with an average sixfold reduction in TFIIB–TBP complex formation at the ML promoter caused by VP16, the ML(mBRE) construct was reduced only twofold. Taken together, these data strongly suggest that the VP16 activation domain can disrupt the TFIIB–BRE interaction.
Figure 3.
The VP16 activation domain in solution disrupts the TFIIB–BRE interaction. Bandshift assay using ML or ML(mBRE) fragments, radiolabeled to equivalent specific activities, incubated with 1 ng of TBP and increasing amounts (8 pg, 40 pg, 0.2 ng, and 1 ng) of TFIIB. GST or GST-VP16 (0.5 μg) was added along with the GTFs. Free probe and TFIIB–TBP–promoter complexes (TB–promoter complex) are indicated. Quantitation as a bar graph is shown below the autoradiogram.
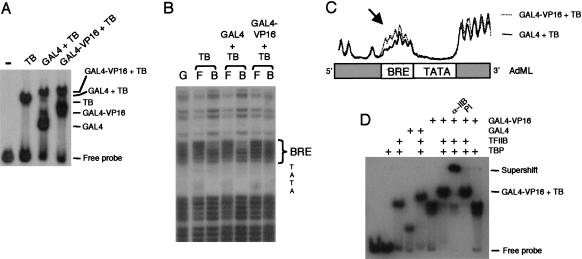
The above experiments showed that the VP16 activation domain that is not tethered to DNA can disrupt TFIIB–TBP complex formation in a BRE-dependent manner. We next sought to determine the effect of a GAL4–VP16 fusion protein on TFIIB–BRE contact in the context of a promoter-bound complex. We therefore performed methylation interference analysis to directly assess TFIIB–BRE contact in complexes that contain TBP, TFIIB, and either GAL4–VP16 or the control GAL4. The complexes were assembled on partially methylated ML core promoter downstream of a single GAL4-recognition site and resolved by native electrophoresis (Fig. 4A). DNA from the intact complexes was isolated, subjected to cleavage with piperidine, and resolved by denaturing electrophoresis alongside a G-track of the same ML promoter fragment (Fig. 4B). In a TBP–TFIIB complex at the wild-type ML promoter, the first three nucleotides of the BRE on the positive strand show interference by methylation, as predicted by the structural and biochemical analysis of the TFIIB–BRE interaction (Fig. 4B; Lagrange et al. 1998; Tsai and Sigler 2000). The same interference was observed in a TBP–TFIIB–ML complex that also contained DNA-bound GAL4. In a GAL4–VP16–TBP–TFIIB complex, however, significantly reduced methylation interference of the BRE was observed (Fig. 4B; trace shown in Fig. 4C). To rule out the possibility that GAL4–VP16 completely displaced TFIIB from the complex, we performed a bandshift in which either anti-TFIIB antibodies or preimmune serum was added to a GAL4–VP16–TFIIB–TBP–promoter complex. The anti-TFIIB antibodies, but not control preimmune serum, supershifted the complex, confirming that TFIIB is, indeed, present (Fig. 4D). Therefore, in the presence of promoter-bound GAL4–VP16, but not GAL4, TFIIB is not engaged in contact with the BRE. Taking these results together with the EMSA data of Figure 3, we conclude that the VP16 transcriptional-activation domain disrupts the TFIIB–BRE interaction.
Figure 4.
Modulation of TFIIB–BRE interaction by an activator within a promoter-bound complex. (A) Bandshift assay showing the formation of the complexes indicated with the radiolabeled and partially methylated ML promoter linked to a single GAL4 DNA-binding site. Amounts of protein used were TBP (2 ng), TFIIB (0.2 ng), GAL4 (1 ng) ,and Gal4–VP16 (1 ng). (B) DNA was recovered from the free probe (F) and the indicated complex (B), cleaved with piperidine, and resolved by denaturing electrophoresis alongside a G-track (G) of the same probe. Positions of the BRE and TATA element are marked. (C) The gel in part B was scanned by PhosphorImager; a trace of the lanes indicated is graphed above a diagram of the BRE and TATA region of the ML core promoter. An arrow indicates the alteration in methylation interference in the BRE region. (D) Bandshift performed as in part A, but with the addition of preimmune serum or anti-TFIIB antibodies. Only the GAL4–VP16 + TFIIB + TBP (TB) complex and free probe are indicated.
Discussion
The BRE was originally identified as an element that had a positive effect on transcription in vitro (Lagrange et al. 1998; Qureshi and Jackson 1998). This effect correlated with an increase in stable TFIIB–TBP–promoter complex formation that was mediated by direct contact of the BRE by TFIIB. In the present study we show that, in both a crude nuclear extract and in transient transfection, the BRE acts to repress basal transcription. The previous study using a eukaryotic system was composed of purified factors (Lagrange et al. 1998). It is therefore possible that purification of the mammalian factors eliminates a BRE-dependent event. Consistent with our results, previous transfection analysis of AdML promoter derivatives found that mutations upstream of the TATA element, that have since been shown to be critical for BRE function, resulted in enhanced transcription from the promoter (Yu and Manley 1984, 1986).
In addition to a mammalian system composed of purified transcription factors, the BRE has also been shown to act as a positive element in an Archaea (Qureshi and Jackson 1998). This raises the possibility that the function of the BRE has altered through the course of evolution, perhaps with increasing complexity of factors. Indeed, the fact that yeast and plant TFIIB do not contain the consensus HTH motif that binds the BRE, suggests a complex evolution of this protein–DNA interaction (Lagrange et al. 1998).
Our finding that an activation domain can disrupt the TFIIB–BRE interaction sheds significant light on our understanding of how transcriptional activators function. First, our data show that an activator modulates TFIIB function within a promoter-bound complex. Second, these results have uncovered a direct effect on the architecture of the preinitiation complex by modulating DNA sequence-specific contacts. Finally, in addition to the well defined activator-driven promotion of interactions within the PIC, our studies reveal that activator-mediated disruption of contacts may also be critical.
How does VP16 disrupt the TFIIB–BRE interaction? VP16 can induce a conformational change in TFIIB (Roberts and Green 1994; Wu and Hampsey 1999), and this could, in turn, affect the ability of TFIIB to interact with DNA. Alternatively, VP16 may directly bind the HTH motif within TFIIB. Indeed, previous deletion mutagenesis and also protease footprinting of the VP16–TFIIB interaction showed that the HTH region of TFIIB engages in contact with VP16 (Roberts et al. 1993; Hori et al. 1995). Further studies will shed light on the specific mechanism by which VP16 can disrupt the TFIIB–BRE interaction. In addition, it will also be interesting to determine if other classes of activator proteins perform the same function.
How disruption of the TFIIB–BRE interaction leads to stimulation of transcription remains to be determined. Our current studies do not reveal the stage in PIC assembly at which disruption of the TFIIB–BRE interaction occurs. It is possible that this could occur simultaneously with TFIIB recruitment into the PIC to leave the BRE sequence available for contact with other factors. Potential candidates could be the TBP-associated factors (TAFs) and other proteins that interact with TBP and regulate PIC formation, such as TFIIA, Mot1, and NC2 (for review, see Pugh 2000). These proteins were presumably not present in the study by Lagrange et al. (1998), which used recombinant TBP in the transcription assays. Moreover, Archaea have not been shown to contain the above TBP-associating factors, perhaps providing an explanation for the nonconserved effect of the TFIIB–BRE interaction.
An alternative scenario is that the activator-mediated disruption of the TFIIB–BRE interaction may contribute to promoter clearance by destabilizing the PIC to release RNA pol II. In this regard, it is interesting to note that upon initiation TFIIB is released from the PIC, but TFIID remains bound to the TATA element (Roberts et al. 1995; Zawel et al. 1995). As a TFIIB–TFIID–promoter complex is stable, the release of TFIIB upon initiation is likely to be an active event. It is possible, therefore, that the BRE regulates the transition of the PIC to elongation complex. Indeed, evidence has been presented that supports a critical role for TFIIB after the assembly of the PIC (Cho and Buratowski 1999; Ranish et al. 1999; Bell and Jackson 2000). These and other possibilities will form the basis of future studies.
Materials and methods
Plasmids and protein purification
The promoter DNA templates G5ML, G5ML(mBRE), and G5ML(m2BRE) contain nucleotides −50 to +22 from the Adenovirus major late promoter cloned downstream of five GAL4 DNA-binding sites in the pCAT basic vector (Promega). G1ML contains the ML core promoter downstream of a single GAL4 DNA-binding site in the vector pGEM3. ML(mBRE) and ML(m2BRE) differ from ML with the substitution of two nucleotides as detailed in Figure 1A. The expression vector for BxGAL4-RII has been described previously (Martin et al. 1990). PGEX, pGEX–VP16 (Lin and Green 1991), pET TFIIB (Ha et al 1993), pET6HISTBP, pET6HIS GAL4, pET6HIS GAL4–VP16, and pTAC GAL4–AH have been described previously (Lin et al. 1988; Hawkes et al. 2000). TFIIB and GAL4–AH were purified as previously described (Ha et al. 1993; Lin et al. 1988). Poly-histidine-tagged TBP, GAL4(1–93), and GAL4(1–93)–VP16 were purified by nickel chelate affinity chromatography (Reece et al. 1993). GST and GST–VP16 were purified as described previously (Lin and Green 1991). Anti-TFIIB antibodies have been described previously (Hawkes and Roberts 1999).
In vitro transcription assay, EMSA, and methylation interference
HeLa cell nuclear extracts were purchased from Computer Cell Culture Centre (Mons, Belgium). In vitro transcription assays were performed as described previously (Lin and Green 1991) except that a CAT primer was used to analyze the transcripts.
Electrophoretic mobility shift assays were performed as described previously (Maldonado et al. 1990). ML and ML(mBRE) promoter fragments were end-labeled with Klenow and [α-32P]dATP. After gel purification, the specific activity of each probe was determined. Binding reactions were assembled in 10 mM Hepes (pH 8), 4 mM MgCl2, 5 mM ammonium sulfate, 8% glycerol, 2% PEG, 55 mM KCl, 5 mM β-mercaptoethanol, 0.2 mM EDTA, and 0.2 mM PMSF. Where indicated, GST or GST–VP16 was added along with TFIIB and TBP. Reactions were incubated at 30°C for 1 h, then loaded onto a 5% nondenaturing polyacrylamide gel run at 100 V for 3 h in 0.5× Tris-Borate-EDTA (pH 8.3). Gels were dried and autoradiographed overnight. For methylation interference, G1ML was labeled on the positive strand by Klenow end fill-in [α-32P]dATP. The radiolabeled promoter fragment was partially methylated with dimethyl sulfate and assembled into complexes as with a regular EMSA (above), except that the reactions were scaled up to use 100,000 cpm of the promoter per reaction. The wet gel was exposed to film, and complexes were excised and treated as described (Ausubel et al. 1995).
Cell culture and transfection
Human embryonic kidney 293 cells were cultured and transfected as described previously (Hawkes and Roberts 1999). Cells were harvested 48 h after transfection, and cell lysates were then used in CAT assays. A Hirt extraction was performed to recover plasmid DNA from transfected cells (Lee et al. 1998). The recovered plasmid was subjected to primer extension using the CAT primer, and the products were resolved by denaturing electrophoresis and autoradiographed.
Acknowledgments
We thank Andy Sharrocks, Paul Shore, and members of the lab for comments on the manuscript. We also thank members of the Division of Gene Expression at Dundee University for helpful comments during the initial stages of this work. We are grateful to Richard Ebright for critique of the data and suggestions. R.E. is supported by a research studentship from the Medical Research Council (UK). This work was funded by the Wellcome Trust (061207/Z/00/Z/CH/TG/dr) and the BBSRC. S.G.E.R. is a Wellcome Trust Senior Research Fellow.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL stefan.roberts@man.ac.uk; FAX 0161-275-5600.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.206901.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short protocols in molecular biology. New York: Wiley; 1995. Methylation and uracil interference assays for analysis of protein–DNA interactions; pp. 12.9–12.11. [Google Scholar]
- Bell SD, Jackson SP. The role of transcription factor B in transcription initiation and promoter clearance in the Archaeon Sulfolobus acidocaldarius. J Biol Chem. 2000;275:12934–12940. doi: 10.1074/jbc.275.17.12934. [DOI] [PubMed] [Google Scholar]
- Bell SD, Kosa PL, Sigler PB, Jackson SP. Orientation of the transcription preinitiation complex in Archaea. Proc Natl Acad Sci. 1999;96:13662–13667. doi: 10.1073/pnas.96.24.13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E-J, Buratowski S. Evidence that transcription factor IIB is required for a post-assembly step in transcription initiation. J Biol Chem. 1999;274:25807–25813. doi: 10.1074/jbc.274.36.25807. [DOI] [PubMed] [Google Scholar]
- Colgan J, Wampler S, Manley JL. Interaction between a transcriptional activator and transcription factor-IIB in vivo. Nature. 1993;362:549–553. doi: 10.1038/362549a0. [DOI] [PubMed] [Google Scholar]
- Ha I, Roberts SGE, Maldonado E, Sun X, Kim L-U, Green MR, Reinberg D. Multiple functional domains of general transcription factor IIB: Distinct interactions with two general transcription factors and RNA polymerase II. Genes & Dev. 1993;7:1021–1032. doi: 10.1101/gad.7.6.1021. [DOI] [PubMed] [Google Scholar]
- Hampsey M. Molecular genetics of the RNA polymerase II general transcription machinery. Microbiol Molec Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes NA, Roberts SGE. The role of human TFIIB in transcription start site selection in vitro and in vivo. J Biol Chem. 1999;274:14337–14343. doi: 10.1074/jbc.274.20.14337. [DOI] [PubMed] [Google Scholar]
- Hawkes NA, Evans R, Roberts SGE. The conformation of TFIIB modulates the response to transcriptional activators in vivo. Curr Biol. 2000;10:273–276. doi: 10.1016/s0960-9822(00)00363-8. [DOI] [PubMed] [Google Scholar]
- Hori R, Pyo M, Carey M. Protease footprinting reveals a surface on transcription factor TFIIB that serves as an interface for activators and coactivators. Proc Natl Acad Sci. 1995;92:6047–6051. doi: 10.1073/pnas.92.13.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Roeder RG. Proline-rich activator CTF1 targets the TFIIB assembly step during transcriptional activation. Proc Natl Acad Sci. 1994;91:4170–4174. doi: 10.1073/pnas.91.10.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH. New core promoter element in RNA polymerase II-dependent transcription: Sequence specific DNA binding by transcription factor IIB. Genes & Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-S, Lee C-H, Chung JH. Studying the recruitment of Sp1 to the β-globin promoter with an in vivo method: Protein position identification with nuclease tail (PIN*POINT) Proc Natl Acad Sci. 1998;95:969–974. doi: 10.1073/pnas.95.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-S, Green MR. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991;64:971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- Lin Y-S, Carey M, Ptashne M, Green MR. GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell. 1988;54:659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- Littlefield O, Korkhin Y, Sigler PB. The structural basis for the oriented assembly of a TBP/TFB/promoter complex. Proc Natl Acad Sci. 1999;96:13668–13673. doi: 10.1073/pnas.96.24.13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado E, Ha I, Cortes P, Weis L, Reinberg D. Role of transcription factors IIA, IID and IIB during formation of a transcription competent complex. Mol Cell Biol. 1990;10:6335–6347. doi: 10.1128/mcb.10.12.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KJ, Lillie JW, Green MR. Evidence for interaction of different eukaryotic transcriptional activators with distinct cellular targets. Nature. 1990;346:147–152. doi: 10.1038/346147a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Pugh BF. Control of gene expression through regulation of the TATA-binding protein. Gene. 2000;255:1–14. doi: 10.1016/s0378-1119(00)00288-2. [DOI] [PubMed] [Google Scholar]
- Qureshi SA, Jackson SP. Sequence-specific DNA binding by the S-shibatae TFIIB homolog, TFB, and its effect on promoter strength. Mol Cell. 1998;1:389–400. doi: 10.1016/s1097-2765(00)80039-8. [DOI] [PubMed] [Google Scholar]
- Ranish JA, Yudkovsky N, Hahn S. Intermediates in the formation and activity of the RNA polymerase II preinitiation complex: Holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes & Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece RJ, Rickles RJ, Ptashne M. Overproduction and single-step purification of Gal4 fusion proteins from Escherichia coli. Gene. 1993;126:105–107. doi: 10.1016/0378-1119(93)90596-u. [DOI] [PubMed] [Google Scholar]
- Roberts SGE. Mechanisms of transcriptional activation and repression. Cell Mol Life Sci. 2000;57:1149–1160. doi: 10.1007/PL00000755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SGE, Green MR. Activator-induced conformational change in general transcription factor TFIIB. Nature. 1994;371:717–720. doi: 10.1038/371717a0. [DOI] [PubMed] [Google Scholar]
- Roberts SGE, Ha I, Maldonado E, Reinberg D, Green MR. Interaction between an acidic activator and transcription factor TFIIB is required for transcriptional activation. Nature. 1993;363:741–744. doi: 10.1038/363741a0. [DOI] [PubMed] [Google Scholar]
- Roberts SGE, Choy B, Walker SS, Lin Y-S, Green MR. A role for activator-mediated TFIIB recruitment in diverse aspects of transcriptional regulation. Curr Biol. 1995;5:508–516. doi: 10.1016/s0960-9822(95)00103-5. [DOI] [PubMed] [Google Scholar]
- Tsai FTF, Sigler PB. Structural basis of preinitiation complex assembly on human Pol II promoters. EMBO J. 2000;19:25–36. doi: 10.1093/emboj/19.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W-H, Hampsey M. An activation-specific role for transcription factor TFIIB in vivo. Proc Natl Acad Sci. 1999;96:2764–2769. doi: 10.1073/pnas.96.6.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Manley JL. Generation and functional analyses for base-substitution mutants of the adenovirus 2 major late promoter. Nucl Acids Res. 1984;12:9309–9321. doi: 10.1093/nar/12.24.9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Structure and function of the S1 nuclease-sensitive site in the Adenovirus Late Promoter. Cell. 1986;45:743–751. doi: 10.1016/0092-8674(86)90788-9. [DOI] [PubMed] [Google Scholar]
- Zawel L, Kumar KP, Reinberg D. Recycling of the general transcription factors during RNA-polymerase-II transcription. Genes & Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]